EPIFBMC: A New Model for Enhancer–Promoter Interaction Prediction
Abstract
1. Introduction
2. Results
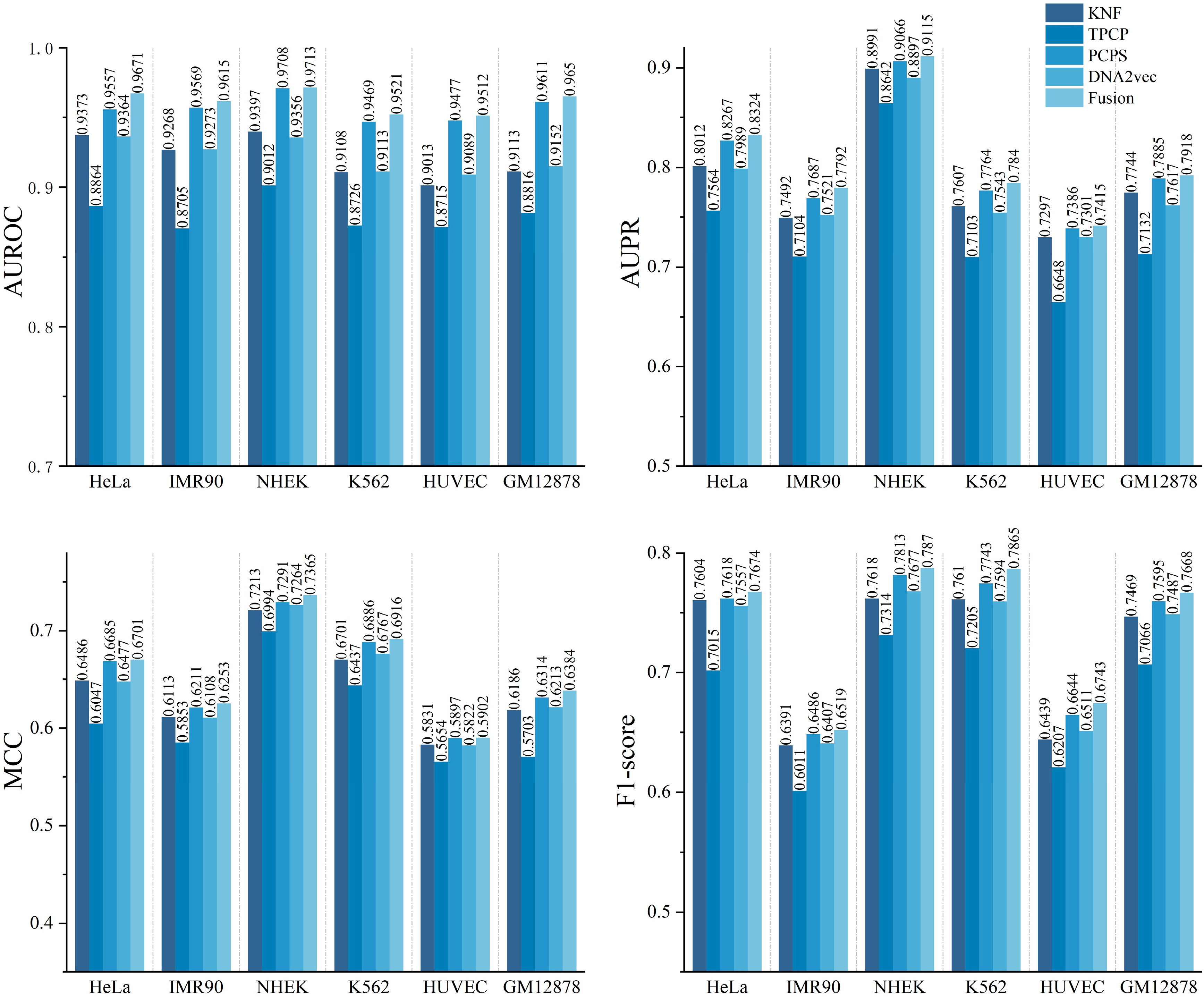
2.1. Performance Evaluation of Different DNA Sequence Features
2.2. Performance Evaluation of Models Based Only on DNA Sequences
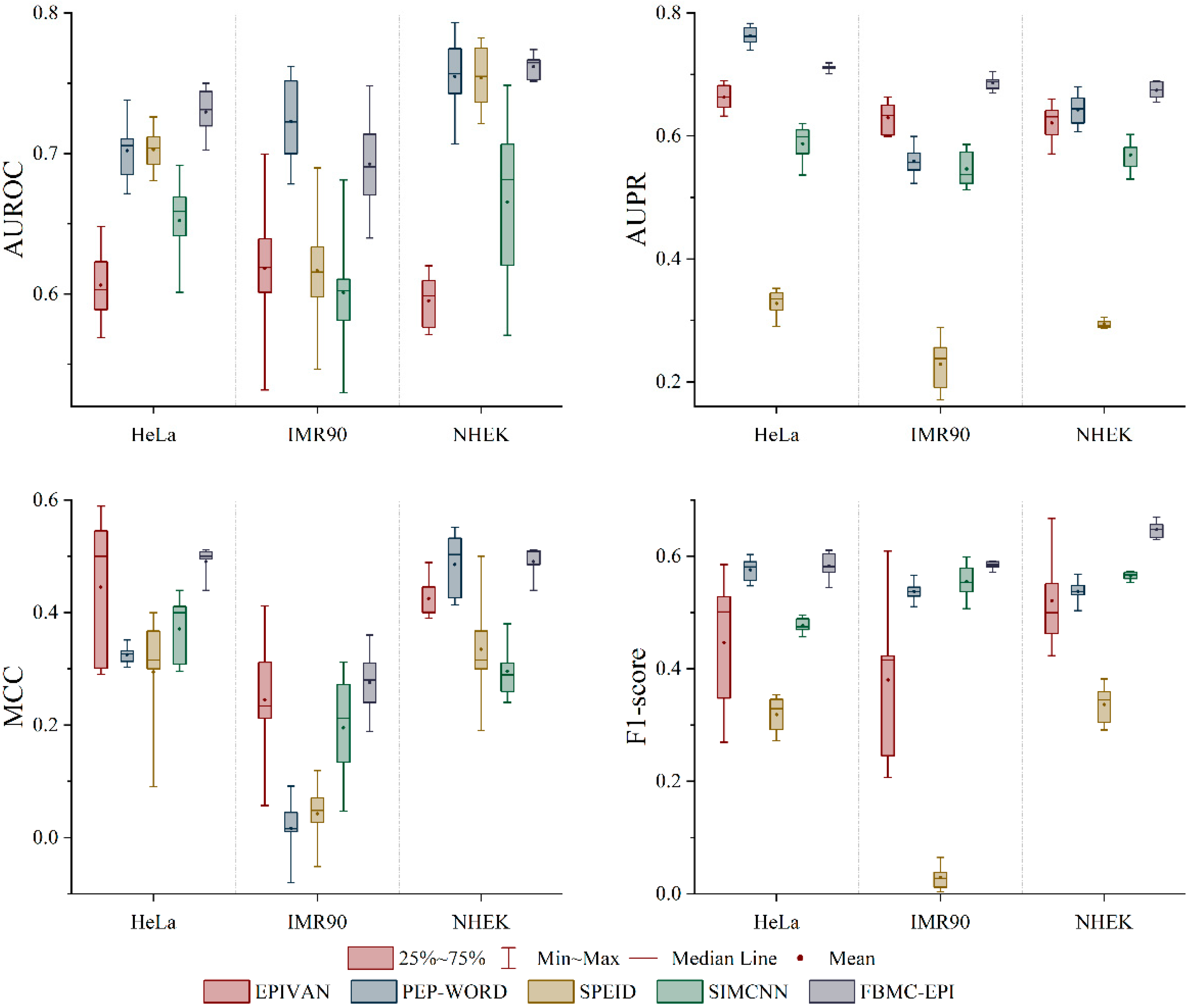
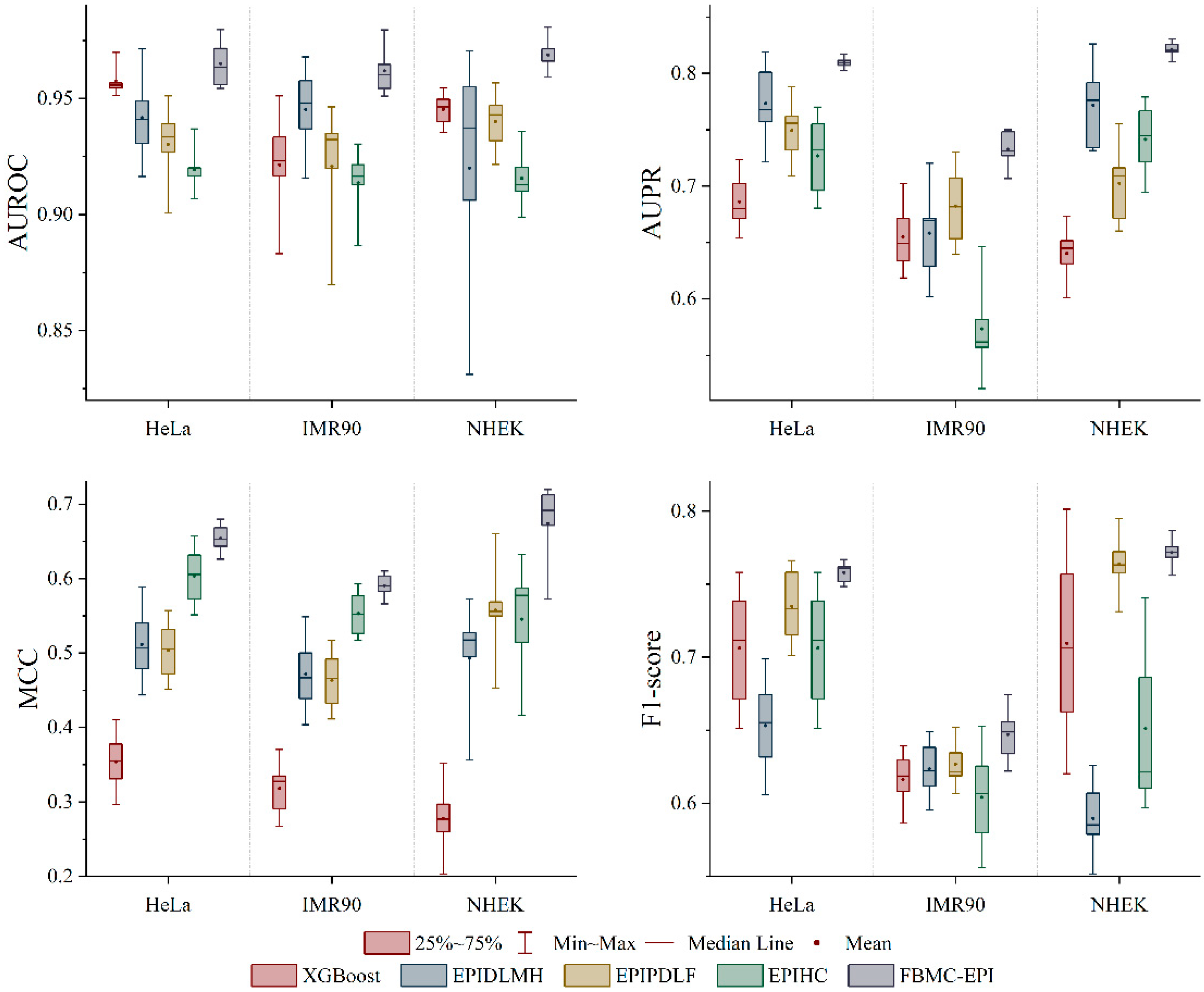
2.3. Performance Comparison with State-of-the-Art Methods
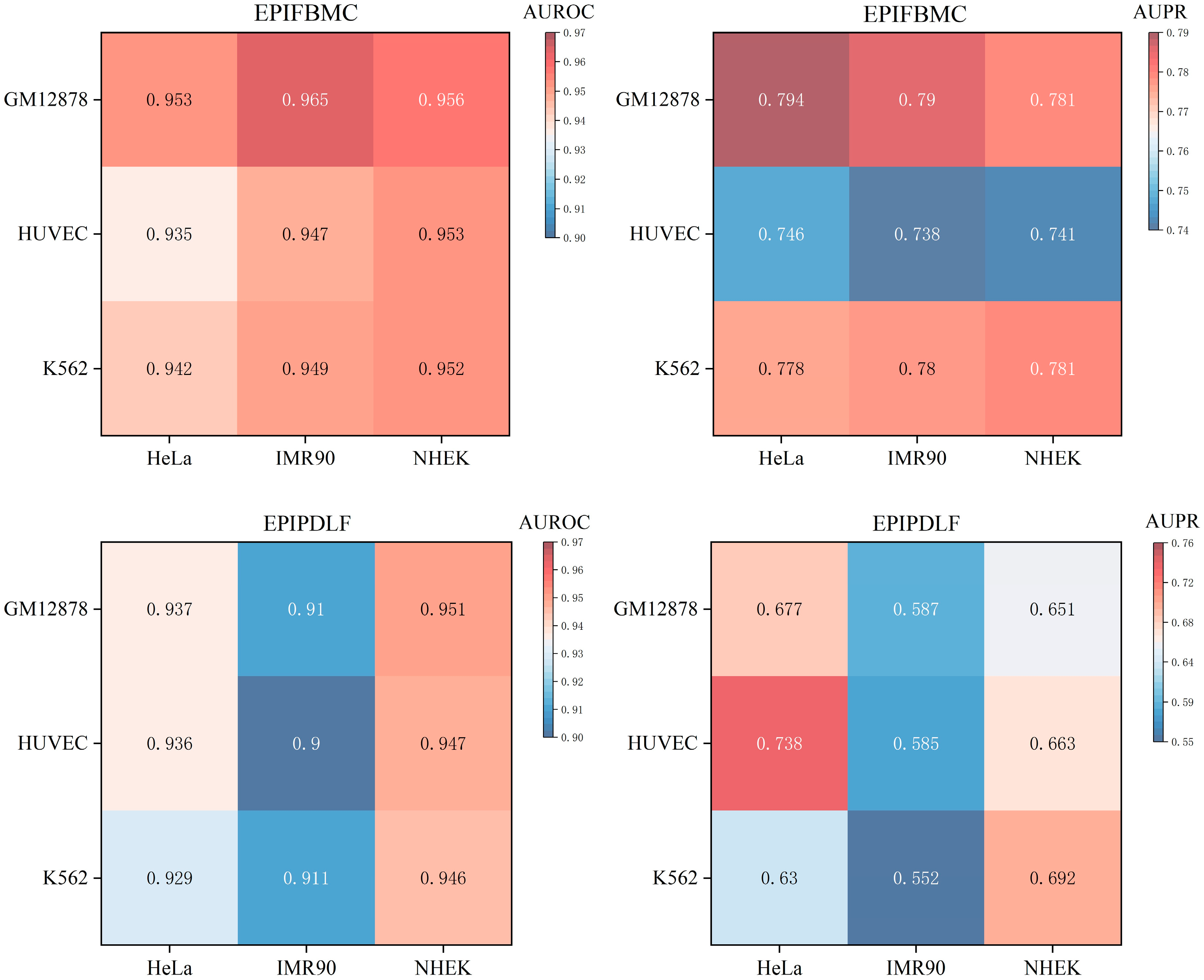
2.4. Cross-Cell-Line Validation
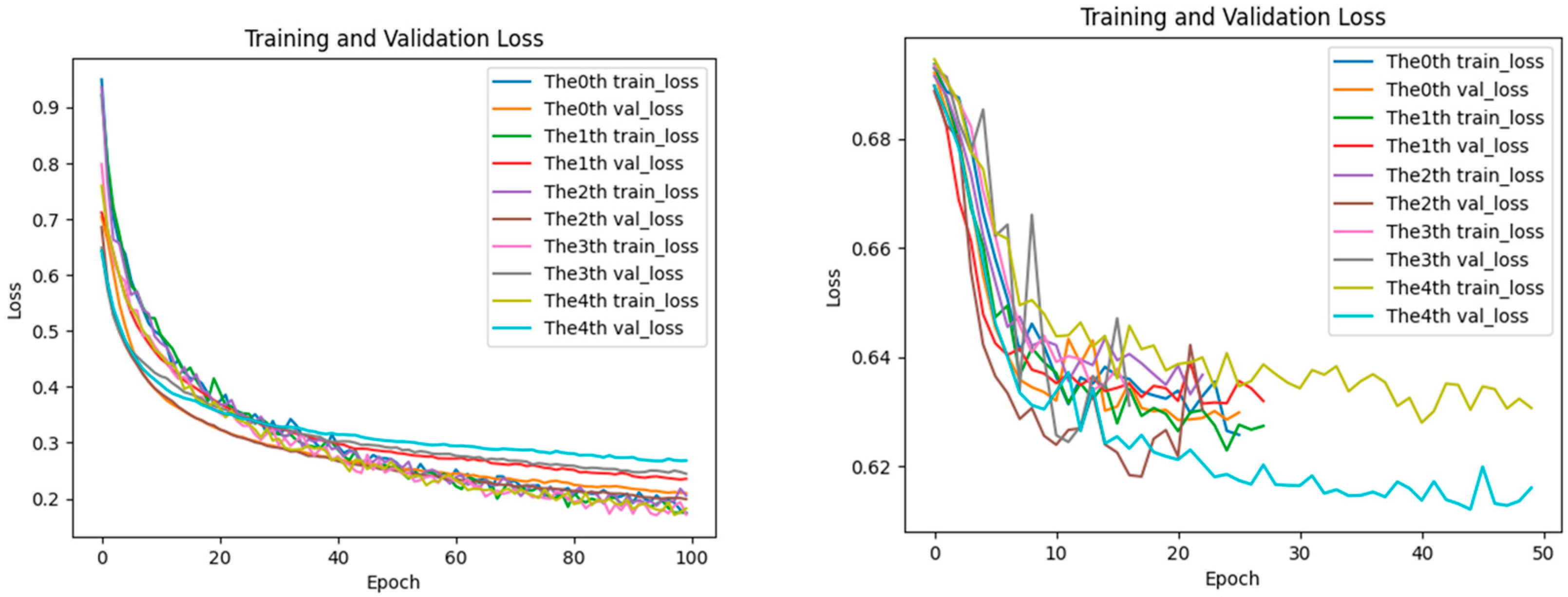
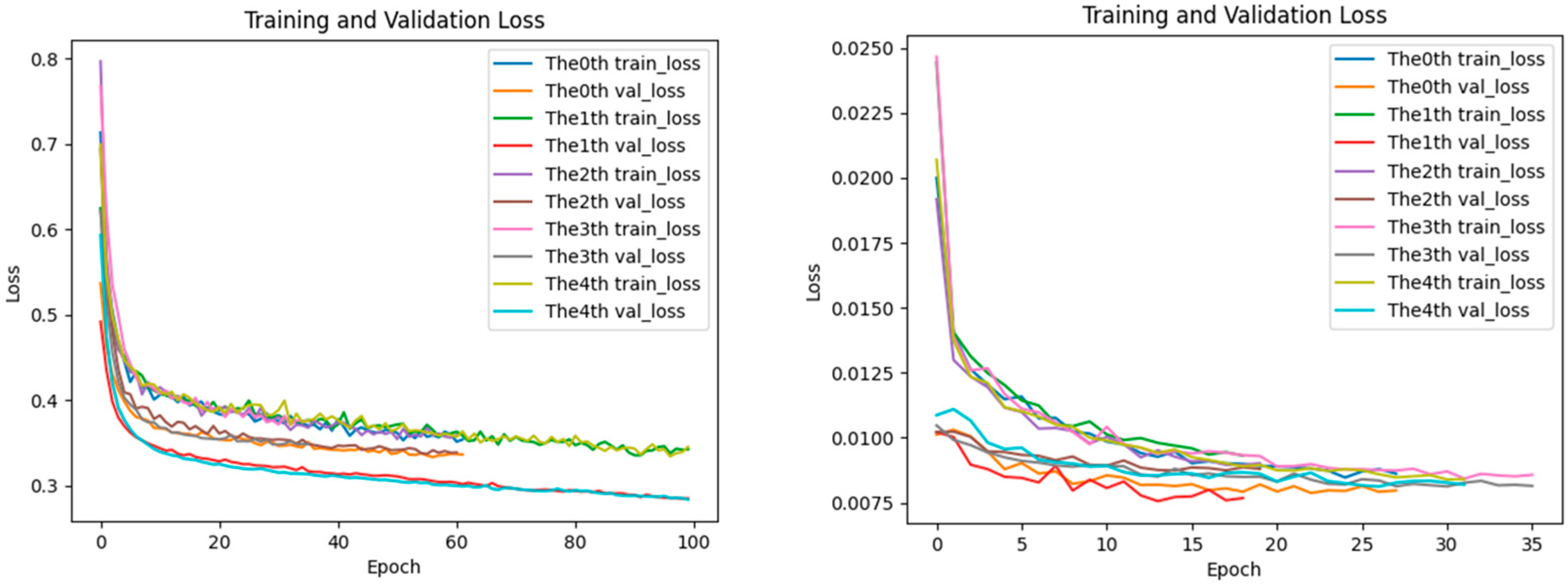
2.5. Analysis of Loss Performance for Training and Testing Datasets
3. Discussion
4. Materials and Methods
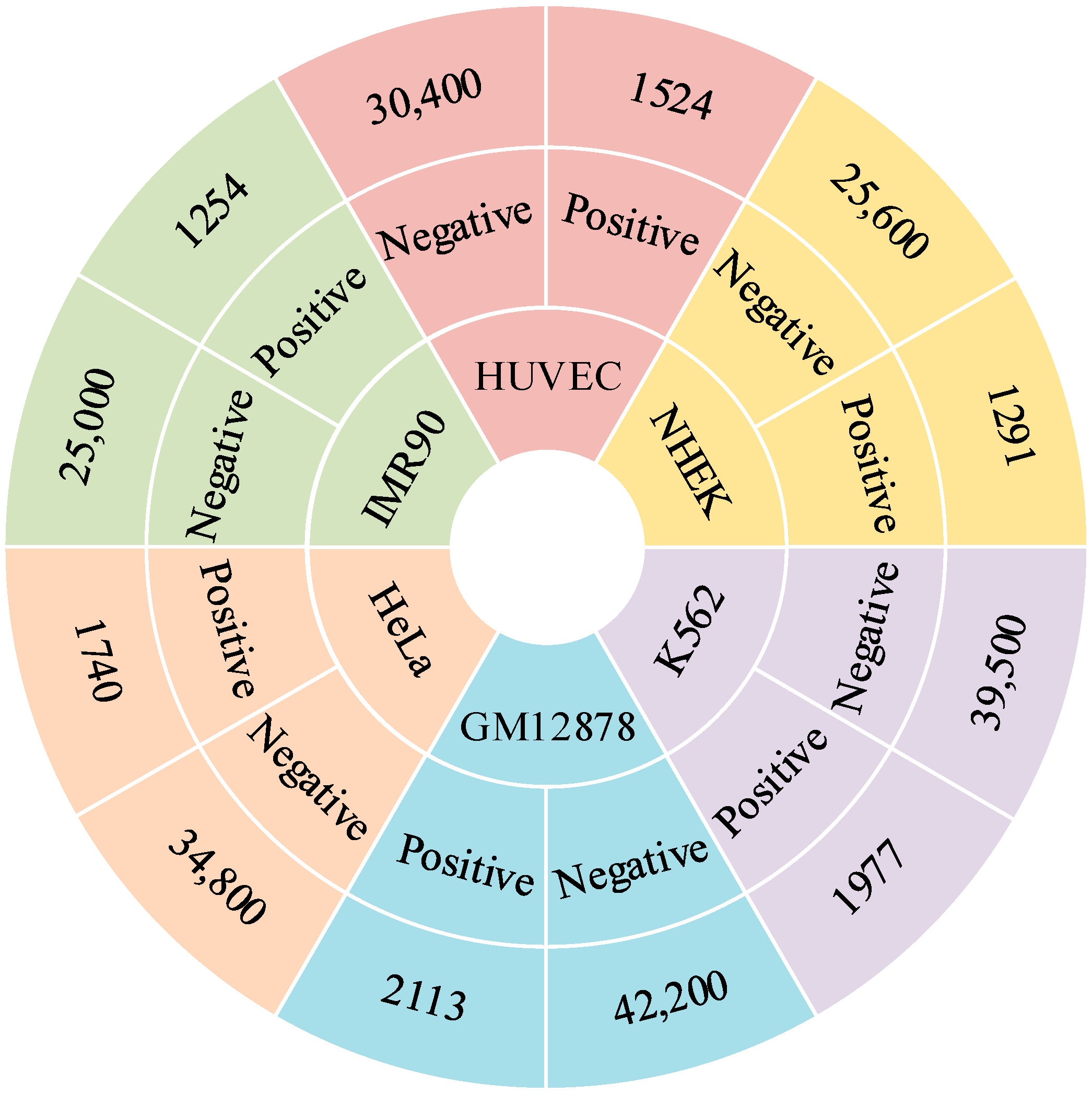
4.1. Benchmark Datasets
4.2. Model Framework
4.2.1. Overall Framework
4.2.2. Four-Encoding
KNF (K-Mer Nucleotide Frequency)
TPCP (Trinucleotide Physicochemical Properties)
PCPS (Position Conservation and Position Special Scoring)
DNA2vec (DNA Sequence Embedding)
4.2.3. BESL (Balanced Ensemble Subset Learning)
4.2.4. MCANet-Model
MCNet (Multi-Scale Convolutional Network)
MANet (Multi Attention Network)
4.3. Effect Evaluation Indicators
4.4. Experimental Environment
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EPIs | Enhancer–Promoter Interactions |
| BESL | Balanced Ensemble Subset Learning |
| Hi-C | high-throughput chromosome conformation capture |
