Cervicovaginal Microbiome and HPV: A Standardized Approach to 16S/ITS NGS and Microbial Community Profiling for Viral Association
Abstract
1. Introduction
2. Results
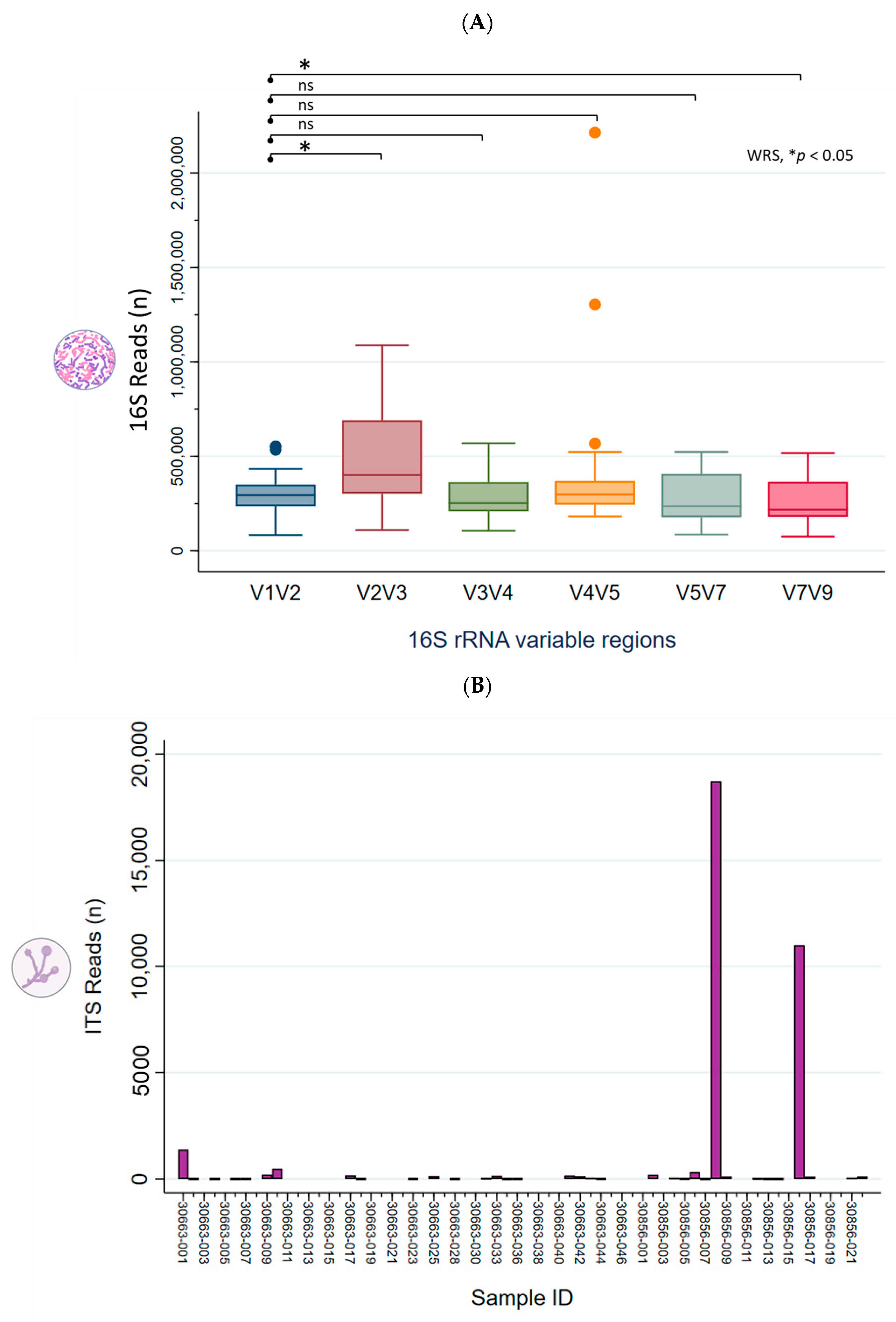
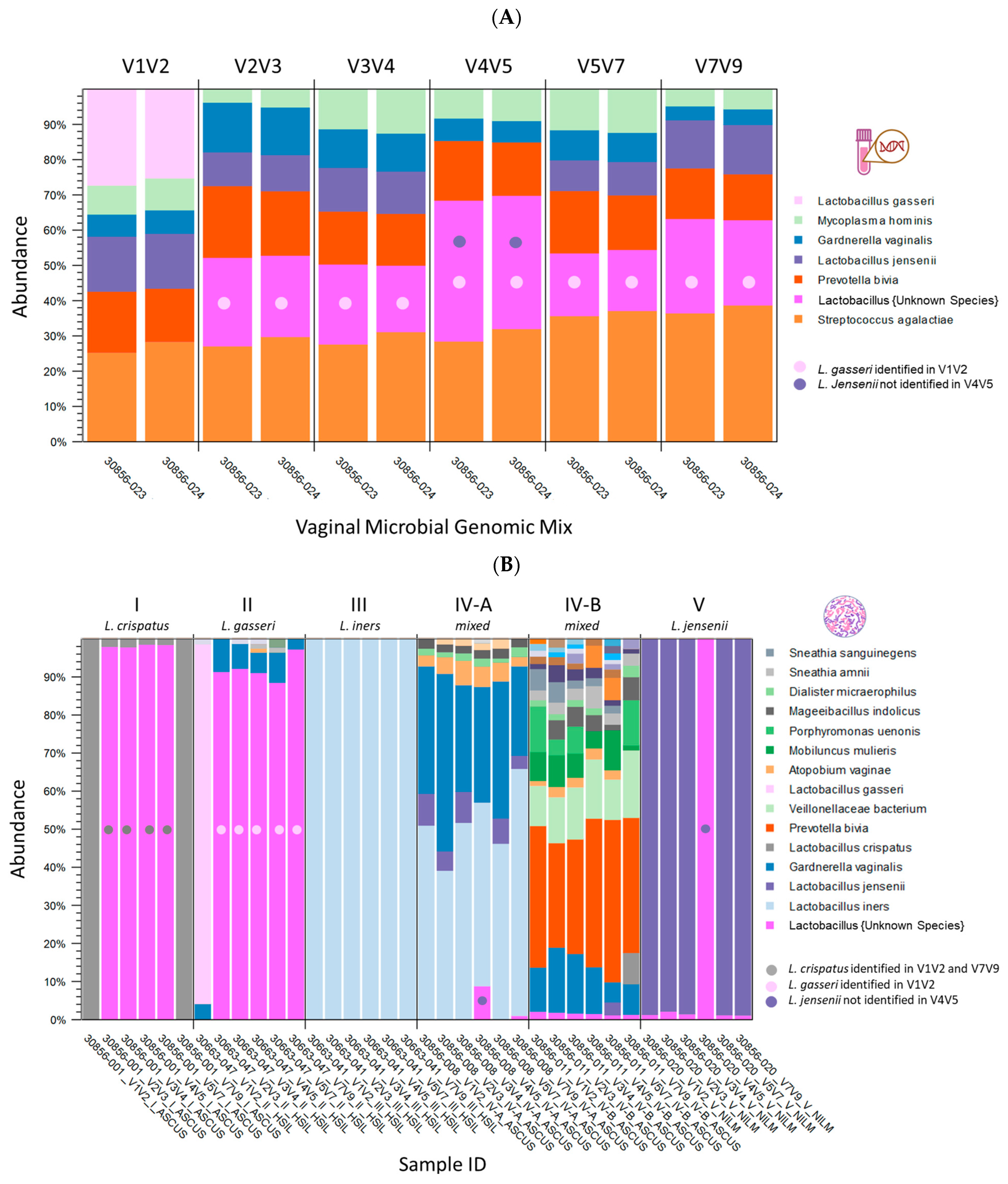
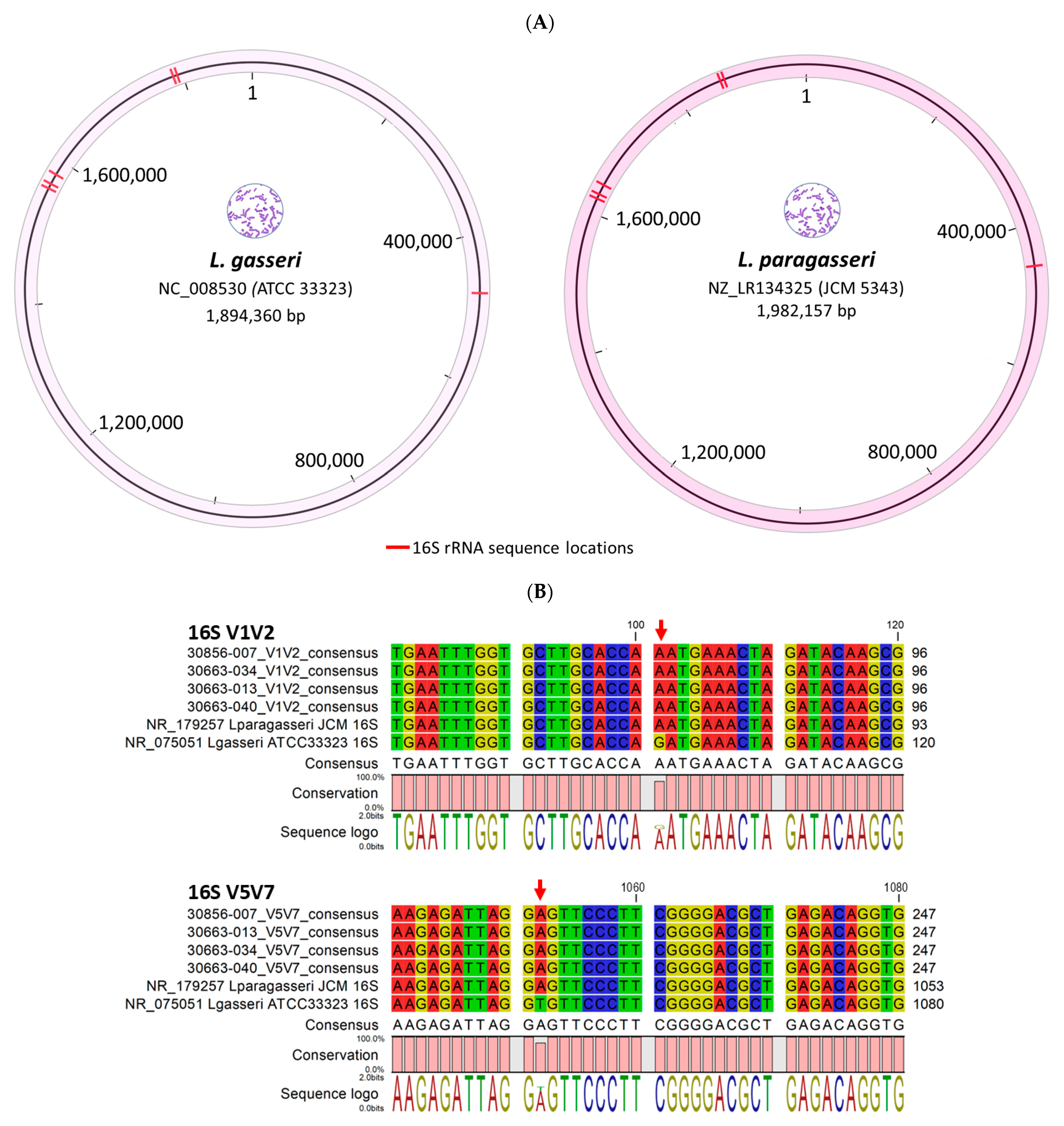
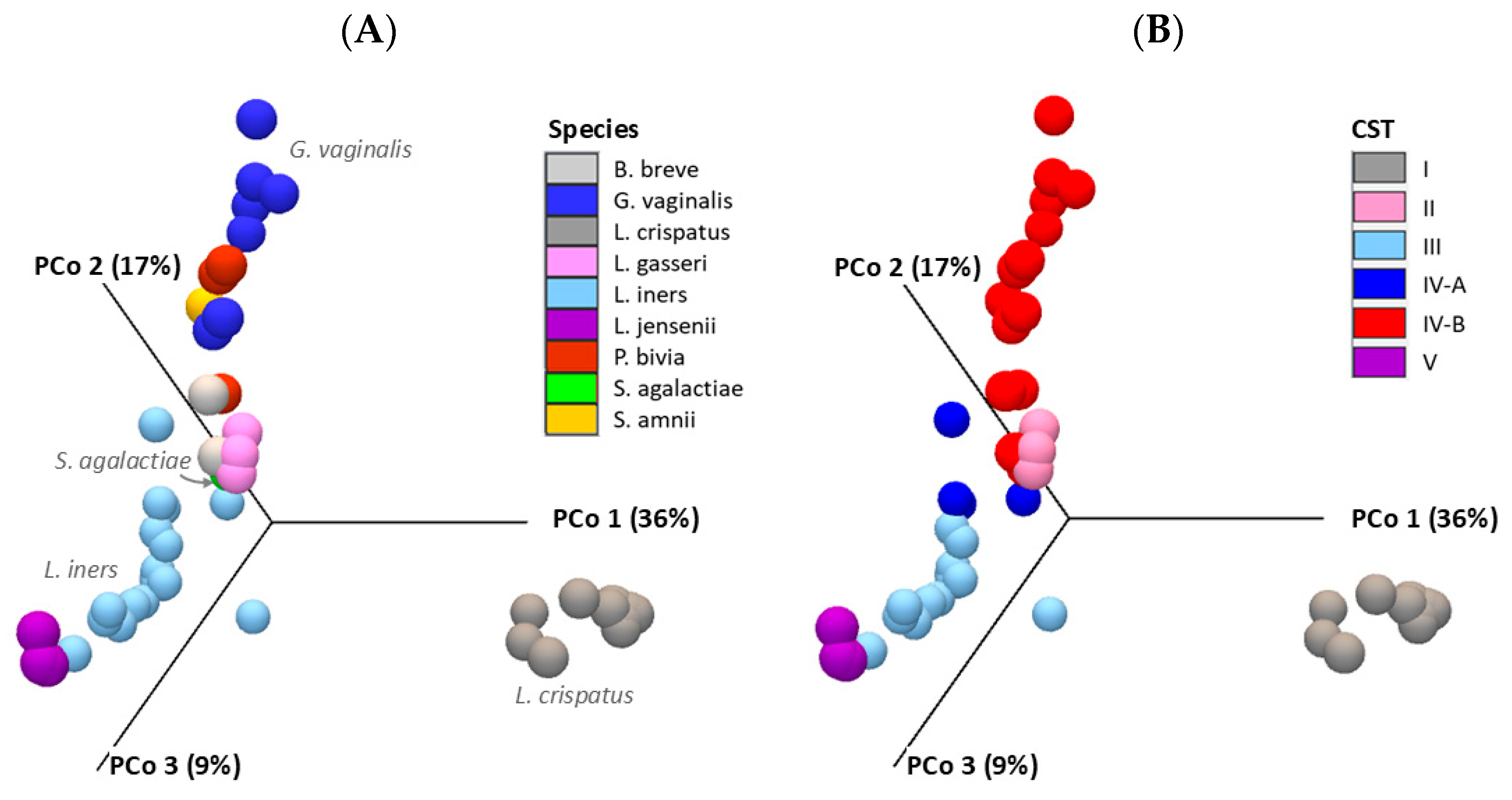
2.1. Taxonomic Classification and Visualization of Cervicovaginal Microbiomes
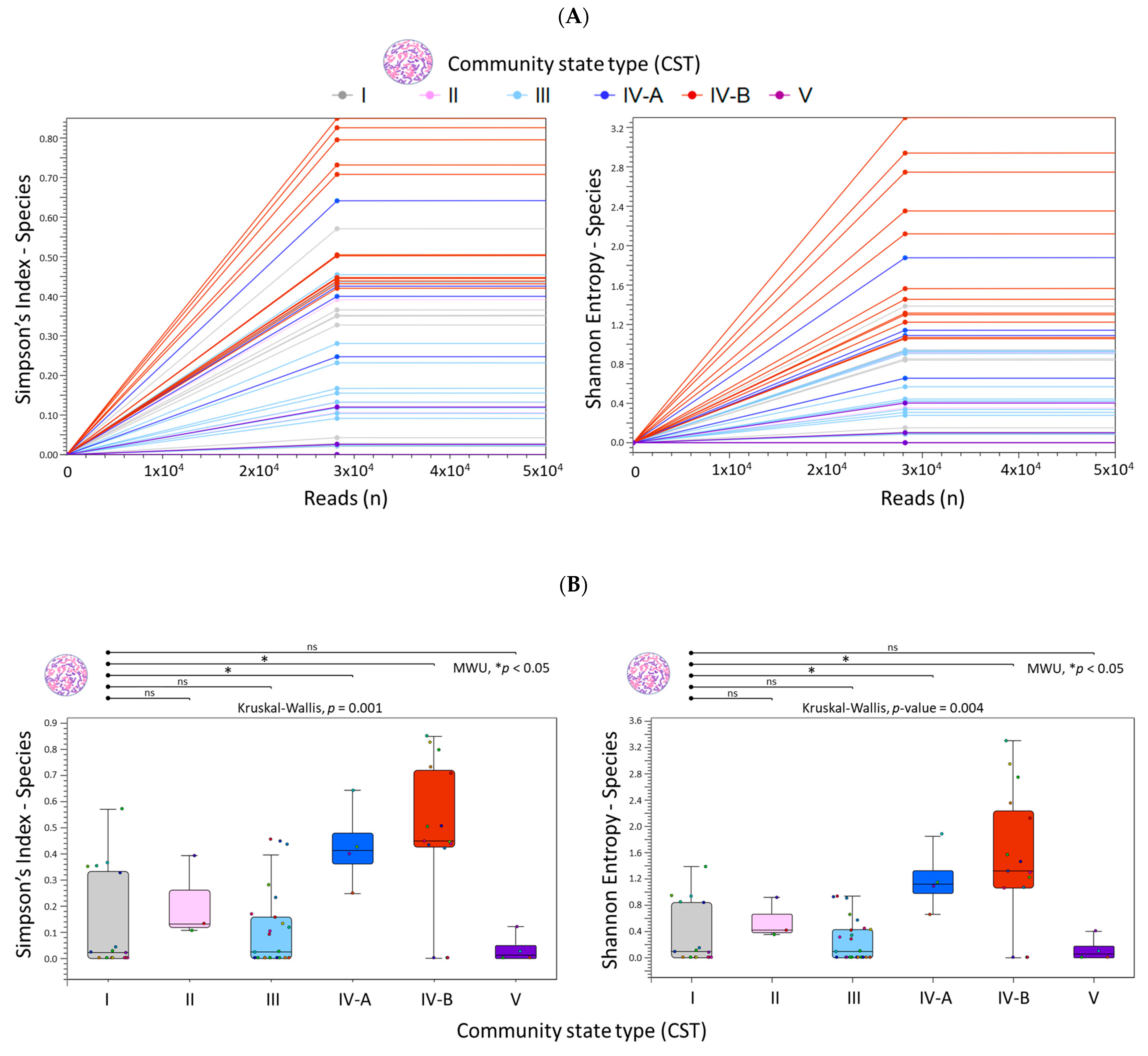
2.2. Diversity Analysis and Visualization of Microbial Communities
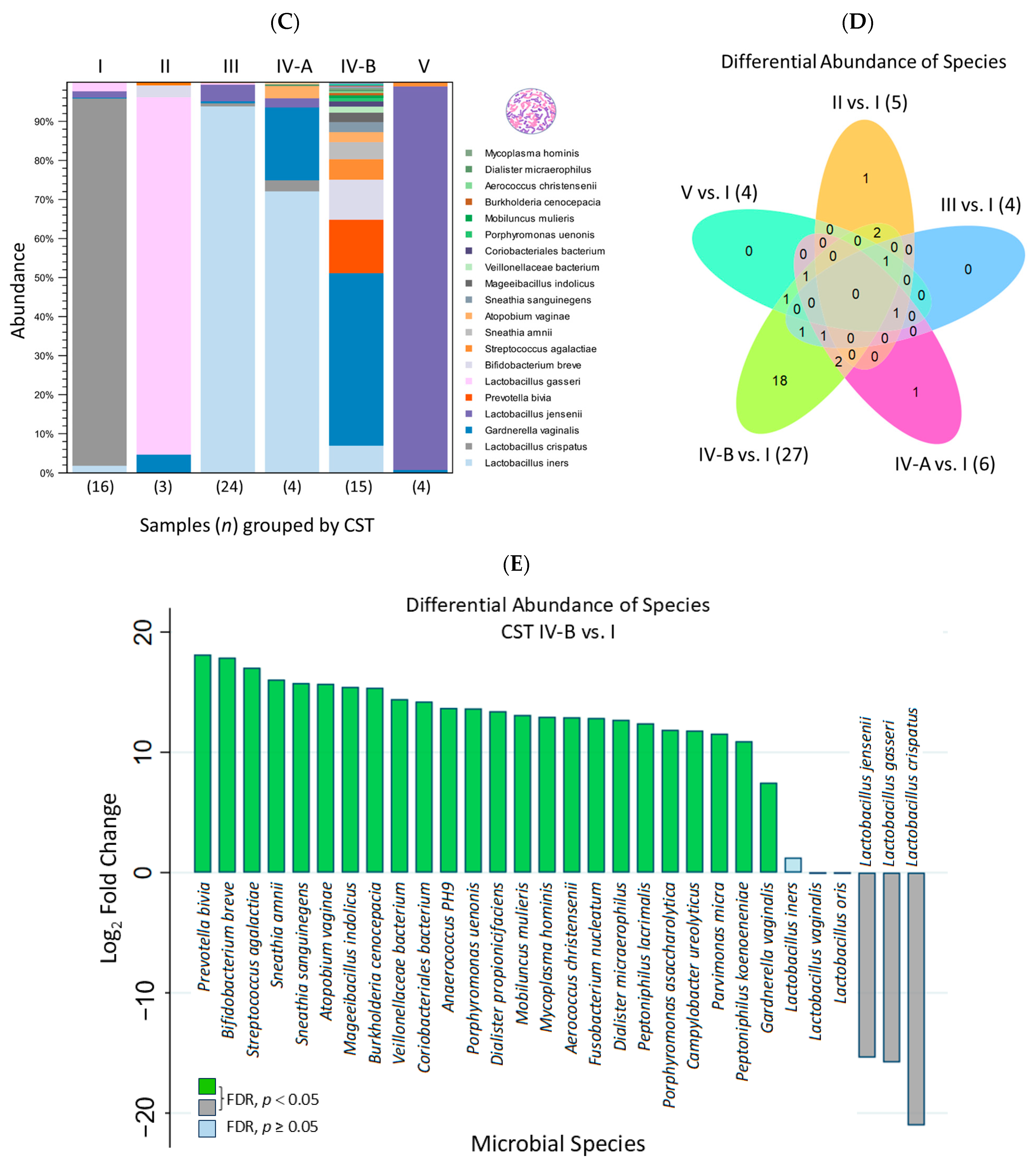
2.3. Differential Abundance Analysis and Visualization Using Hierarchical Clustering Heatmaps
3. Discussion
4. Materials and Methods
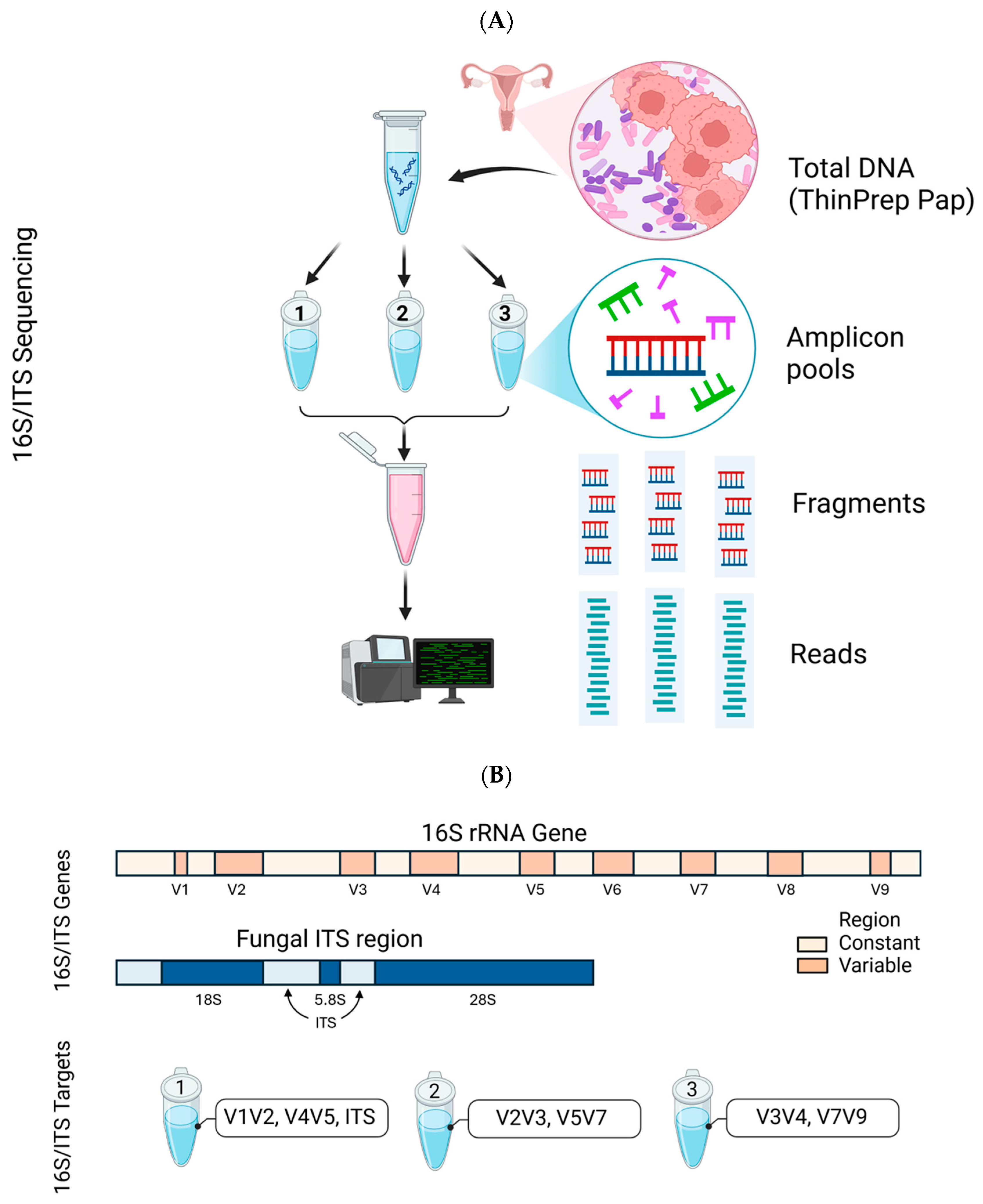
4.1. Clinical Samples, Mock Genomic Standard and Deep Sequencing
4.2. Customized Vaginal Microbiota Reference Database for CLC Workflows
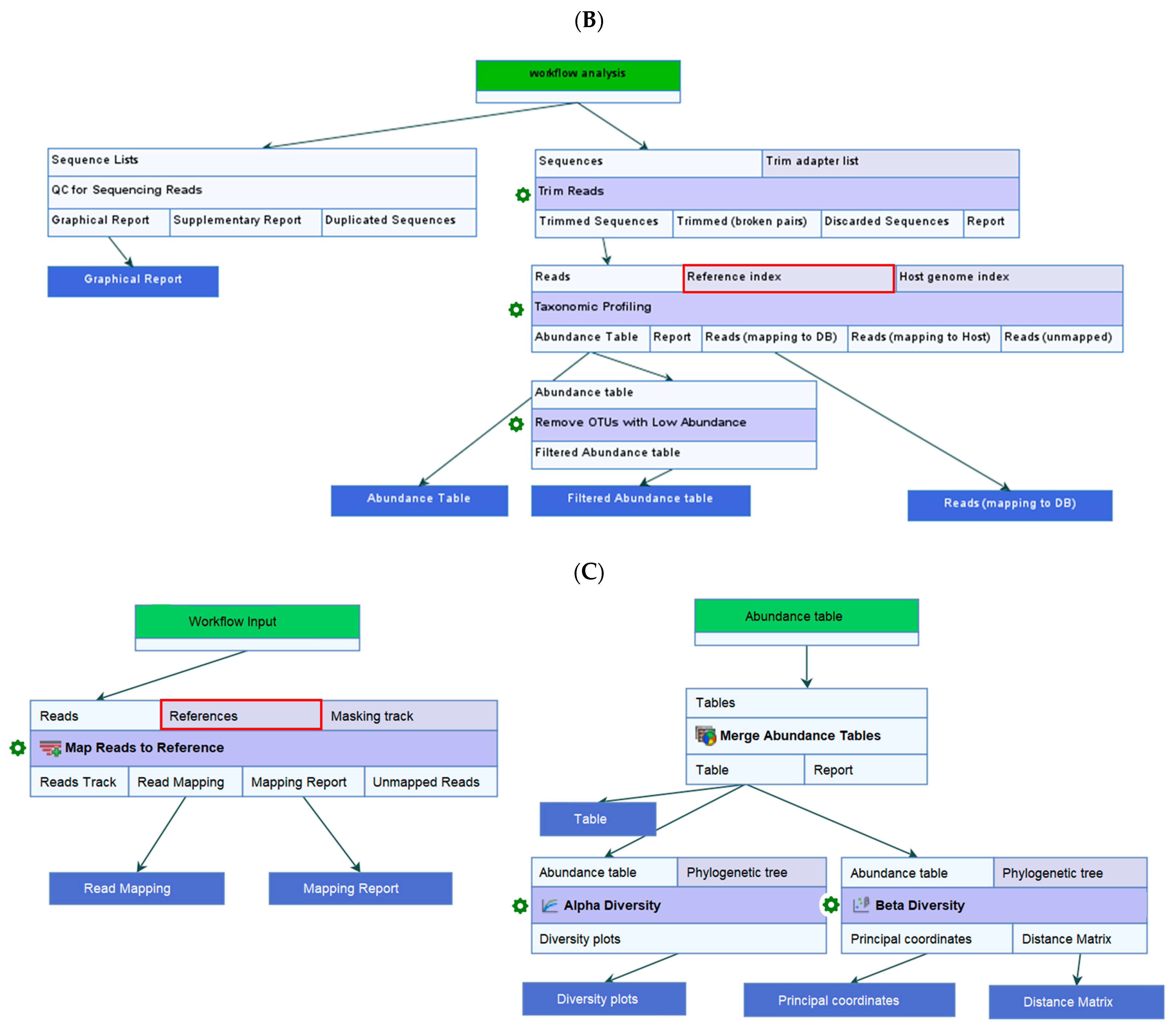
4.3. Data Quality Control (QC), Taxonomic Profiling, and Read Mapping
4.4. Estimate Alpha and Beta Diversities Workflow
4.5. Differential Abundance and Heatmap Analysis of Cervicovaginal Microbiota
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Dunkelberg, W.E. First isolation of Gardnerella vaginalis. J. Clin. Microbiol. 1991, 29, 2911. [Google Scholar] [CrossRef] [PubMed]
- Gardner, H.L.; Dukes, C.D. New etiologic agent in nonspecific bacterial vaginitis. Science 1954, 120, 853. [Google Scholar] [CrossRef]
- Gardner, H.L.; Dukes, C.D. Haemophilus vaginalis vaginitis: A newly defined specific infection previously classified non-specific vaginitis. Am. J. Obs. Gynecol. 1955, 69, 962–976. [Google Scholar] [CrossRef]
- Paramel Jayaprakash, T.; Schellenberg, J.J.; Hill, J.E. Resolution and characterization of distinct cpn60-based subgroups of Gardnerella vaginalis in the vaginal microbiota. PLoS ONE 2012, 7, e43009. [Google Scholar] [CrossRef] [PubMed]
- Gelber, S.E.; Aguilar, J.L.; Lewis, K.L.; Ratner, A.J. Functional and phylogenetic characterization of Vaginolysin, the human-specific cytolysin from Gardnerella vaginalis. J. Bacteriol. 2008, 190, 3896–3903. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Bacterial Vaginosis. Available online: https://www.who.int/news-room/fact-sheets/detail/bacterial-vaginosis (accessed on 6 June 2025).
- Shvartsman, E.; Hill, J.E.; Sandstrom, P.; MacDonald, K.S. Gardnerella Revisited: Species Heterogeneity, Virulence Factors, Mucosal Immune Responses, and Contributions to Bacterial Vaginosis. Infect. Immun. 2023, 91, e0039022. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, S.; Moll, W.M.; Swidsinski, A. Bacterial Vaginosis-Vaginal Polymicrobial Biofilms and Dysbiosis. Dtsch. Arztebl. Int. 2023, 120, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.; Rosca, A.S.; Cools, P.; Vaneechoutte, M.; Cerca, N. Gardnerella vaginalis Enhances Atopobium vaginae Viability in an in vitro Model. Front. Cell Infect. Microbiol. 2020, 10, 83. [Google Scholar] [CrossRef]
