Adaptation of the Protocol for the Isolation of Biotinylated Protein Complexes for Drosophila melanogaster Tissues
Abstract
1. Introduction
2. Results
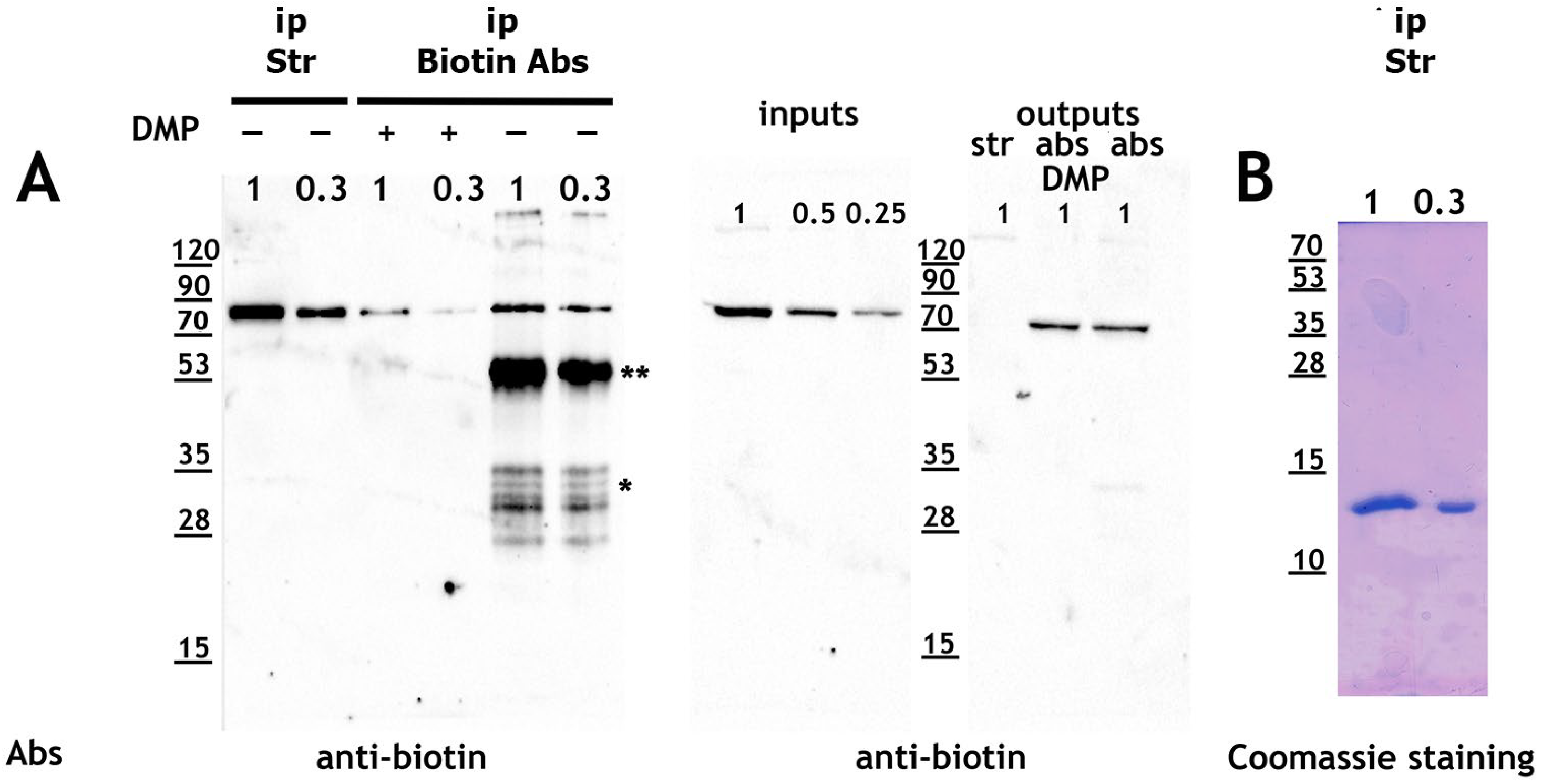
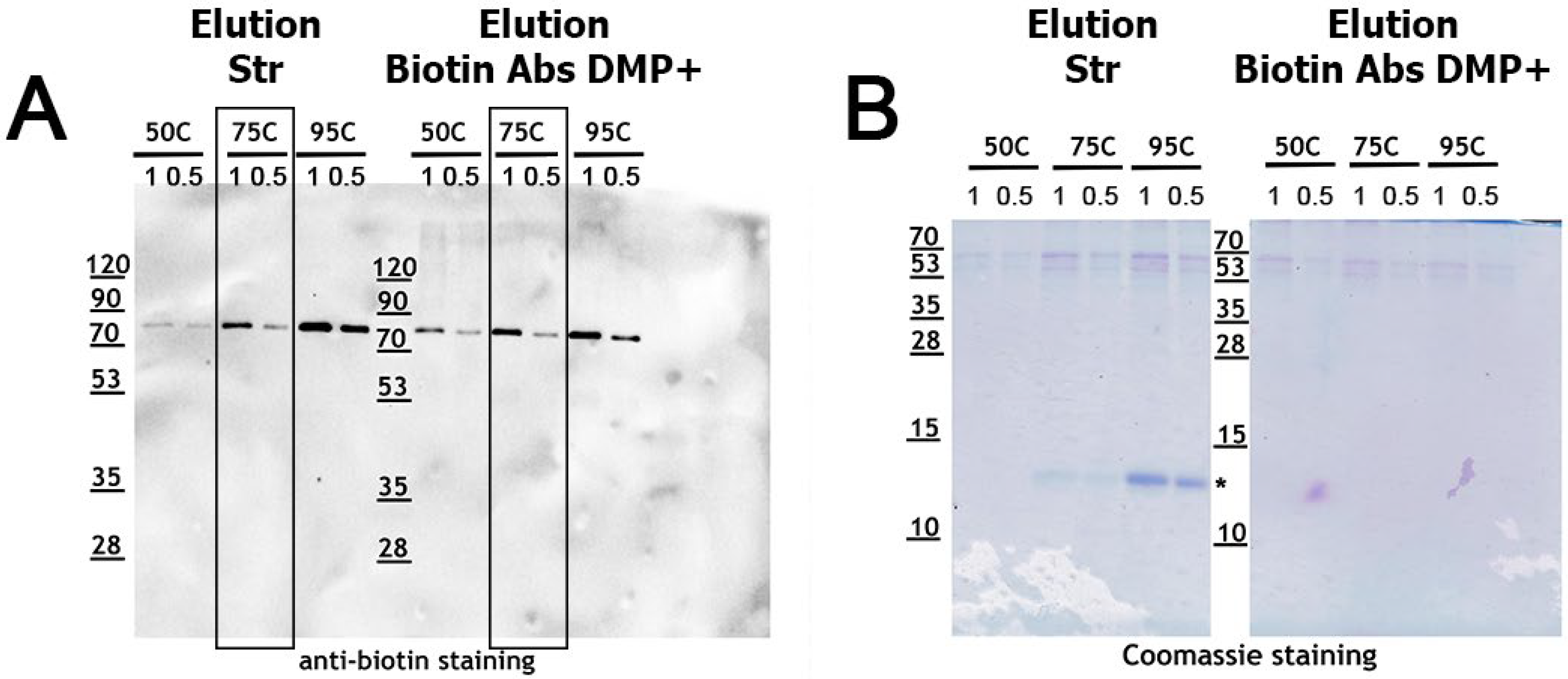
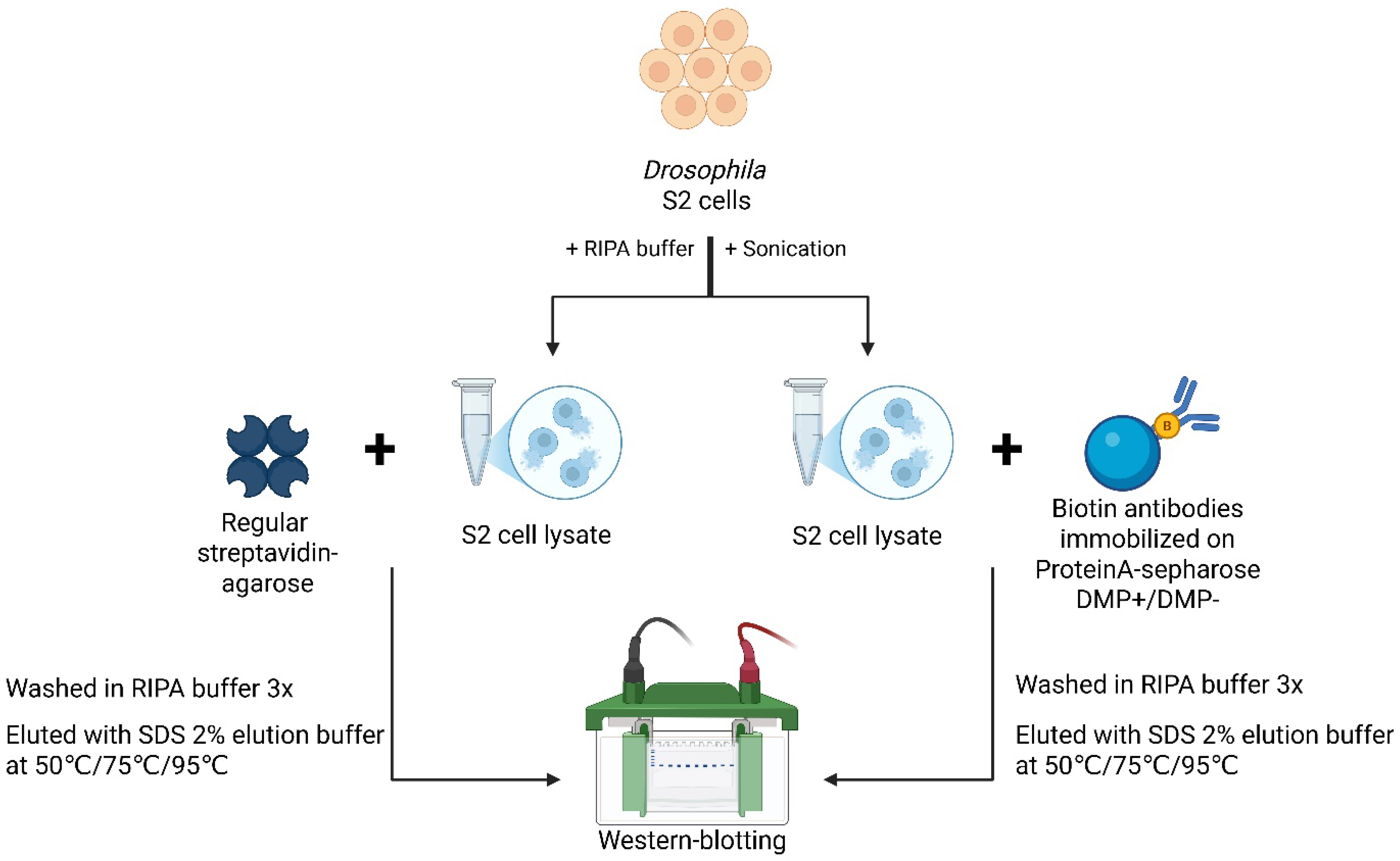
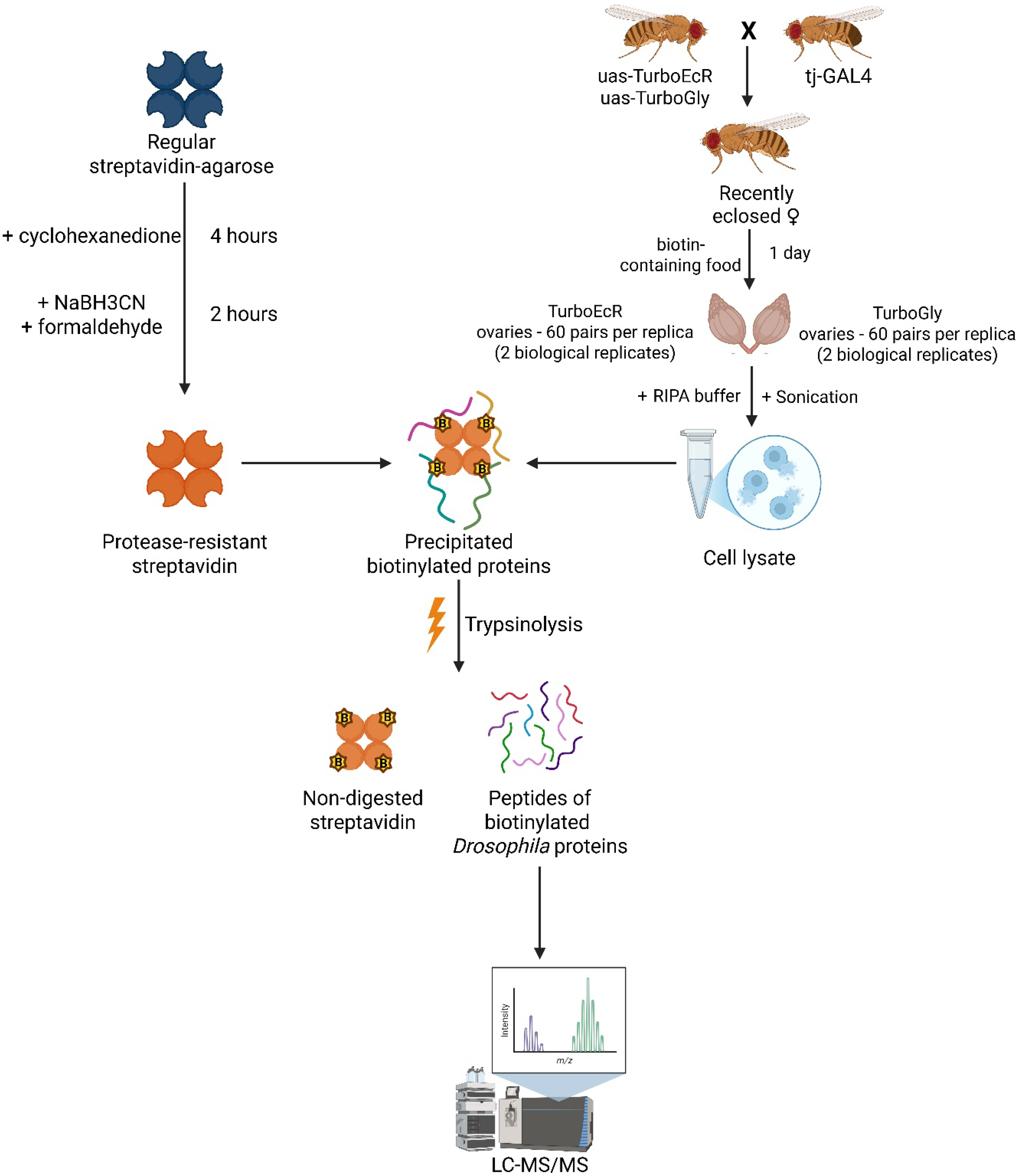
2.1. Direct Comparison of Methods for Precipitation and Elution of Biotinylated Proteins
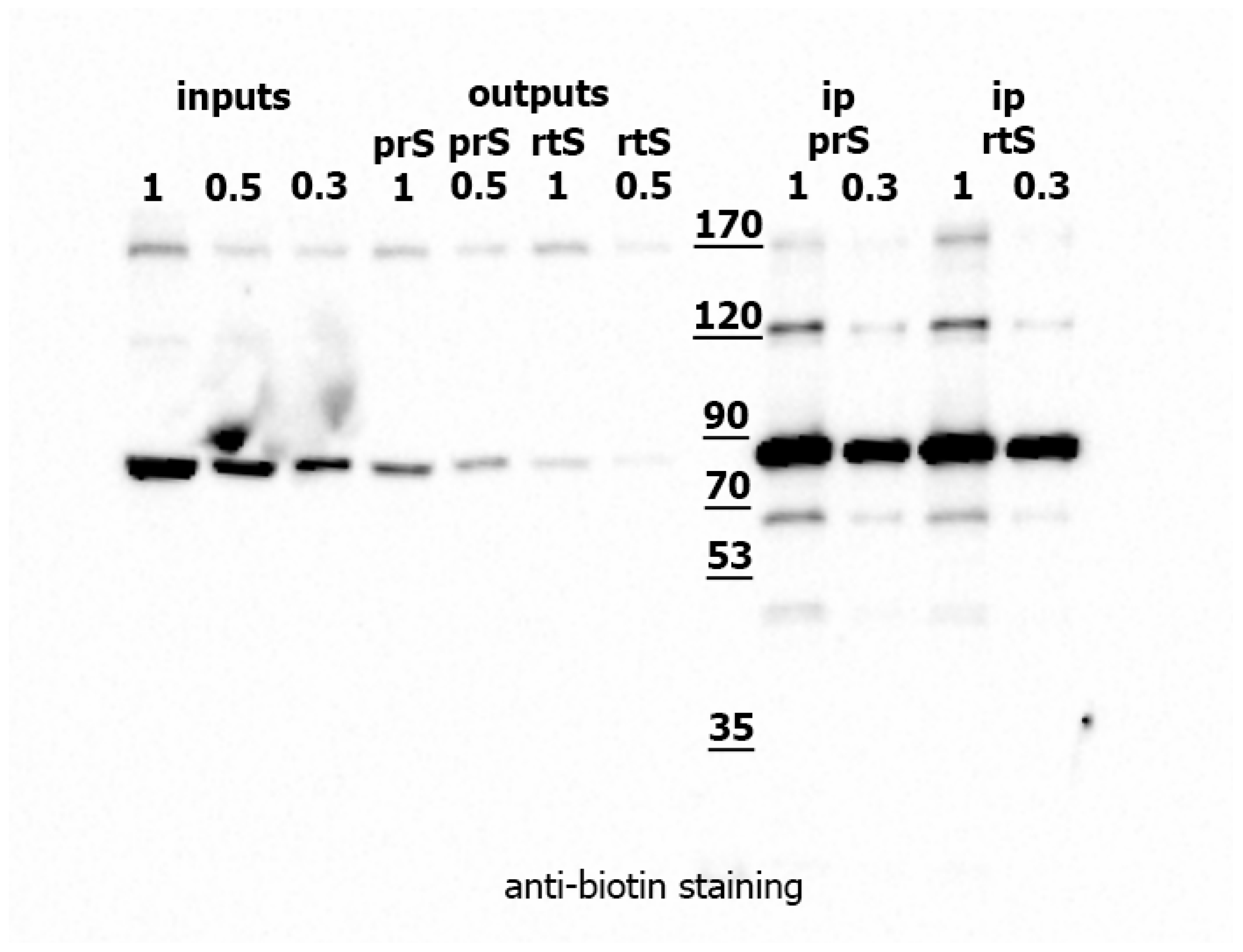
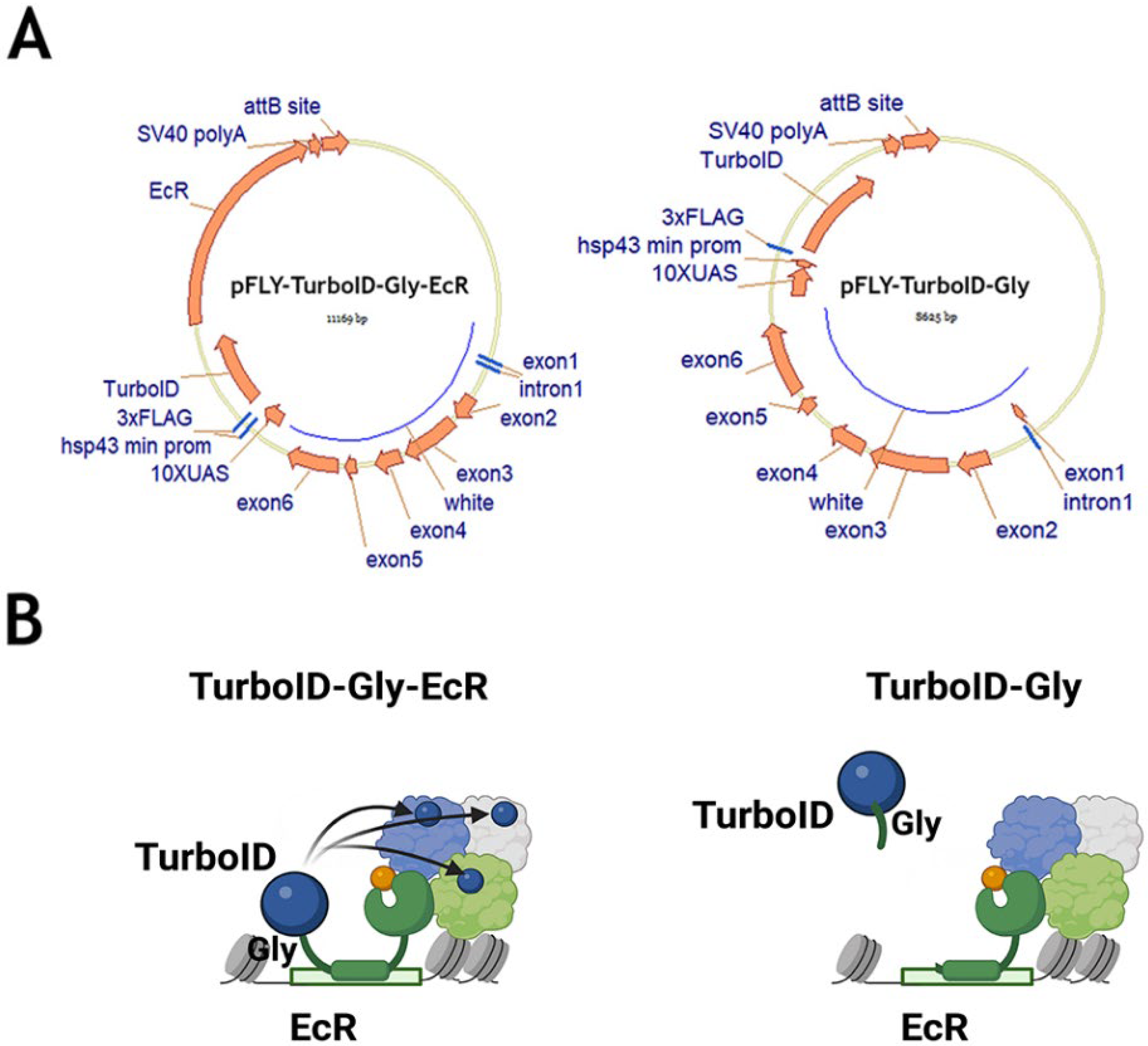
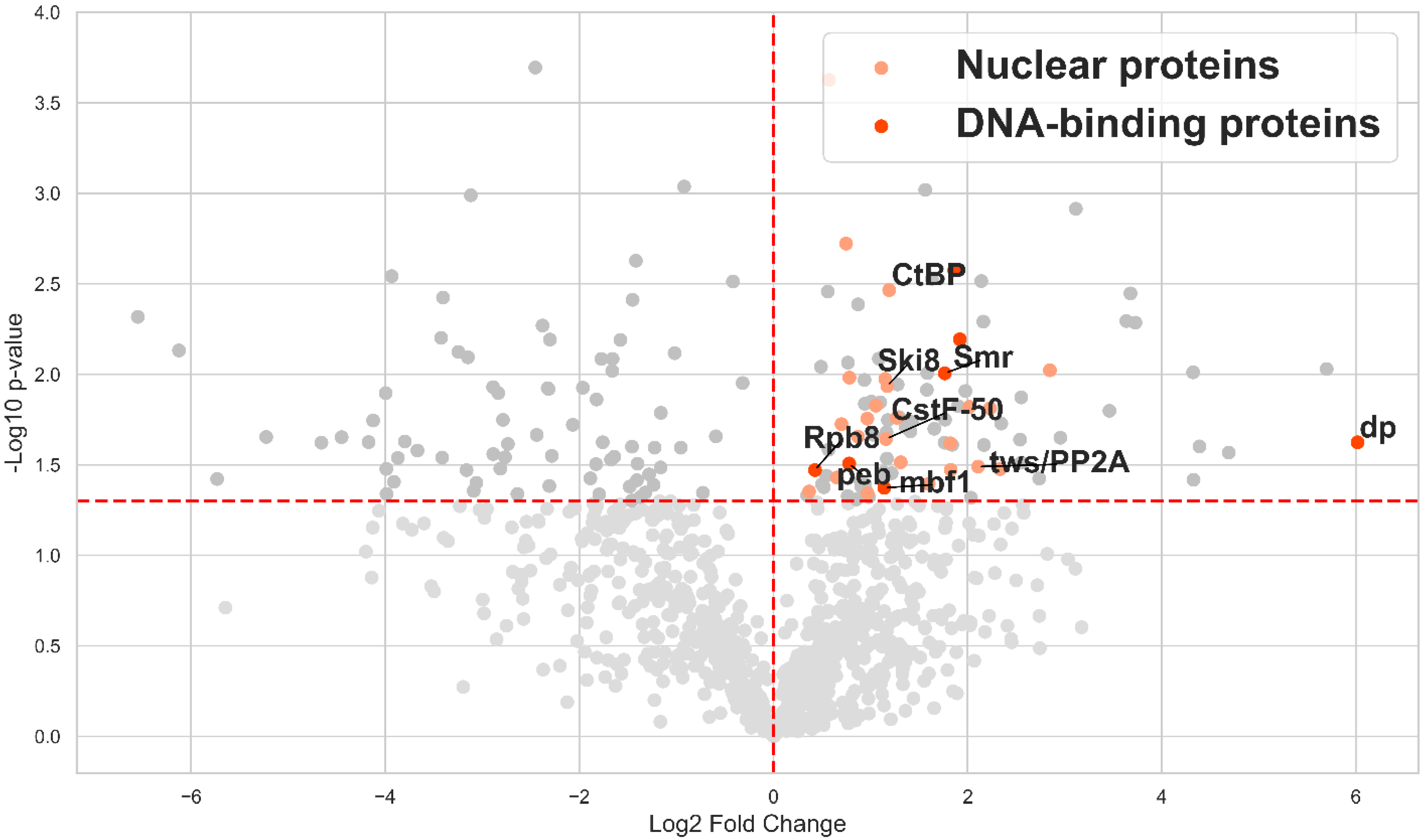
2.2. Identification of EcR Protein Partners in Drosophila Tissues Using the TurboID Biotinylating Technique
3. Discussion
3.1. Pros and Cons of Different Approaches to Enrich Biotinylated Proteins from Drosophila Tissues (With Limited Material Availability)
3.1.1. Protein Extraction
3.1.2. Protein Enrichment and Elution
3.1.3. Trypsinolysis
3.1.4. General Observations
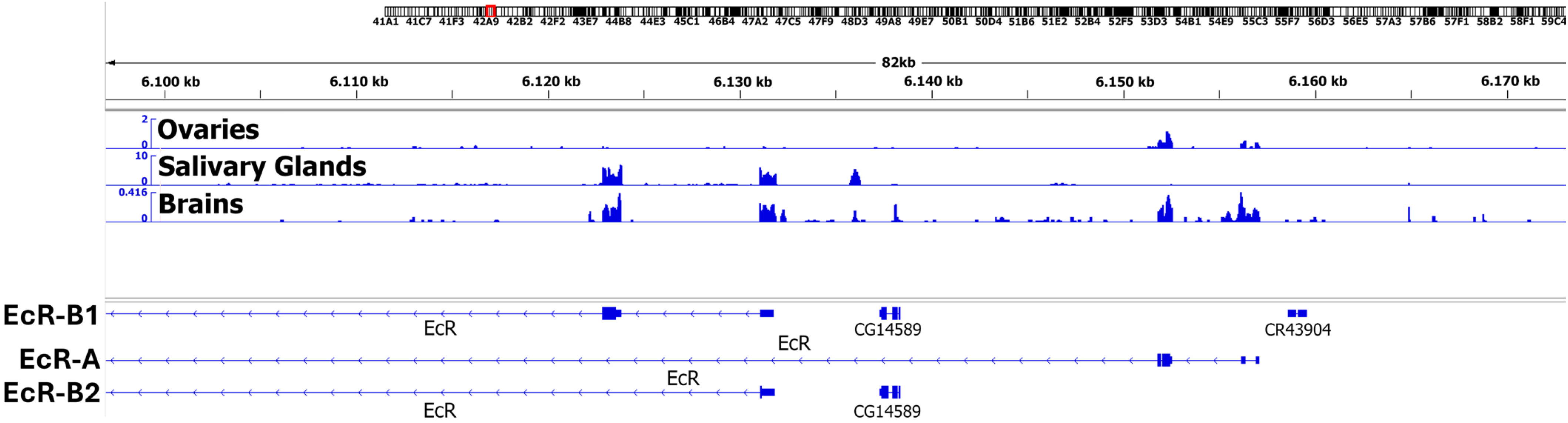
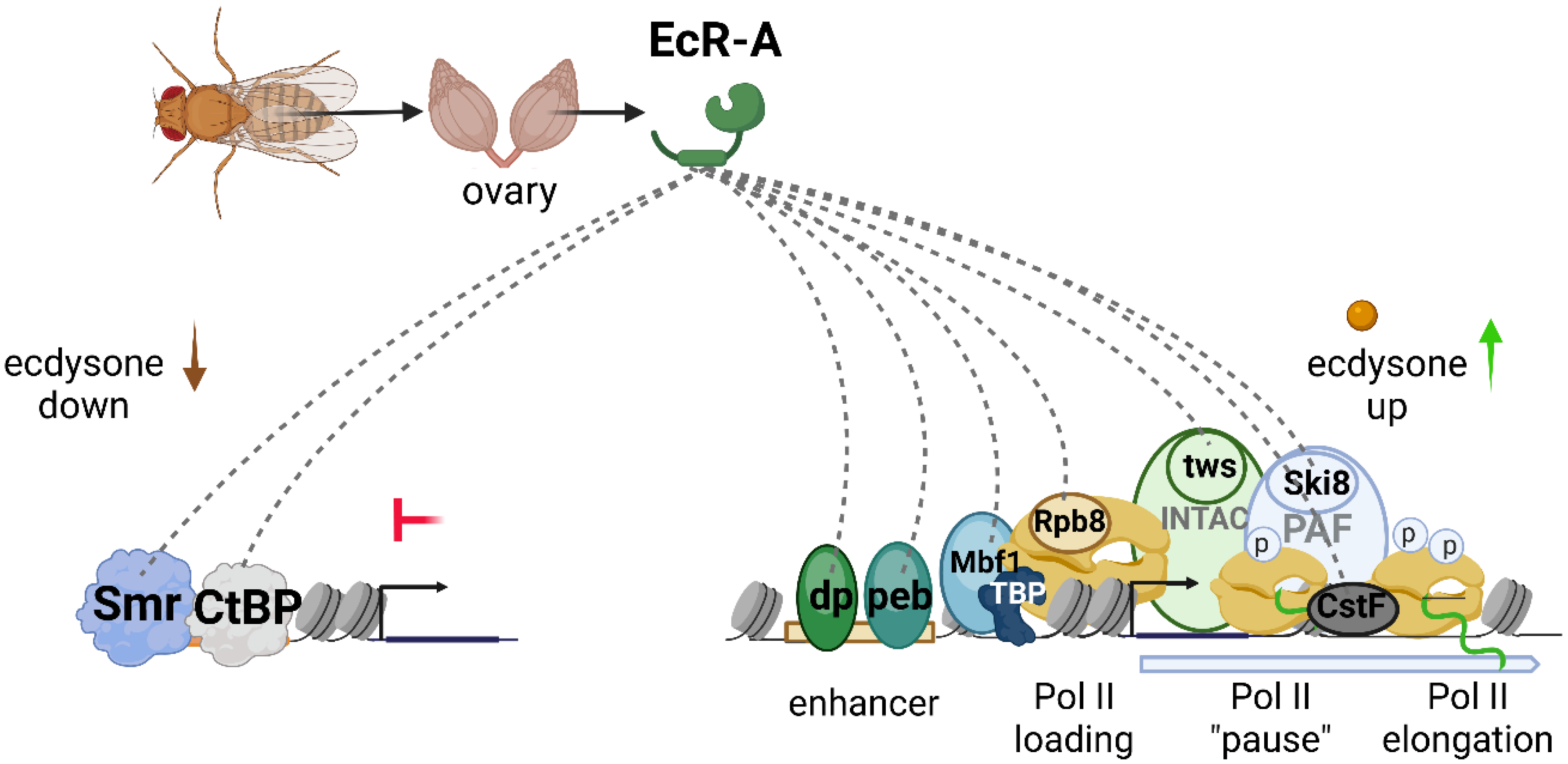
3.2. PB Helped to Identify Previously Unknown Protein Partners of EcR in Drosophila Ovary
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henarejos-Castillo, I.; Sanz, F.J.; Solana-Manrique, C.; Sebastian-Leon, P.; Medina, I.; Remohi, J.; Paricio, N.; Diaz-Gimeno, P. Whole-exome sequencing and Drosophila modelling reveal mutated genes and pathways contributing to human ovarian failure. Reprod. Biol. Endocrinol. 