CRY1 Lysine 151 Regulates Circadian Rhythms Through Ubiquitination-Independent Protein Interactions
Abstract
1. Introduction
2. Results
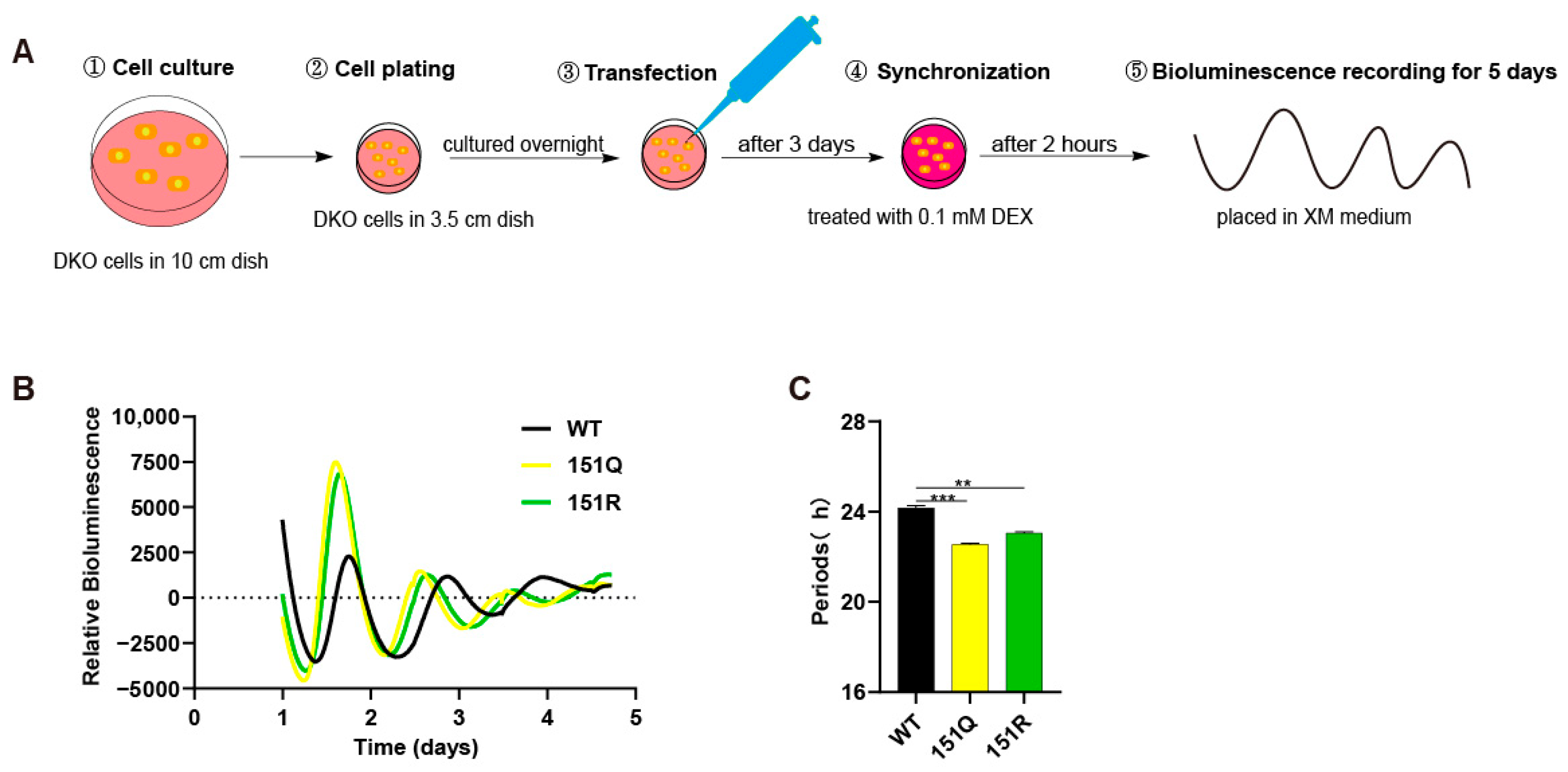
2.1. Impact of CRY1-K151 on Circadian Periodicity
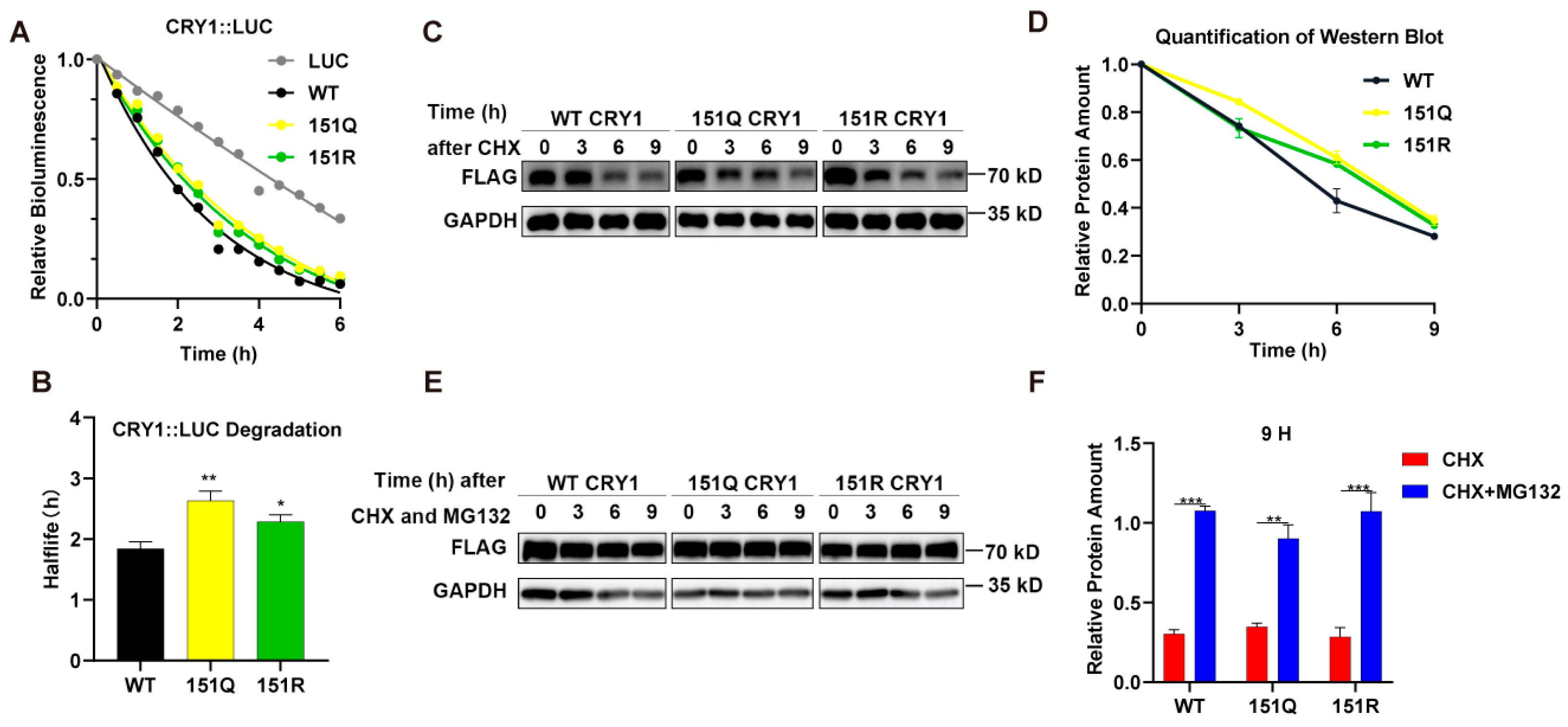
2.2. CRY1-K151 Mutations Delay Initial Protein Degradation Without Altering Proteasome Dependence
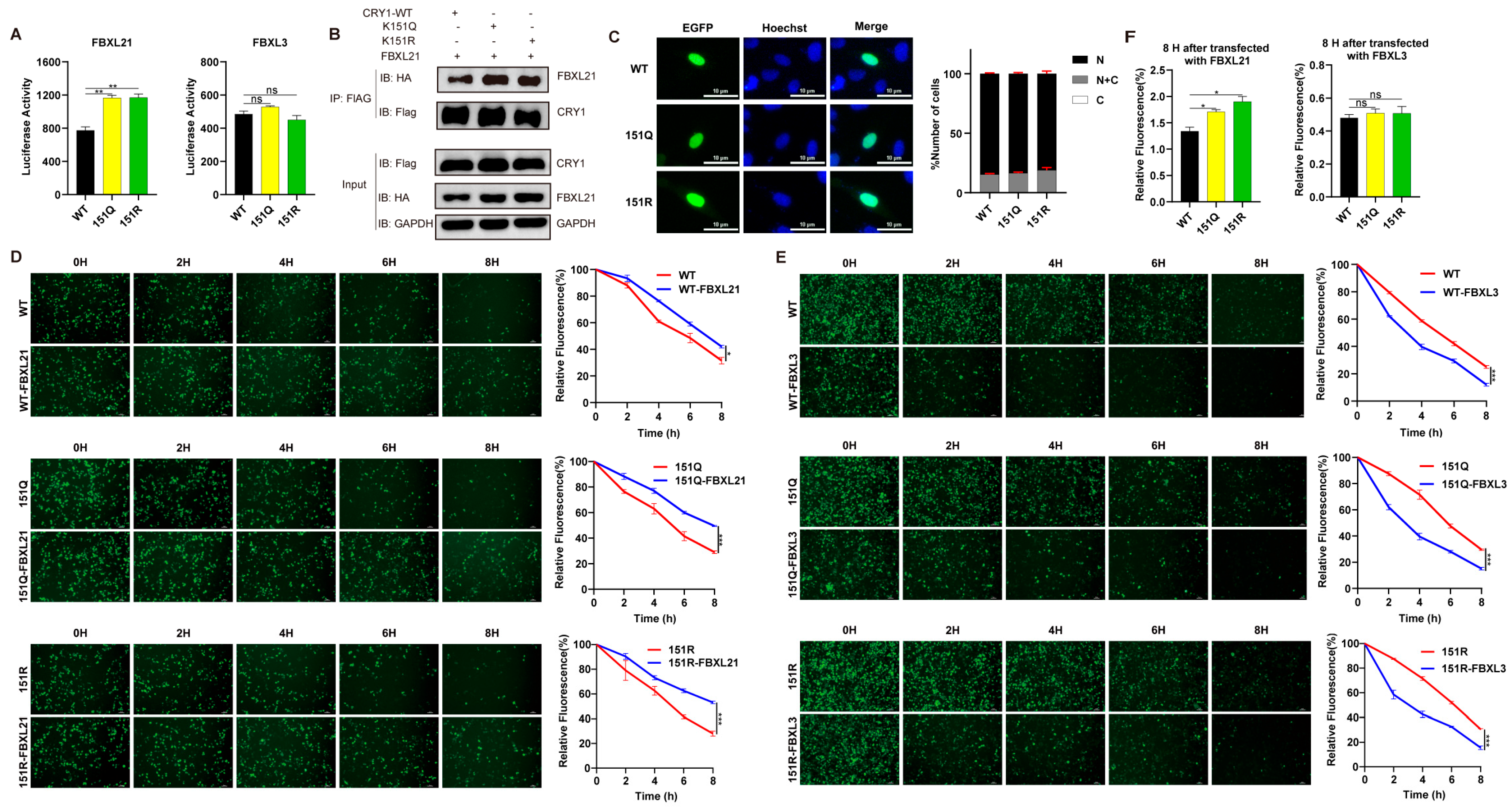
2.3. K151 Mutations Specifically Enhance CRY1’s Interaction with FBXL21
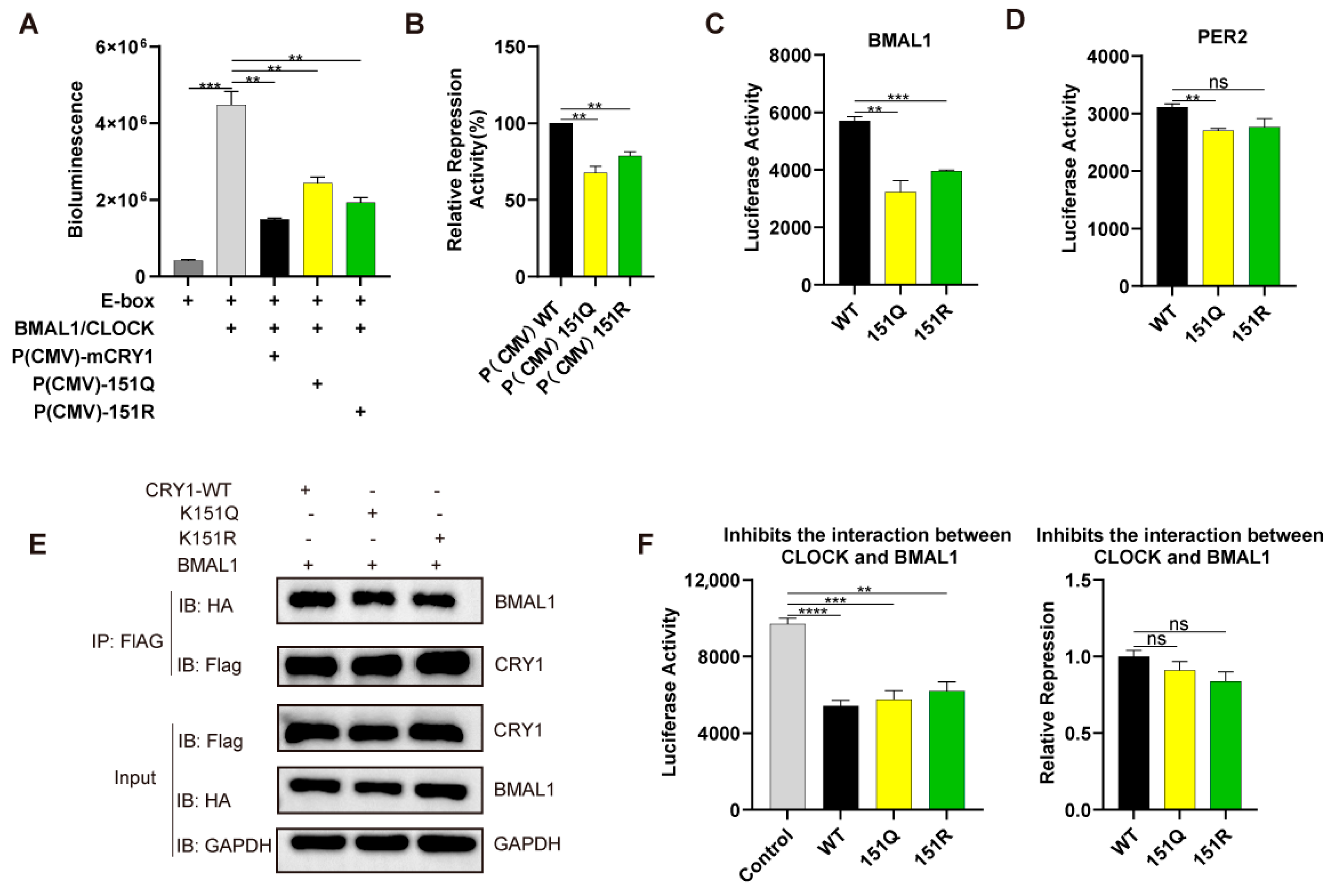
2.4. K151 Mutations Impair CRY1 Transcriptional Repression via Disrupted Core Clock Protein Interactions
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Kinetic Bioluminescence Recording
4.3. Subcellular Localization Assay
4.4. Split-Luciferase Assay
4.5. Luciferase Repression Assay
4.6. Assessment of Protein Stability Using GFP Fluorescence
4.7. Western Blotting
4.8. Luciferase Degradation Assay
4.9. Co-Immunoprecipitation Assay
4.10. Statistical Analyses
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mortimer, T.; Smith, J.G.; Muñoz-Cánoves, P.; Benitah, S.A. Circadian Clock Communication during Homeostasis and Ageing. Nat. Rev. Mol. Cell Biol. 2025, 26, 314–331. [Google Scholar] [CrossRef]