- Janulaitiene, M.; Gegzna, V.; Baranauskiene, L.; Bulavaitė, A.; Simanavicius, M.; Pleckaityte, M. Phenotypic characterization of Gardnerella vaginalis subgroups suggests differences in their virulence potential. PLoS ONE 2018, 13, e0200625. [Google Scholar] [CrossRef] [PubMed]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4680–4687. [Google Scholar] [CrossRef]
- Callahan, B.J.; DiGiulio, D.B.; Goltsman, D.S.A.; Sun, C.L.; Costello, E.K.; Jeganathan, P.; Biggio, J.R.; Wong, R.J.; Druzin, M.L.; Shaw, G.M.; et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc. Natl. Acad. Sci. USA 2017, 114, 9966–9971. [Google Scholar] [CrossRef]
- Gajer, P.; Brotman, R.M.; Bai, G.; Sakamoto, J.; Schütte, U.M.; Zhong, X.; Koenig, S.S.; Fu, L.; Ma, Z.S.; Zhou, X.; et al. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 2012, 4, 132ra52. [Google Scholar] [CrossRef]
- Ma, Z.S.; Li, L. Quantifying the human vaginal community state types (CSTs) with the species specificity index. PeerJ 2017, 5, e3366. [Google Scholar] [CrossRef]
- Du, L.; Dong, X.; Song, J.; Lei, T.; Liu, X.; Lan, Y.; Liu, X.; Wang, J.; Yue, B.; He, M.; et al. Temporal and spatial differences in the vaginal microbiome of Chinese healthy women. PeerJ 2023, 11, e16438. [Google Scholar] [CrossRef] [PubMed]
- Cocomazzi, G.; Contu, V.; De Stefani, S.; Del Pup, L.; Buccheri, M.; Antinori, M.; Parmegiani, L.; De Ruvo, D.; Marino, F.; Virgili, E.; et al. Refining Unfavorable Vaginal Microbial Community in Infertile Women Subjected to Precision Probiotic Intervention: An Exploratory Single-Arm, Prospective, Open-Label, Interventional Study. Microorganisms 2025, 13, 547. [Google Scholar] [CrossRef]
- Molina, M.A.; Andralojc, K.M.; Huynen, M.A.; Leenders, W.P.J.; Melchers, W.J.G. In-depth insights into cervicovaginal microbial communities and hrHPV infections using high-resolution microbiome profiling. NPJ Biofilms Microbiomes 2022, 8, 75. [Google Scholar] [CrossRef]
- Leon-Gomez, P.; Romero, V.I. Human papillomavirus, vaginal microbiota and metagenomics: The interplay between development and progression of cervical cancer. Front. Microbiol. 2025, 15, 1515258. [Google Scholar] [CrossRef]
- Alizhan, D.; Ukybassova, T.; Bapayeva, G.; Aimagambetova, G.; Kongrtay, K.; Kamzayeva, N.; Terzic, M. Cervicovaginal Microbiome: Physiology, Age-Related Changes, and Protective Role Against Human Papillomavirus Infection. J. Clin. Med. 2025, 14, 1521. [Google Scholar] [CrossRef]
- Norenhag, J.; Du, J.; Olovsson, M.; Verstraelen, H.; Engstrand, L.; Brusselaers, N. The vaginal microbiota, human papillomavirus and cervical dysplasia: A systematic review and network meta-analysis. BJOG 2020, 127, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Nicolò, S.; Tanturli, M.; Mattiuz, G.; Antonelli, A.; Baccani, I.; Bonaiuto, C.; Baldi, S.; Nannini, G.; Menicatti, M.; Bartolucci, G.; et al. Vaginal Lactobacilli and Vaginal Dysbiosis-Associated Bacteria Differently Affect Cervical Epithelial and Immune Homeostasis and Anti-Viral Defenses. Int. J. Mol. Sci. 2021, 22, 6487. [Google Scholar] [CrossRef] [PubMed]