2024, 22, 153. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Lee, C.M.; Lee, L.C.; Tung, L.C.; Hsieh-Li, H.M.; Lee-Chen, G.J.; Su, M.T. Mitochondrial dysfunction and oxidative stress contribute to the pathogenesis of spinocerebellar ataxia type 12 (SCA12). J. Biol. Chem. 2011, 286, 21742–21754. [Google Scholar] [CrossRef] [PubMed]
- Pandey, U.B.; Nichols, C.D. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 2011, 63, 411–436. [Google Scholar] [CrossRef]
- Qin, W.; Cho, K.F.; Cavanagh, P.E.; Ting, A.Y. Deciphering molecular interactions by proximity labeling. Nat. Methods 2021, 18, 133–143. [Google Scholar] [CrossRef]
- Samavarchi-Tehrani, P.; Samson, R.; Gingras, A.C. Proximity dependent biotinylation: Key enzymes and adaptation to proteomics approaches. Mol. Cell. Proteom. 2020, 19, 757–773. [Google Scholar] [CrossRef]
- Guo, J.; Guo, S.; Lu, S.; Gong, J.; Wang, L.; Ding, L.; Chen, Q.; Liu, W. The development of proximity labeling technology and its applications in mammals, plants, and microorganisms. Cell Commun. Signal. 2023, 21, 269; Erratum in Cell Commun. Signal. 2024, 22, 403. [Google Scholar] [CrossRef]
- Carnesecchi, J.; Sigismondo, G.; Domsch, K.; Baader, C.E.P.; Rafiee, M.R.; Krijgsveld, J.; Lohmann, I. Multi-level and lineage-specific interactomes of the Hox transcription factor Ubx contribute to its functional specificity. Nat. Commun. 2020, 11, 1388. [Google Scholar] [CrossRef]