- Schrader, L.A.; Ronnekleiv-Kelly, S.M.; Hogenesch, J.B.; Bradfield, C.A.; Malecki, K.M. Circadian Disruption, Clock Genes, and Metabolic Health. J. Clin. Investig. 2024, 134, e170998. [Google Scholar] [CrossRef]
- Nassan, M.; Videnovic, A. Circadian Rhythms in Neurodegenerative Disorders. Nat. Rev. Neurol. 2022, 18, 7–24. [Google Scholar] [CrossRef]
- Gao, Q.; Tang, Z.; Wang, H.; Yamazaki, M.; Jiang, J.; Fu, Y.-H.; Ptacek, L.J.; Zhang, L. Human PERIOD3 Variants Lead to Winter Depression-like Behaviours via Glucocorticoid Signalling. Nat. Metab. 2024, 6, 2267–2280. [Google Scholar] [CrossRef]
- Ashbrook, L.H.; Krystal, A.D.; Fu, Y.-H.; Ptáček, L.J. Genetics of the Human Circadian Clock and Sleep Homeostat. Neuropsychopharmacology 2020, 45, 45–54. [Google Scholar] [CrossRef]
- Jones, C.R.; Campbell, S.S.; Zone, S.E.; Cooper, F.; DeSano, A.; Murphy, P.J.; Jones, B.; Czajkowski, L.; Ptácek, L.J. Familial Advanced Sleep-Phase Syndrome: A Short-Period Circadian Rhythm Variant in Humans. Nat. Med. 1999, 5, 1062–1065. [Google Scholar] [CrossRef] [PubMed]