- Musa, J.; Maiga, M.; Green, S.J.; Magaji, F.A.; Maryam, A.J.; Okolo, M.; Nyam, C.J.; Cosmas, N.T.; Silas, O.A.; Imade, G.E.; et al. Vaginal microbiome community state types and high-risk human papillomaviruses in cervical precancer and cancer in North-central Nigeria. BMC Cancer 2023, 23, 683. [Google Scholar] [CrossRef]
- Mitra, A.; MacIntyre, D.A.; Lee, Y.S.; Smith, A.; Marchesi, J.R.; Lehne, B.; Bhatia, R.; Lyons, D.; Paraskevaidis, E.; Li, J.V.; et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci. Rep. 2015, 5, 16865. [Google Scholar] [CrossRef]
- Sharifian, K.; Shoja, Z.; Jalilvand, S. The interplay between human papillomavirus and vaginal microbiota in cervical cancer development. Virol. J. 2023, 20, 73. [Google Scholar] [CrossRef]
- Wu, M.; Li, H.; Yu, H.; Yan, Y.; Wang, C.; Teng, F.; Fan, A.; Xue, F. Disturbances of Vaginal Microbiome Composition in Human Papillomavirus Infection and Cervical Carcinogenesis: A Qualitative Systematic Review. Front. Oncol. 2022, 12, 941741. [Google Scholar] [CrossRef] [PubMed]
- Garcia, E.M.; Kraskauskiene, V.; Koblinski, J.E.; Jefferson, K.K. Interaction of Gardnerella vaginalis and Vaginolysin with the Apical versus Basolateral Face of a Three-Dimensional Model of Vaginal Epithelium. Infect. Immun. 2019, 87, e00646-18. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.P.; Harris, C.C. Inflammation and cancer: An ancient link with novel potentials. Int. J. Cancer 2007, 121, 2373–2380. [Google Scholar] [CrossRef] [PubMed]
- Okada, F.; Fujii, J. Molecular mechanisms of inflammation-induced carcinogenesis. J. Clin. Biochem. Nutr. 2006, 39, 103–113. [Google Scholar] [CrossRef][Green Version]
- Wu, Y.; Antony, S.; Meitzler, J.L.; Doroshow, J.H. Molecular mechanisms underlying chronic inflammation-associated cancers. Cancer Lett. 2014, 345, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Lonkar, P.; Dedon, P.C. Reactive species and DNA damage in chronic inflammation: Reconciling chemical mechanisms and biological fates. Int. J. Cancer 2011, 128, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Baby, D.; Rajguru, J.P.; Patil, P.B.; Thakkannavar, S.S.; Pujari, V.B. Inflammation and cancer. Ann. Afr. Med. 2019, 18, 121–126. [Google Scholar] [CrossRef]
- Poretsky, R.; Rodriguez-R, L.M.; Luo, C.; Tsementzi, D.; Konstantinidis, K.T. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE 2014, 9, e93827. [Google Scholar] [CrossRef]
- Graspeuntner, S.; Loeper, N.; Künzel, S.; Baines, J.F.; Rupp, J. Selection of validated hypervariable regions is crucial in 16S-based microbiota studies of the female genital tract. Sci. Rep. 2018, 8, 9678. [Google Scholar] [CrossRef] [PubMed]