- Mazina, M.Y.; Ziganshin, R.H.; Magnitov, M.D.; Golovnin, A.K.; Vorobyeva, N.E. Proximity-dependent biotin labelling reveals CP190 as an EcR/Usp molecular partner. Sci. Rep. 2020, 10, 4793. [Google Scholar] [CrossRef]
- Branon, T.C.; Bosch, J.A.; Sanchez, A.D.; Udeshi, N.D.; Svinkina, T.; Carr, S.A.; Feldman, J.L.; Perrimon, N.; Ting, A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. [Google Scholar] [CrossRef]
- Shin, S.; Lee, S.Y.; Kang, M.G.; Jang, D.G.; Kim, J.; Rhee, H.W.; Kim, J.S. Super-resolution proximity labeling with enhanced direct identification of biotinylation sites. Commun. Biol. 2024, 7, 554. [Google Scholar] [CrossRef]
- Udeshi, N.D.; Pedram, K.; Svinkina, T.; Fereshetian, S.; Myers, S.A.; Aygun, O.; Krug, K.; Clauser, K.; Ryan, D.; Ast, T.; et al. Antibodies to biotin enable large-scale detection of biotinylation sites on proteins. Nat. Methods 2017, 14, 1167–1170. [Google Scholar] [CrossRef]
- Kozlova, T.; Thummel, C.S. Steroid regulation of postembryonic development and reproduction in Drosophila. Trends Endocrinol. Metab. 2000, 11, 276–280. [Google Scholar] [CrossRef]
- Evdokimova, A.A.; Kolesnikova, T.D.; Mazina, M.Y.; Krasnov, A.N.; Erokhin, M.; Chetverina, D.; Vorobyeva, N.E. Transcriptional induction by ecdysone in Drosophila salivary glands involves an increase in chromatin accessibility and acetylation. Nucleic Acids Res. 2025, 53, gkaf284. [Google Scholar] [CrossRef]
