Ptip and the Trr-COMPASS-like Complex Regulate Cardiac Progenitor Cell Division in the Drosophila Embryonic Heart Tube
Abstract
1. Introduction
2. Results
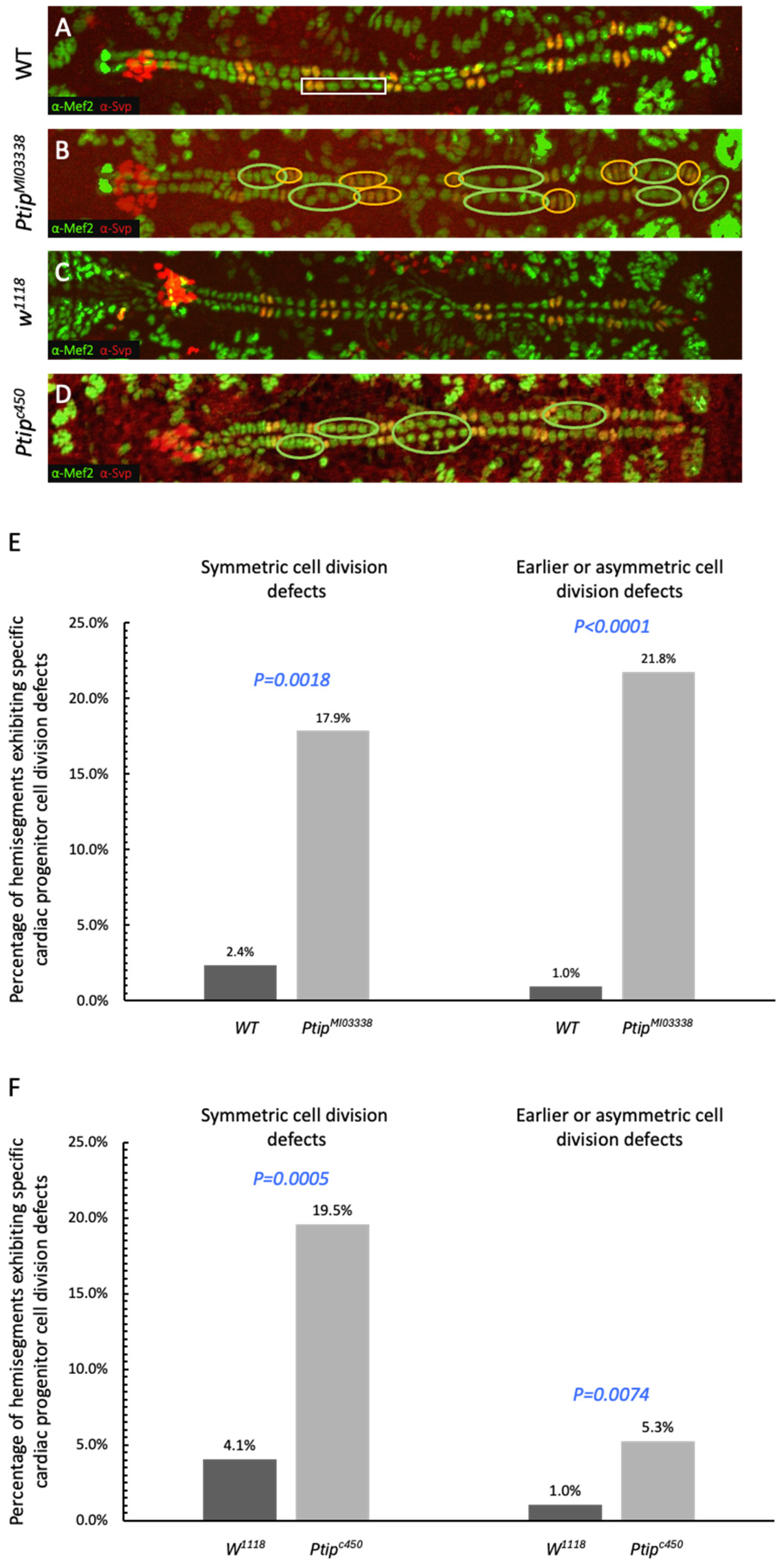
2.1. Drosophila Ptip Regulates Proper Cardiac Cell Division
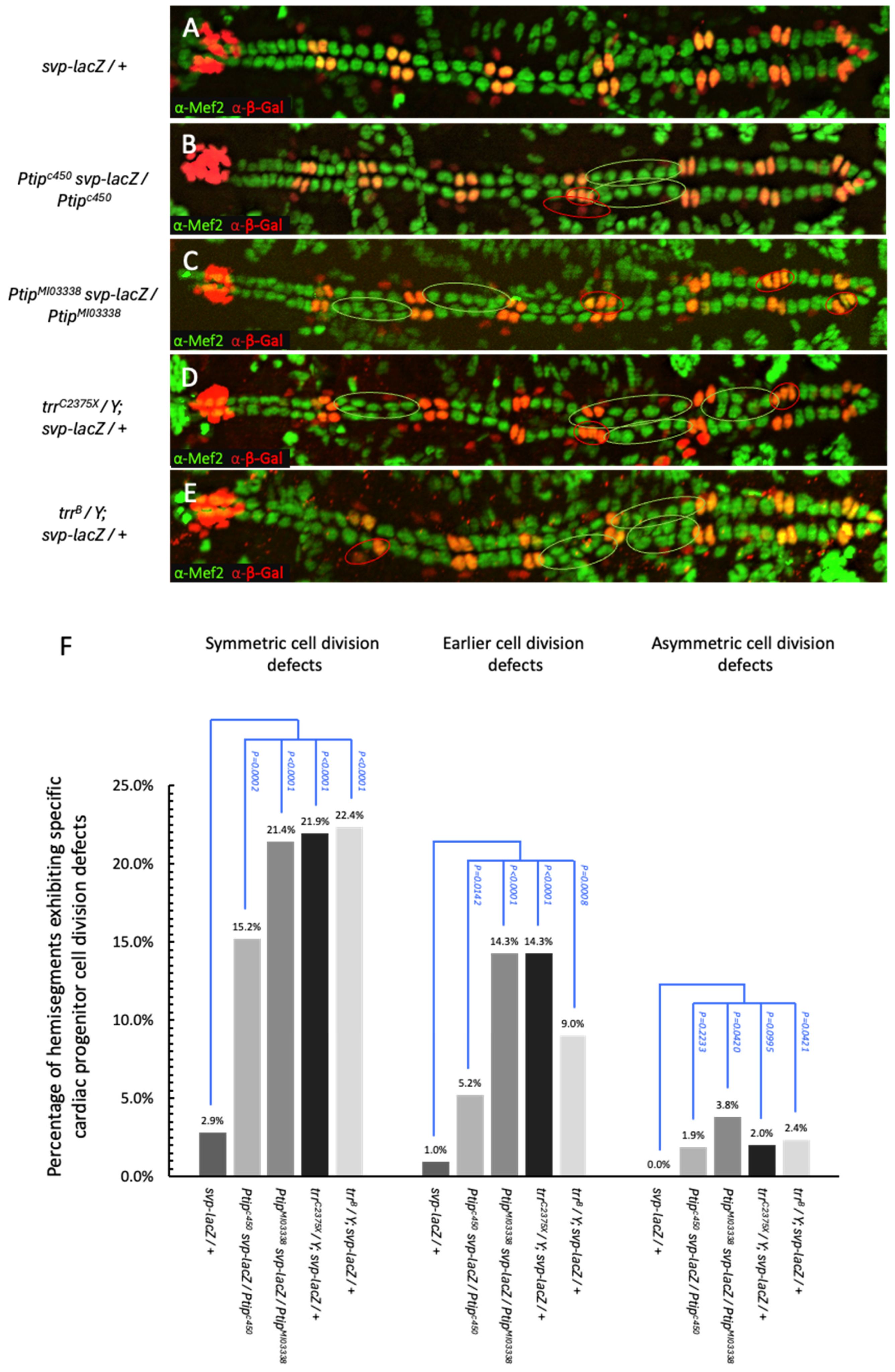
2.2. Ptip and Trr Regulate Tin-Lineage Symmetric and Svp-Lineage Earlier Cardiac Progenitor Cell Divisions
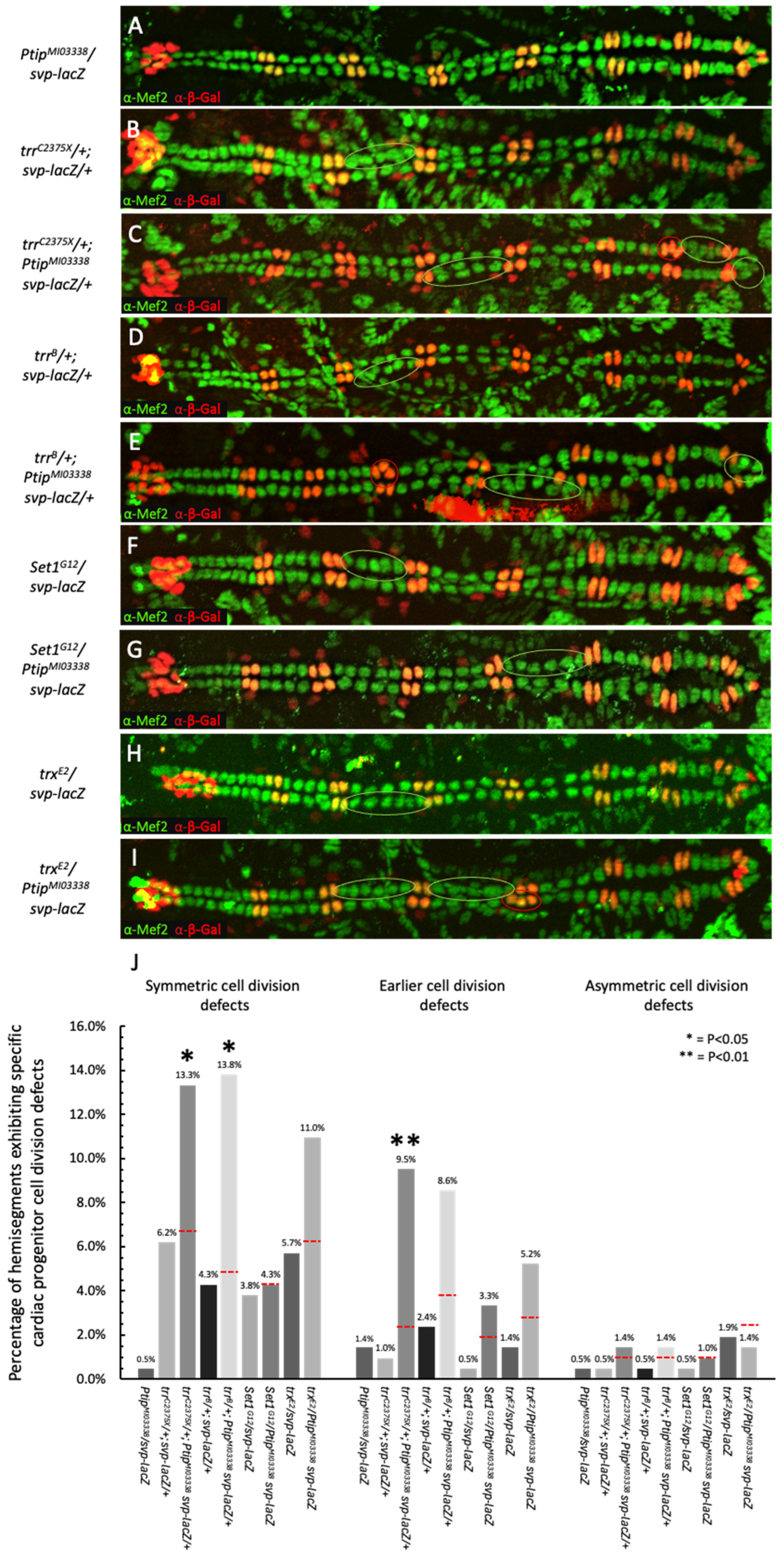
2.3. Genetic Interaction Studies Demonstrate a Synergistic Interaction Between PTIP and Trr in Cardiac Cell Division Regulation
2.4. No Synergistic Genetic Interactions Are Detected Between Ptip and Set1 or Between Ptip and Trx in Cardiac Cell Division Regulation
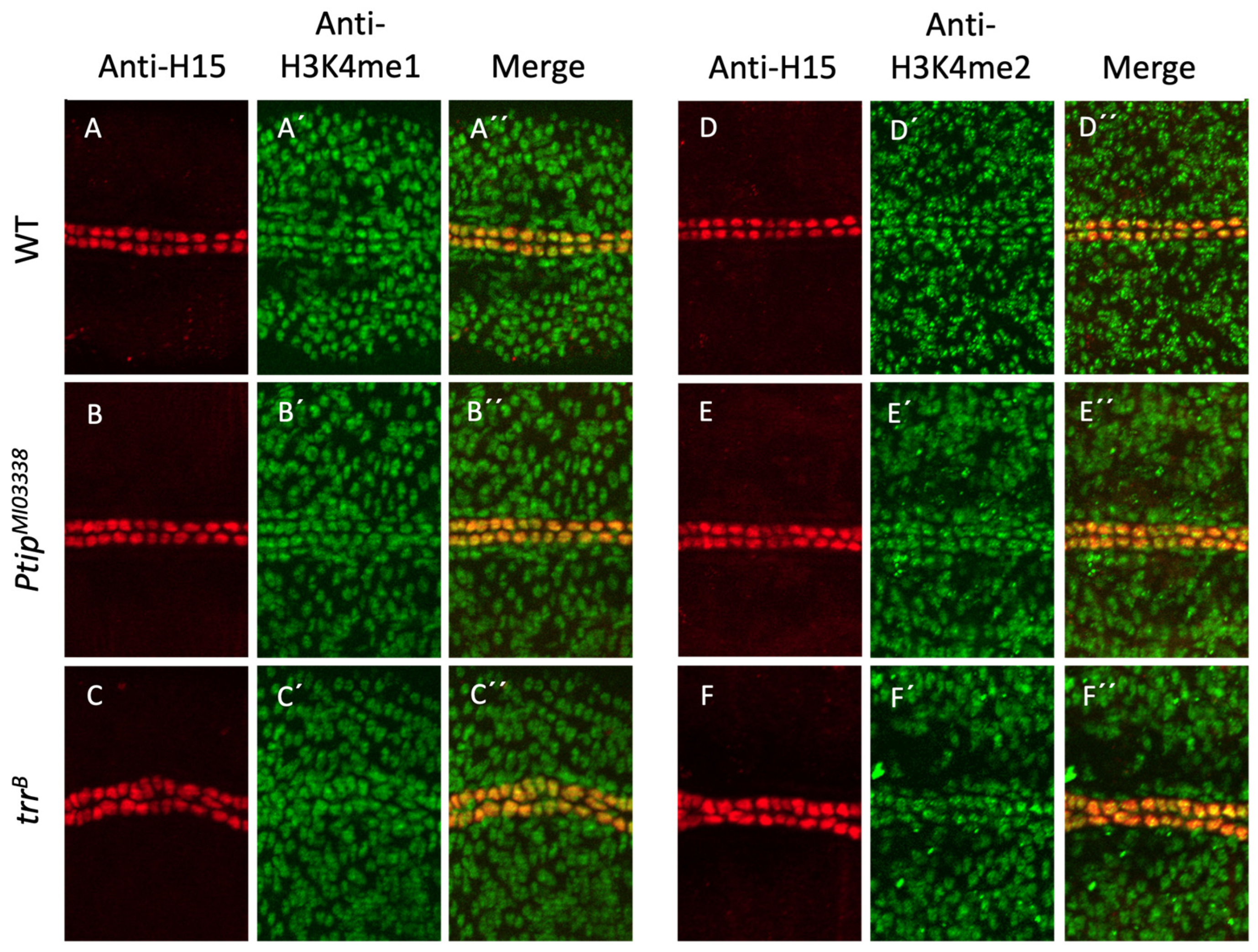
2.5. Global Cardiac Mono- and Di-H3K4 Methylation Is Not Affected by Loss of Ptip or Trr Activity
3. Discussion
4. Materials and Methods
4.1. Drosophila Strains and Genetics
4.2. Immunohistochemistry, Microscopy, and Cell Counting
4.3. Statistical Methods
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, W.; Zhu, J.Y.; Fu, Y.; van de Leemput, J.; Han, Z. Lpt, trr, and Hcf regulate histone mono- and dimethylation that are essential for Drosophila heart development. Dev. Biol. 2022, 490, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Ren, H.; Liu, J.; Cadigan, K.M.; Patel, S.R.; Dressler, G.R. Drosophila ptip is essential for anterior/posterior patterning in development and interacts with the PcG and trxG pathways. Development 2009, 136, 1929–1938. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.R.; Kim, D.; Levitan, I.; Dressler, G.R. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev. Cell 2007, 13, 580–592. [Google Scholar] [CrossRef]
- Cho, Y.W.; Hong, S.; Jin, Q.; Wang, L.; Lee, J.E.; Gavrilova, O.; Ge, K. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab. 2009, 10, 27–39. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Lee, H.; Huang, X.; van de Leemput, J.; Han, Z. Distinct Roles for COMPASS Core Subunits Set1, Trx, and Trr in the Epigenetic Regulation of Drosophila Heart Development. Int. J. Mol. Sci. 2023, 24, 17314. [Google Scholar] [CrossRef]
- Zhu, J.Y.; van de Leemput, J.; Han, Z. Distinct roles of COMPASS subunits to Drosophila heart development. Biol. Open 2024, 13, bio061736. [Google Scholar] [CrossRef]