- Toh, K.L.; Jones, C.R.; He, Y.; Eide, E.J.; Hinz, W.A.; Virshup, D.M.; Ptácek, L.J.; Fu, Y.H. An hPer2 Phosphorylation Site Mutation in Familial Advanced Sleep Phase Syndrome. Science 2001, 291, 1040–1043. [Google Scholar] [CrossRef]
- Webb, J.M.; Abderemane-Ali, F.; Ashbrook, L.; Ma, M.; Nibber, N.; Zou, X.; Yamazaki, M.; Wohler, E.; Sobreira, N.; Minor, D.L.; et al. CACNA1D Is a Circadian Gene and Causes Familial Advanced Sleep Phase. Proc. Natl. Acad. Sci. USA 2025, 122, e2424387122. [Google Scholar] [CrossRef]
- Silvestri, R.; Guarnieri, B. Advanced Sleep Phase Syndrome: Role of Genetics and Aging. Handb. Clin. Neurol. 2025, 206, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Hirano, A.; Shi, G.; Jones, C.R.; Lipzen, A.; Pennacchio, L.A.; Xu, Y.; Hallows, W.C.; McMahon, T.; Yamazaki, M.; Ptáček, L.J.; et al. A Cryptochrome 2 Mutation Yields Advanced Sleep Phase in Humans. eLife 2016, 5, e16695. [Google Scholar] [CrossRef] [PubMed]