- Bulynko, Y.A.; O’Malley, B.W. Nuclear receptor coactivators: Structural and functional biochemistry. Biochemistry 2011, 50, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Rafiee, M.R.; Sigismondo, G.; Kalxdorf, M.; Förster, L.; Brügger, B.; Béthune, J.; Krijgsveld, J. Protease-resistant streptavidin for interaction proteomics. Mol. Syst. Biol. 2020, 16, e9370. [Google Scholar] [CrossRef] [PubMed]
- Robinow, S.; Talbot, W.S.; Hogness, D.S.; Truman, J.W. Programmed cell death in the Drosophila CNS is ecdysone-regulated and coupled with a specific ecdysone receptor isoform. Development 1993, 119, 1251–1259. [Google Scholar] [CrossRef]
- Vorobyeva, N.E.; Erokhin, M.; Chetverina, D.; Krasnov, A.N.; Mazina, M.Y. Su (Hw) primes 66D and 7F Drosophila chorion genes loci for amplification through chromatin decondensation. Sci. Rep. 2021, 11, 16963. [Google Scholar] [CrossRef] [PubMed]
- Kankel, M.W.; Sen, A.; Lu, L.; Theodorou, M.; Dimlich, D.N.; McCampbell, A.; E Henderson, C.; A Shneider, N.; Artavanis-Tsakonas, S. Amyotrophic lateral sclerosis modifiers in Drosophila reveal the phospholipase D pathway as a potential therapeutic target. Genetics 2020, 215, 747–766. [Google Scholar] [CrossRef]