- Cenik, B.K.; Shilatifard, A. COMPASS and SWI/SNF complexes in development and disease. Nat. Rev. Genet. 2021, 22, 38–58. [Google Scholar] [CrossRef]
- Shilatifard, A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef]
- Mohan, M.; Herz, H.M.; Smith, E.R.; Zhang, Y.; Jackson, J.; Washburn, M.P.; Florens, L.; Eissenberg, J.C.; Shilatifard, A. The COMPASS family of H3K4 methylases in Drosophila. Mol. Cell Biol. 2011, 31, 4310–4318. [Google Scholar] [CrossRef]
- Herz, H.M.; Mohan, M.; Garruss, A.S.; Liang, K.; Takahashi, Y.H.; Mickey, K.; Voets, O.; Verrijzer, C.P.; Shilatifard, A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012, 26, 2604–2620. [Google Scholar] [CrossRef]
- Cho, Y.W.; Hong, T.; Hong, S.; Guo, H.; Yu, H.; Kim, D.; Guszczynski, T.; Dressler, G.R.; Copeland, T.D.; Kalkum, M.; et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007, 282, 20395–20406. [Google Scholar] [CrossRef] [PubMed]
- Ardehali, M.B.; Mei, A.; Zobeck, K.L.; Caron, M.; Lis, J.T.; Kusch, T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011, 30, 2817–2828. [Google Scholar] [CrossRef] [PubMed]
- Hallson, G.; Hollebakken, R.E.; Li, T.; Syrzycka, M.; Kim, I.; Cotsworth, S.; Fitzpatrick, K.A.; Sinclair, D.A.; Honda, B.M. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics 2012, 190, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.L.; Westfield, G.H.; Takahashi, Y.H.; Cook, M.; Gao, X.; Woodfin, A.R.; Lee, J.S.; Morgan, M.A.; Jackson, J.; Smith, E.R.; et al. Context dependency of Set1/COMPASS-mediated histone H3 Lys4 trimethylation. Genes Dev. 2014, 28, 115–120. [Google Scholar] [CrossRef]
- Hanson, R.D.; Hess, J.L.; Yu, B.D.; Ernst, P.; van Lohuizen, M.; Berns, A.; van der Lugt, N.M.; Shashikant, C.S.; Ruddle, F.H.; Seto, M.; et al. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. USA 1999, 96, 14372–14377. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Ganapathi, M.; Leblanc, B.; Portoso, M.; Jaschek, R.; Tolhuis, B.; van Lohuizen, M.; Tanay, A.; Cavalli, G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009, 7, e13. [Google Scholar] [CrossRef]
- Yu, B.D.; Hanson, R.D.; Hess, J.L.; Horning, S.E.; Korsmeyer, S.J. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. USA 1998, 95, 10632–10636. [Google Scholar] [CrossRef]