| 3C | chromosome conformation capture |
| 4C | circular chromosome conformation capture |
| ChIA-PET | paired-end tag chromatin interaction analysis |
References
- Wang, H.; Huang, B.; Wang, J. Predict long-range enhancer regulation based on protein–protein interactions between transcription factors. Nucleic Acids Res. 2021, 49, 10347–10368. [Google Scholar] [CrossRef]
- Williamson, I.; Hill, R.E.; Bickmore, W.A. Enhancers: From Developmental Genetics to the Genetics of Common Human Disease. Dev. Cell 2011, 21, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Smemo, S.; Campos, L.C.; Moskowitz, I.P.; Krieger, J.E.; Pereira, A.C.; Nobrega, M.A. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum. Mol. Genet. 2012, 21, 3255–3263. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; Van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Dekker, J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef]
- Pucéat, M. Capturing chromosome conformation. Methods Mol. Biol. (Clifton N.J.) 2021, 2157, 1–7. [Google Scholar] [CrossRef]
- Simonis, M.; Klous, P.; Splinter, E.; Moshkin, Y.; Willemsen, R.; De Wit, E.; De Laat, W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 2006, 38, 1348–1354. [Google Scholar] [CrossRef]
- Dostie, J.; Richmond, T.A.; Arnaout, R.A.; Selzer, R.R.; Lee, W.L.; Honan, T.A.; Rubio, E.D.; Krumm, A.; Lamb, J.; Nusbaum, C.; et al. Chromosome conformation capture carbon copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006, 16, 1299–1309. [Google Scholar] [CrossRef]
- Fullwood, M.J.; Liu, M.H.; Pan, Y.F.; Liu, J.; Xu, H.; Mohamed, Y.B.; Ruan, Y. An oestrogen-receptor-α-bound human chromatin interactome. Nature 2009, 462, 58–64. [Google Scholar] [CrossRef]
- Zhang, Y.; An, L.; Xu, J.; Zhang, B.; Zheng, W.J.; Hu, M.; Yue, F. Enhancing hi-C data resolution with deep convolutional neural network HiCPlus. Nat. Commun. 2018, 9, 750. [Google Scholar] [CrossRef]
- Wei, L.; He, W.; Malik, A.; Su, R.; Cui, L.; Manavalan, B. Computational prediction and interpretation of cell-specific replication origin sites from multiple eukaryotes by exploiting stacking framework. Brief. Bioinform. 2021, 22, bbaa275. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Li, H.; Xu, K.; Hong, H.; Jiang, S.; Du, G.; Bo, X. Computational methods for the prediction of chromatin interaction and organization using sequence and epigenomic profiles. Brief. Bioinform. 2021, 22, bbaa405. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, R.; Singh, S.; Ma, J. Exploiting sequence-based features for predicting enhancer–promoter interactions. Bioinformatics 2017, 33, i252–i260. [Google Scholar] [CrossRef]
- Cao, F.; Fullwood, M.J. Inflated performance measures in enhancer–promoter interaction-prediction methods. Nat. Genet. 2019, 51, 1196–1198. [Google Scholar] [CrossRef]