- Chen, C.L.; Perrimon, N. Proximity-dependent labeling methods for proteomic profiling in living cells. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e272. [Google Scholar] [CrossRef]
- Hofmann, K.; Wood, S.W.; Brinton, C.C.; Montibeller, J.A.; Finn, F.M. Iminobiotin affinity columns and their application to retrieval of streptavidin. Proc. Natl. Acad. Sci. USA 1980, 77, 4666–4668. [Google Scholar] [CrossRef]
- Slavoff, S.A.; Chen, I.; Choi, Y.A.; Ting, A.Y. Expanding the substrate tolerance of biotin ligase through exploration of enzymes from diverse species. J. Am. Chem. Soc. 2008, 130, 1160–1162. [Google Scholar] [CrossRef]
- Barshop, W.D.; Kim, H.J.; Fan, X.; Sha, J.; Rayatpisheh, S.; Wohlschlegel, J.A. Chemical derivatization of affinity matrices provides protection from tryptic proteolysis. J. Proteome Res. 2019, 18, 3586–3596. [Google Scholar] [CrossRef] [PubMed]
- Schiapparelli, L.M.; McClatchy, D.B.; Liu, H.H.; Sharma, P.; Yates, J.R., III; Cline, H.T. Direct detection of biotinylated proteins by mass spectrometry. J. Proteome Res. 2014, 13, 3966–3978. [Google Scholar] [CrossRef] [PubMed]
- Wong, X.; Cutler, J.A.; Hoskins, V.E.; Gordon, M.; Madugundu, A.K.; Pandey, A.; Reddy, K.L. Mapping the micro-proteome of the nuclear lamina and lamina-associated domains. Life Sci. Alliance 2021, 4, e202000774. [Google Scholar] [CrossRef] [PubMed]