- Yu, B.D.; Hess, J.L.; Horning, S.E.; Brown, G.A.; Korsmeyer, S.J. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 1995, 378, 505–508. [Google Scholar] [CrossRef]
- Farmer, A.J.; Katariya, R.; Islam, S.; Rayhan, M.S.A.; Inlow, M.H.; Ahmad, S.M.; Schwab, K.R. Trithorax is an essential regulator of cardiac Hox gene expression and anterior-posterior patterning of the Drosophila embryonic heart tube. Biol. Open 2025, 14, bio061919. [Google Scholar] [CrossRef]
- Ji, W.; Ferdman, D.; Copel, J.; Scheinost, D.; Shabanova, V.; Brueckner, M.; Khokha, M.K.; Ment, L.R. De novo damaging variants associated with congenital heart diseases contribute to the connectome. Sci. Rep. 2020, 10, 7046. [Google Scholar] [CrossRef]
- Jinxiu, L.; Shuimei, L.; Ming, X.; Jonathan, L.C.; Xiangju, L.; Wenyuan, D. Wiedemann-steiner syndrome with a de novo mutation in KMT2A: A case report. Medicine 2020, 99, e19813. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.; Brueckner, M. Genetics and Genomics of Congenital Heart Disease. Circ. Res. 2017, 120, 923–940. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.; Choi, M.; Wakimoto, H.; Ma, L.; Jiang, J.; Overton, J.D.; Romano-Adesman, A.; Bjornson, R.D.; Breitbart, R.E.; Brown, K.K.; et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature 2013, 498, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Eissenberg, J.C.; Shilatifard, A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 2010, 339, 240–249. [Google Scholar] [CrossRef]
- Froimchuk, E.; Jang, Y.; Ge, K. Histone H3 lysine 4 methyltransferase KMT2D. Gene 2017, 627, 337–342. [Google Scholar] [CrossRef]
- Lavery, W.J.; Barski, A.; Wiley, S.; Schorry, E.K.; Lindsley, A.W. KMT2C/D COMPASS complex-associated diseases [KCDCOM-ADs]: An emerging class of congenital regulopathies. Clin. Epigenet. 2020, 12, 10. [Google Scholar] [CrossRef]
- Hannibal, M.C.; Buckingham, K.J.; Ng, S.B.; Ming, J.E.; Beck, A.E.; McMillin, M.J.; Gildersleeve, H.I.; Bigham, A.W.; Tabor, H.K.; Mefford, H.C.; et al. Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. Am. J. Med. Genet. Part A 2011, 155, 1511–1516. [Google Scholar] [CrossRef]
- Ng, S.B.; Bigham, A.W.; Buckingham, K.J.; Hannibal, M.C.; McMillin, M.J.; Gildersleeve, H.I.; Beck, A.E.; Tabor, H.K.; Cooper, G.M.; Mefford, H.C.; et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010, 42, 790–793. [Google Scholar] [CrossRef]