- Weinroth, M.D.; Belk, A.D.; Dean, C.; Noyes, N.; Dittoe, D.K.; Rothrock, M.J.; Ricke, S.C.; Myer, P.R.; Henniger, M.T.; Ramírez, G.A.; et al. Considerations and best practices in animal science 16S ribosomal RNA gene sequencing microbiome studies. J. Anim. Sci. 2022, 100, skab346. [Google Scholar] [CrossRef] [PubMed]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K. Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing. mSphere 2021, 6, e01202-20. [Google Scholar] [CrossRef]
- Regueira-Iglesias, A.; Balsa-Castro, C.; Blanco-Pintos, T.; Tomás, I. Critical review of 16S rRNA gene sequencing workflow in microbiome studies: From primer selection to advanced data analysis. Mol. Oral. Microbiol. 2023, 38, 347–399. [Google Scholar] [CrossRef]
- Hill, J.; Albert, A. Resolution and cooccurrence patterns of Gardnerella leopoldii, G. swidsinskii, G. piotii, and G. vaginalis within the vaginal microbiome. Infect. Immun. 2019, 87, e00532-19. [Google Scholar] [CrossRef] [PubMed]
- Qiagen: QIAseq 16S/ITS Screening Panels and Index Kits. Available online: https://www.qiagen.com/us/products/next-generation-sequencing/metagenomics/targeted-metagenomics/qiaseq-16s-its-screening-panels-and-index-kits (accessed on 6 June 2025).
- American Type Culture Collection (ATCC). Vaginal Microbiome Genomic Mix. Available online: https://www.atcc.org/products/msa-1007 (accessed on 6 June 2025).
- Qiagen Digital Insights. CLC Genomics Workbench 25.0.1. Available online: https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Introduction_CLC_Genomics_Workbench.html (accessed on 6 June 2025).
- Qiagen Digital Insights. CLC Microbial Genomics Module 25.0.1. Available online: https://resources.qiagenbioinformatics.com/manuals/clcmgm/current/index.php?manual=Introduction.html (accessed on 8 May 2025).
- Tanizawa, Y.; Tada, I.; Kobayashi, H.; Endo, A.; Maeno, S.; Toyoda, A.; Arita, M.; Nakamura, Y.; Sakamoto, M.; Ohkuma, M.; et al. Lactobacillus paragasseri sp. nov., a sister taxon of Lactobacillus gasseri, based on whole-genome sequence analyses. Int. J. Syst. Evol. Microbiol. 2018, 68, 3512–3517. [Google Scholar] [CrossRef] [PubMed]
- Ene, A.; Stegman, N.; Wolfe, A.; Putonti, C. Genomic insights into Lactobacillus gasseri and Lactobacillus paragasseri. PeerJ. 2022, 10, e13479. [Google Scholar] [CrossRef] [PubMed]
- Katiraei, S.; Anvar, Y.; Hoving, L.; Berbée, J.F.P.; van Harmelen, V.; Willems van Dijk, K. Evaluation of Full-Length Versus V4-Region 16S rRNA Sequencing for Phylogenetic Analysis of Mouse Intestinal Microbiota After a Dietary Intervention. Curr. Microbiol. 2022, 79, 276. [Google Scholar] [CrossRef] [PubMed]
- Graham, A.S.; Patel, F.; Little, F.; van der Kouwe, A.; Kaba, M.; Holmes, M.J. Using short-read 16S rRNA sequencing of multiple variable regions to generate high-quality results to a species level. Front. Bioinform. 2025, 5, 1484113. [Google Scholar] [CrossRef]