- Whalen, S.; Truty, R.M.; Pollard, K.S. Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat. Genet. 2016, 48, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Siahpirani, A.F.; Chasman, D.; Knaack, S.; Ay, F.; Stewart, R.; Wilson, M.; Sridharan, R. A predictive modeling approach for cell line-specific long-range regulatory interactions. Nucleic Acids Res. 2016, 44, 1977–1978. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Chen, C.; Teng, L.; Tan, K. Global view of enhancer-promoter interactome in human cells. Proc. Natl. Acad. Sci. USA 2014, 111, E2191–E2199. [Google Scholar] [CrossRef]
- Singh, S.; Yang, Y.; Póczos, B.; Ma, J. Predicting enhancer-promoter interaction from genomic sequence with deep neural networks. Quant. Biol. 2019, 7, 122–137. [Google Scholar] [CrossRef]
- Hong, Z.; Zeng, X.; Wei, L.; Liu, X. Identifying enhancer–promoter interactions with neural network based on pre-trained DNA vectors and attention mechanism. Bioinformatics 2020, 36, 1037–1043. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, M.; Zhu, M. RAEPI: Predicting enhancer-promoter interactions based on restricted attention mechanism. Interdiscip. Sci. Comput. Life Sci. 2025, 17, 153–165. [Google Scholar] [CrossRef]