- Ang, S.Y.; Uebersohn, A.; Spencer, C.I.; Huang, Y.; Lee, J.E.; Ge, K.; Bruneau, B.G. KMT2D regulates specific programs in heart development via histone H3 lysine 4 di-methylation. Development 2016, 143, 810–821. [Google Scholar] [CrossRef]
- Daniel, J.A.; Santos, M.A.; Wang, Z.; Zang, C.; Schwab, K.R.; Jankovic, M.; Filsuf, D.; Chen, H.T.; Gazumyan, A.; Yamane, A.; et al. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science 2010, 329, 917–923. [Google Scholar] [CrossRef]
- Kim, D.; Patel, S.R.; Xiao, H.; Dressler, G.R. The role of PTIP in maintaining embryonic stem cell pluripotency. Stem Cells 2009, 27, 1516–1523. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, G.M.; Patel, S.R.; Kim, D.; Tessarollo, L.; Dressler, G.R. Altering a histone H3K4 methylation pathway in glomerular podocytes promotes a chronic disease phenotype. PLoS Genet. 2010, 6, e1001142. [Google Scholar] [CrossRef] [PubMed]
- Schwab, K.R.; Patel, S.R.; Dressler, G.R. Role of PTIP in class switch recombination and long-range chromatin interactions at the immunoglobulin heavy chain locus. Mol. Cell Biol. 2011, 31, 1503–1511. [Google Scholar] [CrossRef]
- Cho, E.A.; Prindle, M.J.; Dressler, G.R. BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol. Cell Biol. 2003, 23, 1666–1673. [Google Scholar] [CrossRef]
- Deschamps, J.; van Nes, J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 2005, 132, 2931–2942. [Google Scholar] [CrossRef]
- Mu, W.; Wang, W.; Schimenti, J.C. An allelic series uncovers novel roles of the BRCT domain-containing protein PTIP in mouse embryonic vascular development. Mol. Cell Biol. 2008, 28, 6439–6451. [Google Scholar] [CrossRef]
- Stein, A.B.; Goonewardena, S.N.; Jones, T.A.; Prusick, P.J.; Bazzi, A.A.; Belyavskaya, J.M.; McCoskey, M.M.; Dandar, R.A. The PTIP-Associated Histone Methyltransferase Complex Prevents Stress-Induced Maladaptive Cardiac Remodeling. PLoS ONE 2015, 10, e0127839. [Google Scholar] [CrossRef]
- Stein, A.B.; Jones, T.A.; Herron, T.J.; Patel, S.R.; Day, S.M.; Noujaim, S.F.; Milstein, M.L.; Klos, M.; Furspan, P.B.; Jalife, J.; et al. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J. Clin. Investig. 2011, 121, 2641–2650. [Google Scholar] [CrossRef]