- Yheskel, M.; Sidoli, S.; Secombe, J. Proximity labeling reveals a new in vivo network of interactors for the histone demethylase KDM5. Epigenetics Chromatin 2023, 16, 8. [Google Scholar] [CrossRef]
- Liu, Q.X.; Jindra, M.; Ueda, H.; Hiromi, Y.; Hirose, S. Drosophila MBF1 is a co-activator for Tracheae Defective and contributes to the formation of tracheal and nervous systems. Development 2003, 130, 719–728. [Google Scholar] [CrossRef]
- Mazina, M.Y.; Kovalenko, E.V.; Derevyanko, P.K.; Nikolenko, J.V.; Krasnov, A.N.; Vorobyeva, N.E. One signal stimulates different transcriptional activation mechanisms. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2018, 1861, 178–189. [Google Scholar] [CrossRef]
- Chen, F.X.; Woodfin, A.R.; Gardini, A.; Rickels, R.A.; Marshall, S.A.; Smith, E.R.; Shiekhattar, R.; Shilatifard, A. PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell 2015, 162, 1003–1015. [Google Scholar] [CrossRef]
- Hu, S.; Peng, L.; Song, A.; Ji, Y.X.; Cheng, J.; Wang, M.; Chen, F.X. INTAC endonuclease and phosphatase modules differentially regulate transcription by RNA polymerase II. Mol. Cell 2023, 83, 1588–1604. [Google Scholar] [CrossRef]
- Janssens, V.; Goris, J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001, 353, 417–439. [Google Scholar] [CrossRef]