- Zhuang, Z.; Shen, X.; Pan, W. A simple convolutional neural network for prediction of enhancer–promoter interactions with DNA sequence data. Bioinformatics 2019, 35, 2899–2906. [Google Scholar] [CrossRef]
- Liu, S.; Xu, X.; Yang, Z.; Zhao, X.; Liu, S.; Zhang, W. EPIHC: Improving enhancer-promoter interaction prediction by using hybrid features and communicative learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 3435–3443. [Google Scholar] [CrossRef] [PubMed]
- Min, X.; Ye, C.; Liu, X.; Zeng, X. Predicting enhancer-promoter interactions by deep learning and matching heuristic. Brief. Bioinform. 2021, 22, bbaa254. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Li, Y.; Ding, Y.; Yu, L. EPIPDLF: A pre-trained deep learning framework for predicting enhancer-promoter interactions. Bioinformatics 2025, 41, btae716. [Google Scholar] [CrossRef]
- Belokopytova, P.S.; Nuriddinov, M.A.; Mozheiko, E.A.; Fishman, D.; Fishman, V. Quantitative prediction of enhancer–promoter interactions. Genome Res. 2020, 30, 72–84. [Google Scholar] [CrossRef]
- Mao, W.; Kostka, D.; Chikina, M. Modeling enhancer-promoter interactions with attention-based neural networks. BioRxiv 2017, 219667. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Yang, Y.; Zhang, Y.; Ma, J. Predicting CTCF-mediated chromatin loops using CTCF-MP. Bioinformatics 2018, 34, i133–i141. [Google Scholar] [CrossRef]