- Bodmer, R.; Frasch, M. Chapter 1.2–Development and Aging of the Drosophila Heart. In Heart Development and Regeneration; Rosenthal, N., Harvey, R.P., Eds.; Academic Press: Boston, MA, USA, 2010; pp. 47–86. [Google Scholar]
- Ahmad, S.M.; Tansey, T.R.; Busser, B.W.; Nolte, M.T.; Jeffries, N.; Gisselbrecht, S.S.; Rusan, N.M.; Michelson, A.M. Two forkhead transcription factors regulate the division of cardiac progenitor cells by a Polo-dependent pathway. Dev. Cell 2012, 23, 97–111. [Google Scholar] [CrossRef]
- Hasan, M.R.; Kump, A.J.; Stepaniak, E.C.; Panta, M.; Shashidhar, K.; Katariya, R.; Sabbir, M.K.; Schwab, K.R.; Inlow, M.H.; Chen, Y.; et al. Genome-Wide Expression Profiling and Phenotypic Analysis of Downstream Targets Identify the Fox Transcription Factor Jumeau as a Master Regulator of Cardiac Progenitor Cell Division. Int. J. Mol. Sci. 2024, 25, 12933. [Google Scholar] [CrossRef]
- Kump, A.J.; Panta, M.; Schwab, K.R.; Inlow, M.H.; Ahmad, S.M. The Drosophila Forkhead/Fox transcription factor Jumeau mediates specific cardiac progenitor cell divisions by regulating expression of the kinesin Nebbish. Sci. Rep. 2021, 11, 3221. [Google Scholar] [CrossRef]
- Ahmad, S.M.; Busser, B.W.; Huang, D.; Cozart, E.J.; Michaud, S.; Zhu, X.; Jeffries, N.; Aboukhalil, A.; Bulyk, M.L.; Ovcharenko, I.; et al. Machine learning classification of cell-specific cardiac enhancers uncovers developmental subnetworks regulating progenitor cell division and cell fate specification. Development 2014, 141, 878–888. [Google Scholar] [CrossRef]
- Ahmad, S.M. Conserved signaling mechanisms in Drosophila heart development. Dev. Dyn. 2017, 246, 641–656. [Google Scholar] [CrossRef]
- Meganathan, K.; Sotiriadou, I.; Natarajan, K.; Hescheler, J.; Sachinidis, A. Signaling molecules, transcription growth factors and other regulators revealed from in-vivo and in-vitro models for the regulation of cardiac development. Int. J. Cardiol. 2015, 183, 117–128. [Google Scholar] [CrossRef]
- Cripps, R.M.; Olson, E.N. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev. Biol. 2002, 246, 14–28. [Google Scholar] [CrossRef]