- Vaneechoutte, M. Lactobacillus iners, the unusual suspect. Res. Microbiol. 2017, 168, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Vaneechoutte, M. The human vaginal microbial community. Res. Microbiol. 2017, 168, 811–825. [Google Scholar] [CrossRef]
- France, M.T.; Mendes-Soares, H.; Forney, L.J. Genomic Comparisons of Lactobacillus crispatus and Lactobacillus iners Reveal Potential Ecological Drivers of Community Composition in the Vagina. Appl. Environ. Microbiol. 2016, 82, 7063–7073. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.C.; Hill, J.E. Quantification, isolation and characterization of Bifidobacterium from the vaginal microbiomes of reproductive aged women. Anaerobe 2017, 47, 145–156. [Google Scholar] [CrossRef]
- Di Paola, M.; Sani, C.; Clemente, A.M.; Iossa, A.; Perissi, E.; Castronovo, G.; Tanturli, M.; Rivero, D.; Cozzolino, F.; Cavalieri, D.; et al. Characterization of cervico-vaginal microbiota in women developing persistent high-risk Human Papillomavirus infection. Sci. Rep. 2017, 7, 10200. [Google Scholar] [CrossRef]
- Zhou, Z.W.; Long, H.Z.; Cheng, Y.; Luo, H.Y.; Wen, D.D.; Gao, L.C. From Microbiome to Inflammation: The Key Drivers of Cervical Cancer. Front. Microbiol. 2021, 12, 767931. [Google Scholar] [CrossRef]
- Horrocks, V.; Hind, C.K.; Wand, M.E.; Fady, P.E.; Chan, J.; Hopkins, J.C.; Houston, G.L.; Tribe, R.M.; Sutton, J.M.; Mason, A.J. Nuclear Magnetic Resonance Metabolomics of Symbioses between Bacterial Vaginosis-Associated Bacteria. mSphere 2022, 7, e0016622. [Google Scholar] [CrossRef] [PubMed]
- Haggerty, C.L.; Totten, P.A.; Tang, G.; Astete, S.G.; Ferris, M.J.; Norori, J.; Bass, D.C.; Martin, D.H.; Taylor, B.D.; Ness, R.B. Identification of novel microbes associated with pelvic inflammatory disease and infertility. Sex. Transm. Infect. 2016, 92, 441–446. [Google Scholar] [CrossRef]
- Peng, Y.; Tang, Q.; Wu, S.; Zhao, C. Associations of Atopobium, Garderella, Megasphaera, Prevotella, Sneathia, and Streptococcus with human papillomavirus infection, cervical intraepithelial neoplasia, and cancer: A systematic review and meta-analysis. BMC Infect. Dis. 2025, 25, 708. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Chen, Y.; Gao, J.; Wu, M.; Li, C.; Wang, Z.; Huang, N.; Cui, L.; Du, M.; Ying, C. Characterization of Vaginal Microbiota in Women with Recurrent Spontaneous Abortion That Can Be Modified by Drug Treatment. Front. Cell Infect. Microbiol. 2021, 11, 680643. [Google Scholar] [CrossRef] [PubMed]
- Gruwier, L.; Sprenkels, A.; Hulsbosch, S.; Vankeerberghen, A.; Cartuyvels, R. Sneathia amnii bacteraemia and chorioamnionitis leading to second trimester abortion: A case report. Access Microbiol. 2021, 3, 000290. [Google Scholar] [CrossRef] [PubMed]
- Austin, M.N.; Rabe, L.K.; Srinivasan, S.; Fredricks, D.N.; Wiesenfeld, H.C.; Hillier, S.L. Mageeibacillus indolicus gen. nov., sp. nov.: A novel bacterium isolated from the female genital tract. Anaerobe 2015, 32, 37–42. [Google Scholar] [CrossRef]