- Gong, H.; Chen, Z.; Tang, Y.; Li, M.; Zhang, S.; Zhang, X.; Chen, Y. Computational methods for identifying enhancer-promoter interactions. Quant. Biol. 2023, 11, 122–142. [Google Scholar] [CrossRef]
- Lv, H.; Dao, F.Y.; Zulfiqar, H.; Su, W.; Ding, H.; Liu, L.; Lin, H. A sequence-based deep learning approach to predict CTCF-mediated chromatin loop. Brief. Bioinform. 2021, 22, bbab031. [Google Scholar] [CrossRef]
- Zhang, K.; Pan, X.; Yang, Y.; Shen, H.B. CRIP: Predicting circRNA–RBP-binding sites using a codon-based encoding and hybrid deep neural networks. RNA 2019, 25, 1604–1615. [Google Scholar] [CrossRef]
- Wang, Z.; Lei, X. Prediction of RBP binding sites on circRNAs using an LSTM-based deep sequence learning architecture. Brief. Bioinform. 2021, 22, bbab342. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, P.; Li, C.; Li, F.; Xiang, D.; Chen, Y.Z.; Song, J. iLearnPlus: A comprehensive and automated machine-learning platform for nucleic acid and protein sequence analysis, prediction and visualization. Nucleic Acids Res. 2021, 49, e60. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, H.; Wu, H. iPro-WAEL: A comprehensive and robust framework for identifying promoters in multiple species. Nucleic Acids Res. 2022, 50, 10278–10289. [Google Scholar] [CrossRef]
- Manavalan, B.; Basith, S.; Shin, T.H.; Wei, L.; Lee, G. Meta-4mCpred: A sequence-based meta-predictor for accurate DNA 4mC site prediction using effective feature representation. Mol. Ther. Nucleic Acids 2019, 16, 733–744. [Google Scholar] [CrossRef]