- Olson, E.N. Gene regulatory networks in the evolution and development of the heart. Science 2006, 313, 1922–1927. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; May, H.; Kazmierczak, K.; Liang, J.; Nguyen, N.; Hill, J.A.; Gillette, T.G.; Szczesna-Cordary, D.; Chang, A.N. The MYPT2-regulated striated muscle-specific myosin light chain phosphatase limits cardiac myosin phosphorylation in vivo. J. Biol. Chem. 2024, 300, 105652. [Google Scholar] [CrossRef] [PubMed]
- Venken, K.J.; Schulze, K.L.; Haelterman, N.A.; Pan, H.; He, Y.; Evans-Holm, M.; Carlson, J.W.; Levis, R.W.; Spradling, A.C.; Hoskins, R.A.; et al. MiMIC: A highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 2011, 8, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.L.; Vadla, G.P.; Lu, J.J.; Ahmad, V.; Klein, T.J.; Liu, L.F.; Glazer, P.M.; Xu, T.; Chabu, C.Y. Cooperation between oncogenic Ras and wild-type p53 stimulates STAT non-cell autonomously to promote tumor radioresistance. Commun. Biol. 2021, 4, 374. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, H.; Cai, Y.; Wang, H.; Niu, K.; Wu, X.; Ma, H.; Yang, Y.; Tong, W.; Liu, F.; et al. Epigenetic drift of H3K27me3 in aging links glycolysis to healthy longevity in Drosophila. Elife 2018, 7, e35368. [Google Scholar] [CrossRef]
- Alvarez, A.D.; Shi, W.; Wilson, B.A.; Skeath, J.B. Pannier and pointedP2 act sequentially to regulate Drosophila heart development. Development 2003, 130, 3015–3026. [Google Scholar] [CrossRef]
- Gajewski, K.; Choi, C.Y.; Kim, Y.; Schulz, R.A. Genetically distinct cardial cells within the Drosophila heart. Genesis 2000, 28, 36–43. [Google Scholar] [CrossRef]
- Han, Z.; Bodmer, R. Myogenic cells fates are antagonized by Notch only in asymmetric lineages of the Drosophila heart, with or without cell division. Development 2003, 130, 3039–3051. [Google Scholar] [CrossRef] [PubMed]
- Lo, P.C.; Frasch, M. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mech. Dev. 2001, 104, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Ward, E.J.; Skeath, J.B. Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development 2000, 127, 4959–4969. [Google Scholar] [CrossRef] [PubMed]
- Mlodzik, M.; Hiromi, Y.; Weber, U.; Goodman, C.S.; Rubin, G.M. The Drosophila seven-up gene, a member of the steroid receptor gene superfamily, controls photoreceptor cell fates. Cell 1990, 60, 211–224. [Google Scholar] [CrossRef]