- Gao, J.; Peng, Y.; Jiang, N.; Shi, Y.; Ying, C. High-Throughput Sequencing-Based Analysis of Changes in the Vaginal Microbiome during the Disease Course of Patients with Bacterial Vaginosis: A Case-Control Study. Biology 2022, 11, 1797. [Google Scholar] [CrossRef]
- Ma, X.; Wen, G.; Zhao, Z.; Lu, L.; Li, T.; Gao, N.; Han, G. Alternations in the human skin, gut and vaginal microbiomes in perimenopausal or postmenopausal Vulvar lichen sclerosus. Sci. Rep. 2024, 14, 8429. [Google Scholar] [CrossRef]
- Zhu, H.; Yip, H.C.; Cheung, M.K.; Chan, H.C.; Ng, C.; Lau, E.H.L.; Yeung, Z.W.C.; Wong, E.W.Y.; Leung, L.; Qu, X.; et al. Convergent dysbiosis of upper aerodigestive microbiota between patients with esophageal and oral cavity squamous cell carcinoma. Int. J. Cancer 2023, 152, 1903–1915. [Google Scholar] [CrossRef]
- Chen, I.; Kelkar, Y.D.; Gu, Y.; Zhou, J.; Qiu, X.; Wu, H. High-dimensional linear state space models for dynamic microbial interaction networks. PLoS ONE 2017, 12, e0187822. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.; Cerca, N. Influence of Biofilm Formation by Gardnerella vaginalis and Other Anaerobes on Bacterial Vaginosis. J. Infect. Dis. 2015, 212, 1856–1861. [Google Scholar] [CrossRef]
- Obregon, M.; Khan, A. Cardiac Tamponade Caused by Campylobacter ureolyticus Purulent Effusion. Cureus 2024, 16, e56051. [Google Scholar] [CrossRef] [PubMed]
- Maki, J.J.; Howard, M.; Connelly, S.; Pettengill, M.A.; Hardy, D.J.; Cameron, A. Species Delineation and Comparative Genomics within the Campylobacter ureolyticus Complex. J. Clin. Microbiol. 2023, 61, e0004623. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.Y.; Cho, J.H.; Shin, Y.; Paek, J.; Park, I.S.; Kim, J.S.; Kim, W.; Ma, J.Y.; Park, S.J.; Chang, Y.H. Peptoniphilus rhinitidis sp. nov., isolated from specimens of chronic rhinosinusitis. Anaerobe 2014, 30, 30–34. [Google Scholar] [CrossRef]
- Beye, M.; Bakour, S.; Le Dault, E.; Rathored, J.; Michelle, C.; Cadoret, F.; Raoult, D.; Fournier, P.E. Peptoniphilus lacydonensis sp. nov., a new human-associated species isolated from a patient with chronic refractory sinusitis. New Microbes New Infect. 2018, 23, 61–69. [Google Scholar] [CrossRef]
- Tabatabaei, N.; Eren, A.M.; Barreiro, L.B.; Yotova, V.; Dumaine, A.; Allard, C.; Fraser, W.D. Vaginal microbiome in early pregnancy and subsequent risk of spontaneous preterm birth: A case-control study. BJOG 2019, 126, 349–358. [Google Scholar] [CrossRef]
- Carlstein, C.; Marie Søes, L.; Jørgen Christensen, J. Aerococcus christensenii as Part of Severe Polymicrobial Chorioamnionitis in a Pregnant Woman. Open Microbiol. J. 2016, 10, 27–31. [Google Scholar] [CrossRef]
- Aziz, M.; Hartley, K.; Chadha, D. Recurrent Cerebral Embolic Infarcts in a Patient with a Mechanical Valve: A Rare Case of Infective Endocarditis Caused by Parvimonas micra. Cureus 2024, 16, e71521. [Google Scholar] [CrossRef] [PubMed]