- Patke, A.; Murphy, P.J.; Onat, O.E.; Krieger, A.C.; Özçelik, T.; Campbell, S.S.; Young, M.W. Mutation of the Human Circadian Clock Gene CRY1 in Familial Delayed Sleep Phase Disorder. Cell 2017, 169, 203–215.e13. [Google Scholar] [CrossRef]
- Lucio-Enríquez, K.R.; Rubio-Valles, M.; Ramos-Jiménez, A.; Pérez-León, J.A. Human Melanopsin (OPN4) Gene Polymorphisms: A Systematic Review. Front. Neurosci. 2025, 19, 1581266. [Google Scholar] [CrossRef]
- Webb, J.M.; Ma, M.; Yin, C.; Ptáček, L.J.; Fu, Y.-H. An Excitatory Peri-Tegmental Reticular Nucleus Circuit for Wake Maintenance. Proc. Natl. Acad. Sci. USA 2022, 119, e2203266119. [Google Scholar] [CrossRef]
- Pellegrino, R.; Kavakli, I.H.; Goel, N.; Cardinale, C.J.; Dinges, D.F.; Kuna, S.T.; Maislin, G.; Van Dongen, H.P.A.; Tufik, S.; Hogenesch, J.B.; et al. A Novel BHLHE41 Variant Is Associated with Short Sleep and Resistance to Sleep Deprivation in Humans. Sleep 2014, 37, 1327–1336. [Google Scholar] [CrossRef]
- Zhang, E.E.; Kay, S.A. Clocks Not Winding down: Unravelling Circadian Networks. Nat. Rev. Mol. Cell Biol. 2010, 11, 764–776. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, E.E. Phosphorylation Regulating the Ratio of Intracellular CRY1 Protein Determines the Circadian Period. Front. Neurol. 