- Gervais, L.; van den Beek, M.; Josserand, M.; Salle, J.; Stefanutti, M.; Perdigoto, C.N.; Skorski, P.; Mazouni, K.; Marshall, O.J.; Brand, A.H.; et al. Stem Cell Proliferation Is Kept in Check by the Chromatin Regulators Kismet/CHD7/CHD8 and Trr/MLL3/4. Dev. Cell 2019, 49, 556–573.e556. [Google Scholar] [CrossRef]
- Graves, H.K.; Jangam, S.; Tan, K.L.; Pignata, A.; Seto, E.S.; Yamamoto, S.; Wangler, M.F. A Genetic Screen for Genes That Impact Peroxisomes in Drosophila Identifies Candidate Genes for Human Disease. G3 2020, 10, 69–77. [Google Scholar] [CrossRef]
- Kanda, H.; Nguyen, A.; Chen, L.; Okano, H.; Hariharan, I.K. The Drosophila ortholog of MLL3 and MLL4, trithorax related, functions as a negative regulator of tissue growth. Mol. Cell Biol. 2013, 33, 1702–1710. [Google Scholar] [CrossRef]
- Prudencio, P.; Guilgur, L.G.; Sobral, J.; Becker, J.D.; Martinho, R.G.; Navarro-Costa, P. The Trithorax group protein dMLL3/4 instructs the assembly of the zygotic genome at fertilization. EMBO Rep. 2018, 19, e45728. [Google Scholar] [CrossRef]
- Hu, D.; Gao, X.; Morgan, M.A.; Herz, H.M.; Smith, E.R.; Shilatifard, A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell Biol. 2013, 33, 4745–4754. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Wang, C.; Xu, S.; Cho, Y.W.; Wang, L.; Feng, X.; Baldridge, A.; Sartorelli, V.; Zhuang, L.; Peng, W.; et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2013, 2, e01503. [Google Scholar] [CrossRef] [PubMed]
- Starnes, L.M.; Su, D.; Pikkupeura, L.M.; Weinert, B.T.; Santos, M.A.; Mund, A.; Soria, R.; Cho, Y.W.; Pozdnyakova, I.; Kubec Hojfeldt, M.; et al. A PTIP-PA1 subcomplex promotes transcription for IgH class switching independently from the associated MLL3/MLL4 methyltransferase complex. Genes Dev. 2016, 30, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Ozturk-Colak, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; Goutte-Gattat, D.; Jenkins, V.K.; Matthews, B.B.; Millburn, G.; Dos Santos, G.; Tabone, C.J.; et al. FlyBase: Updates to the Drosophila genes and genomes database. Genetics 2024, 227, iyad211. [Google Scholar] [CrossRef]
- Zheng, X. An Introductory Guide to Using Bloomington Drosophila Stock Center and FlyBase for Aging Research. Cells 2024, 13, 1192. [Google Scholar] [CrossRef]
- Kennison, J.A.; Tamkun, J.W. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. USA 1988, 85, 8136–8140. [Google Scholar] [CrossRef]