- Itagaki, S.; Kenzaka, T. Pyogenic Spondylitis Caused by Parvimonas micra: A Case Report. Cureus 2023, 15, e48665. [Google Scholar] [CrossRef] [PubMed]
- Higashi, D.L.; Krieger, M.C.; Qin, H.; Zou, Z.; Palmer, E.A.; Kreth, J.; Merritt, J. Who is in the driver’s seat? Parvimonas micra: An understudied pathobiont at the crossroads of dysbiotic disease and cancer. Environ. Microbiol. Rep. 2023, 15, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Du, J.; Dou, Y.; Chen, R.; Li, Y.; Zhao, L.; Liu, H.; Zhang, K. The Associations of Genital Mycoplasmas with Female Infertility and Adverse Pregnancy Outcomes: A Systematic Review and Meta-analysis. Reprod. Sci. 2021, 28, 3013–3031. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Yin, J.; Wang, W.; Zhao, F.; Ding, X.; Yu, H.; Zhang, X.; Wang, B. The associations between low abundance of Mycoplasma hominis and female fecundability: A pregnancy-planning cohort study. BMC Microbiol. 2022, 22, 121. [Google Scholar] [CrossRef] [PubMed]
- Periaiah, P.; Antony, T.; Samuel, S. Identification of Burkholderia cepacia Complex: Comparing Conventional, Automated, and Molecular Methods in a Tertiary Care Center. Cureus 2024, 16, e70847. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, S.; Sajjan, U.S. Host evasion by Burkholderia cenocepacia. Front. Cell Infect. Microbiol. 2012, 1, 25. [Google Scholar] [CrossRef]
- Hugon, P.; Mishra, A.K.; Robert, C.; Raoult, D.; Fournier, P.E. Non-contiguous finished genome sequence and description of Anaerococcus vaginalis. Stand. Genom. Sci. 2012, 6, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Jumas-Bilak, E.; Jean-Pierre, H.; Carlier, J.P.; Teyssier, C.; Bernard, K.; Gay, B.; Campos, J.; Morio, F.; Marchandin, H. Dialister micraerophilus sp. nov. and Dialister propionicifaciens sp. nov., isolated from human clinical samples. Int. J. Syst. Evol. Microbiol. 2005, 55 Pt 6, 2471–2478. [Google Scholar] [CrossRef] [PubMed]
- Eastment, M.C.; Balkus, J.E.; Richardson, B.A.; Srinivasan, S.; Kimani, J.; Anzala, O.; Schwebke, J.; Fiedler, T.L.; Fredricks, D.N.; McClelland, R.S. Association Between Vaginal Bacterial Microbiota and Vaginal Yeast Colonization. J. Infect. Dis. 2021, 223, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, H.; Tadera, K.; Omori, K.; Nomura, T.; Shigemoto, N.; Ohge, H. A case of bacteremia caused by Dialister micraerophilus with Enterocloster clostridioformis and Eggerthella lenta in a patient with pyometra. BMC Infect. Dis. 2024, 24, 128. [Google Scholar] [CrossRef]
- WHO: Group B Streptococcus Infection Causes an Estimated 150,000 Preventable Stillbirths and Infant Deaths Every Year. Available online: https://www.who.int/news/item/05-11-2017-group-b-streptococcus-infection-causes-an-estimated-150-000-preventable-stillbirths-and-infant-deaths-every-year (accessed on 6 June 2025).