- Kai, Y.; Andricovich, J.; Zeng, Z.; Zhu, J.; Tzatsos, A.; Peng, W. Predicting CTCF-mediated chromatin interactions by integrating genomic and epigenomic features. Nat. Commun. 2018, 9, 4221. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, X.; Sun, H.; Gao, B.; Liu, X. GATv2EPI: Predicting enhancer–promoter interactions with a dynamic graph attention network. Genes 2024, 15, 1511. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wei, Y.; Tan, Z.; Zhang, Q.; Sun, J.; Zhao, Q. Predicting circRNA-RBP Binding Sites Using a Hybrid Deep Neural Network. Interdiscip. Sci. Comput. Life Sci. 2024, 16, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Orenstein, Y.; Wang, Y.; Berger, B. RCK: Accurate and efficient inference of sequence-and structure-based protein–RNA binding models from RNAcompete data. Bioinformatics 2016, 32, i351–i359. [Google Scholar] [CrossRef]
- Jin, J.; Yu, Y.; Wei, L. Mouse4mC-BGRU: Deep learning for predicting DNA N4-methylcytosine sites in mouse genome. Methods 2022, 204, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Yun, C.M. Correction: A deep learning model for predicting risks of crop pests and diseases from sequential environmental data. Plant Methods 2024, 20, 24. [Google Scholar] [CrossRef]


| Model/Cell Line | HeLa | IMR90 | NHEK |