- Moreno-Morcillo, M.; Minvielle-Sébastia, L.; Mackereth, C.; Fribourg, S. Hexameric architecture of CstF supported by CstF-50 homodimerization domain structure. RNA 2011, 17, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Sekar, A.; Leiblich, A.; Wainwright, S.M.; Mendes, C.C.; Sarma, D.; Hellberg, J.E.E.U.; Gandy, C.; Goberdhan, D.C.I.; Hamdy, F.C.; Wilson, C.; et al. Rbf/E2F1 control growth and endoreplication via steroid-independent Ecdysone Receptor signalling in Drosophila prostate-like secondary cells. PLoS Genet. 2023, 19, e1010815. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, N.K.; Cinar, B.; Mukhopadhyay, L.; Lutchman, M.; Ferdinand, A.S.; Kim, J.; Chung, L.W.K.; Adam, R.M.; Ray, S.K.; Leiter, A.B.; et al. The zinc finger protein ras-responsive element binding protein-1 is a coregulator of the androgen receptor: Implications for the role of the Ras pathway in enhancing androgenic signaling in prostate cancer. Mol. Endocrinol. 2007, 21, 2056–2070. [Google Scholar] [CrossRef]
- Erokhin, M.; Gorbenko, F.; Lomaev, D.; Mazina, M.Y.; Mikhailova, A.; Garaev, A.K.; Parshikov, A.; Vorobyeva, N.E.; Georgiev, P.; Schedl, P.; et al. Boundaries potentiate polycomb response element-mediated silencing. BMC Biol. 2021, 19, 113. [Google Scholar] [CrossRef] [PubMed]
- Vorobyeva, N.E.; Soshnikova, N.V.; Nikolenko, J.V.; Kuzmina, J.L.; Nabirochkina, E.N.; Georgieva, S.G.; Shidlovskii, Y.V. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc. Natl. Acad. Sci. USA 2009, 106, 11049–11054. [Google Scholar] [CrossRef]