- Yuan, X.Y.; Liu, H.Z.; Liu, J.F.; Sun, Y.; Song, Y. Pathogenic mechanism, detection methods and clinical significance of group B Streptococcus. Future Microbiol. 2021, 16, 671–685. [Google Scholar] [CrossRef] [PubMed]
- ACOG Committee Opinion. Prevention of Group B Streptococcal Early-Onset Disease in Newborns No. 797. February 2020. Available online: https://www.acog.org/clinical/clinical-guidance/committee-opinion/articles/2020/02/prevention-of-group-b-streptococcal-early-onset-disease-in-newborns (accessed on 6 June 2025).
- Raabe, V.N.; Shane, A.L. Group B Streptococcus (Streptococcus agalactiae). Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Takeda, S.; Tanaka, Y.; Takeichi, Y.; Hirata, H.; Tabuchi, A. A rare case of right-sided infective endocarditis caused by group B Streptococcus complicated with septic knee arthritis and subcutaneous abscess in the lower extremity. Acute Med. Surg. 2019, 7, e456. [Google Scholar] [CrossRef]
- Virtanen, S.; Saqib, S.; Kanerva, T.; Ventin-Holmberg, R.; Nieminen, P.; Holster, T.; Kalliala, I.; Salonen, A. Metagenome-validated combined amplicon sequencing and text mining-based annotations for simultaneous profiling of bacteria and fungi: Vaginal microbiota and mycobiota in healthy women. Microbiome 2024, 12, 273. [Google Scholar] [CrossRef]
- Qiagen: QIAamp DNA Kits for DNA Extraction. Available online: https://www.qiagen.com/us/products/discovery-and-translational-research/dna-rna-purification/dna-purification/genomic-dna/qiaamp-dna-kits?catno=51304 (accessed on 31 July 2025).
- He, Z.; Huo, X.; Piao, J. Rapid preparation of Candida genomic DNA: Combined use of enzymatic digestion and thermal disruption. AMB Express 2023, 13, 1. [Google Scholar] [CrossRef]
- Nagpal, S.; Mande, S.S.; Hooda, H.; Dutta, U.; Taneja, B. EnsembleSeq: A workflow towards real-time, rapid, and simultaneous multi-kingdom-amplicon sequencing for holistic and resource-effective microbiome research at scale. Microbiol. Spectr. 2024, 12, e0415023. [Google Scholar] [CrossRef] [PubMed]
- Pierre, J.F.; Phillips, G.J.; Chandra, L.C.; Rendina, D.N.; Thomas-Gosain, N.F.; Lubach, G.R.; Lyte, M.; Coe, C.L.; Gosain, A. Lyticase Facilitates Mycobiome Resolution Without Disrupting Microbiome Fidelity in Primates. J. Surg. Res. 2021, 267, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Gargis, A.S.; Kalman, L.; Lubin, I.M. Assuring the Quality of Next-Generation Sequencing in Clinical Microbiology and Public Health Laboratories. J. Clin. Microbiol. 2016, 54, 2857–2865. [Google Scholar] [CrossRef]
- Jones-Kellett, A.E.; McNichol, J.C.; Raut, Y.; Cain, K.R.; Ribalet, F.; Armbrust, E.V.; Follows, M.J.; Fuhrman, J.A. Amplicon sequencing with internal standards yields accurate picocyanobacteria cell abundances as validated with flow cytometry. ISME Commun. 2024, 4, ycae115. [Google Scholar] [CrossRef]
- Marotz, C.A.; Sanders, J.G.; Zuniga, C.; Zaramela, L.S.; Knight, R.; Zengler, K. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome 2018, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Qiagen: Microbial DNA qPCR Assay. Available online: https://www.qiagen.com/us/products/discovery-and-translational-research/pcr-qpcr-dpcr/qpcr-assays-and-instruments/microbial-dna-qpcr-assays-and-panels/microbial-dna-qpcr-assays (accessed on 31 July 2025).
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic. Acids. Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Shen-Gunther, J.; Xia, Q.; Cai, H.; Wang, Y. HPV DeepSeq: An Ultra-Fast Method of NGS Data Analysis and Visualization Using Automated Workflows and a Customized Papillomavirus Database in CLC Genomics Workbench. Pathogens 2021, 10, 1026. [Google Scholar] [CrossRef] [PubMed]
- Qiagen: Qiasymphony DSP DNA Kit. Available online: https://www.qiagen.com/us/products/diagnostics-and-clinical-research/sample-processing/qiasymphony-dsp-dna-kits-us (accessed on 31 July 2025).
- Qiagen: Qiaxpert System. Available online: https://www.qiagen.com/us/products/instruments-and-automation/quality-control-fragment-analysis/qiaxpert-system?catno=9002340 (accessed on 31 July 2025).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen-Gunther, J.; Xia, Q.; Cai, H.; Wang, Y. Cervicovaginal Microbiome and HPV: A Standardized Approach to 16S/ITS NGS and Microbial Community Profiling for Viral Association. Int. J. Mol. Sci. 2025, 26, 8090. https://doi.org/10.3390/ijms26168090
Shen-Gunther J, Xia Q, Cai H, Wang Y. Cervicovaginal Microbiome and HPV: A Standardized Approach to 16S/ITS NGS and Microbial Community Profiling for Viral Association. International Journal of Molecular Sciences. 2025; 26(16):8090. https://doi.org/10.3390/ijms26168090
Chicago/Turabian StyleShen-Gunther, Jane, Qingqing Xia, Hong Cai, and Yufeng Wang. 2025. "Cervicovaginal Microbiome and HPV: A Standardized Approach to 16S/ITS NGS and Microbial Community Profiling for Viral Association" International Journal of Molecular Sciences 26, no. 16: 8090. https://doi.org/10.3390/ijms26168090
APA StyleShen-Gunther, J., Xia, Q., Cai, H., & Wang, Y. (2025). Cervicovaginal Microbiome and HPV: A Standardized Approach to 16S/ITS NGS and Microbial Community Profiling for Viral Association. International Journal of Molecular Sciences, 26(16), 8090. https://doi.org/10.3390/ijms26168090