|---|---|---|---|
| EPIFBMC | 5563 | 5614 | 1406 |
| SIMCNN | 16,615 | 12,156 | 12,038 |
| SPEID | 24,137 | 20,407 | 20,476 |
| PEP-WORD | 21,034 | 18,374 | 17,512 |
| EPIVAN | 16,128 | 12,467 | 12,168 |
| Model/Cell Line | HeLa | IMR90 | NHEK |
|---|---|---|---|
| EPIFBMC | 12,944 | 9101 | 4314 |
| EPIPDLF | 21,417 | 18,179 | 17,245 |
| EPIHC | 15,481 | 12,067 | 10,375 |
| EPIDLMH | 20,131 | 17,647 | 16,842 |
| XGBoost | 19,522 | 18,822 | 16,379 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bao, C.; Wang, G.; Sheng, G.; Chen, Y. EPIFBMC: A New Model for Enhancer–Promoter Interaction Prediction. Int. J. Mol. Sci. 2025, 26, 8035. https://doi.org/10.3390/ijms26168035
Bao C, Wang G, Sheng G, Chen Y. EPIFBMC: A New Model for Enhancer–Promoter Interaction Prediction. International Journal of Molecular Sciences. 2025; 26(16):8035. https://doi.org/10.3390/ijms26168035
Chicago/Turabian StyleBao, Chengfeng, Gang Wang, Guojun Sheng, and Yu Chen. 2025. "EPIFBMC: A New Model for Enhancer–Promoter Interaction Prediction" International Journal of Molecular Sciences 26, no. 16: 8035. https://doi.org/10.3390/ijms26168035
APA StyleBao, C., Wang, G., Sheng, G., & Chen, Y. (2025). EPIFBMC: A New Model for Enhancer–Promoter Interaction Prediction. International Journal of Molecular Sciences, 26(16), 8035. https://doi.org/10.3390/ijms26168035