- Ahmad, S.M.; Bhattacharyya, P.; Jeffries, N.; Gisselbrecht, S.S.; Michelson, A.M. Two Forkhead transcription factors regulate cardiac progenitor specification by controlling the expression of receptors of the fibroblast growth factor and Wnt signaling pathways. Development 2016, 143, 306–317. [Google Scholar] [CrossRef]
- Leal, S.M.; Qian, L.; Lacin, H.; Bodmer, R.; Skeath, J.B. Neuromancer1 and Neuromancer2 regulate cell fate specification in the developing embryonic CNS of Drosophila melanogaster. Dev. Biol. 2009, 325, 138–150. [Google Scholar] [CrossRef]
- Manly, B.F.J. Randomization, Bootstrap and Monte Carlo Methods in Biology, 3rd ed.; Chapman & Hall/CRC: Boca Raton, FL, USA, 2007; p. 455. [Google Scholar]
- Phipson, B.; Smyth, G.K. Permutation P-values should never be zero: Calculating exact P-values when permutations are randomly drawn. Stat. Appl. Genet. Mol. Biol. 2010, 9, 1–16. [Google Scholar] [CrossRef]
- Winkler, A.M.; Ridgway, G.R.; Webster, M.A.; Smith, S.M.; Nichols, T.E. Permutation inference for the general linear model. Neuroimage 2014, 92, 381–397. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farmer, A.J.; Inlow, M.H.; Ahmad, S.M.; Schwab, K.R. Ptip and the Trr-COMPASS-like Complex Regulate Cardiac Progenitor Cell Division in the Drosophila Embryonic Heart Tube. Int. J. Mol. Sci. 2025, 26, 7954. https://doi.org/10.3390/ijms26167954
Farmer AJ, Inlow MH, Ahmad SM, Schwab KR. Ptip and the Trr-COMPASS-like Complex Regulate Cardiac Progenitor Cell Division in the Drosophila Embryonic Heart Tube. International Journal of Molecular Sciences. 2025; 26(16):7954. https://doi.org/10.3390/ijms26167954
Chicago/Turabian StyleFarmer, Adam J., Mark H. Inlow, Shaad M. Ahmad, and Kristopher R. Schwab. 2025. "Ptip and the Trr-COMPASS-like Complex Regulate Cardiac Progenitor Cell Division in the Drosophila Embryonic Heart Tube" International Journal of Molecular Sciences 26, no. 16: 7954. https://doi.org/10.3390/ijms26167954
APA StyleFarmer, A. J., Inlow, M. H., Ahmad, S. M., & Schwab, K. R. (2025). Ptip and the Trr-COMPASS-like Complex Regulate Cardiac Progenitor Cell Division in the Drosophila Embryonic Heart Tube. International Journal of Molecular Sciences, 26(16), 7954. https://doi.org/10.3390/ijms26167954