- Markstein, M.; Pitsouli, C.; Villalta, C.; Celniker, S.E.; Perrimon, N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 2008, 40, 476–483. [Google Scholar] [CrossRef]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shokodko, I.A.; Ziganshin, R.H.; Vorobyeva, N.E. Adaptation of the Protocol for the Isolation of Biotinylated Protein Complexes for Drosophila melanogaster Tissues. Int. J. Mol. Sci. 2025, 26, 8009. https://doi.org/10.3390/ijms26168009
Shokodko IA, Ziganshin RH, Vorobyeva NE. Adaptation of the Protocol for the Isolation of Biotinylated Protein Complexes for Drosophila melanogaster Tissues. International Journal of Molecular Sciences. 2025; 26(16):8009. https://doi.org/10.3390/ijms26168009
Chicago/Turabian StyleShokodko, Igor A., Rustam H. Ziganshin, and Nadezhda E. Vorobyeva. 2025. "Adaptation of the Protocol for the Isolation of Biotinylated Protein Complexes for Drosophila melanogaster Tissues" International Journal of Molecular Sciences 26, no. 16: 8009. https://doi.org/10.3390/ijms26168009
APA StyleShokodko, I. A., Ziganshin, R. H., & Vorobyeva, N. E. (2025). Adaptation of the Protocol for the Isolation of Biotinylated Protein Complexes for Drosophila melanogaster Tissues. International Journal of Molecular Sciences, 26(16), 8009. https://doi.org/10.3390/ijms26168009