2016, 7, 159. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, Z.; Cao, J.; Dong, Y.; Chen, Y. Perfecting the Life Clock: The Journey from PTO to TTFL. Int. J. Mol. Sci. 2023, 24, 2402. [Google Scholar] [CrossRef] [PubMed]
- Rosensweig, C.; Green, C.B. Periodicity, Repression, and the Molecular Architecture of the Mammalian Circadian Clock. Eur. J. Neurosci. 2020, 51, 139–165. [Google Scholar] [CrossRef] [PubMed]
- St John, P.C.; Hirota, T.; Kay, S.A.; Doyle, F.J. Spatiotemporal Separation of PER and CRY Posttranslational Regulation in the Mammalian Circadian Clock. Proc. Natl. Acad. Sci. USA 2014, 111, 2040–2045. [Google Scholar] [CrossRef]
- Cao, X.; Yang, Y.; Selby, C.P.; Liu, Z.; Sancar, A. Molecular Mechanism of the Repressive Phase of the Mammalian Circadian Clock. Proc. Natl. Acad. Sci. USA 2021, 118, e2021174118. [Google Scholar] [CrossRef]
- Börding, T.; Abdo, A.N.; Maier, B.; Gabriel, C.; Kramer, A. Generation of Human CRY1 and CRY2 Knockout Cells Using Duplex CRISPR/Cas9 Technology. Front. Physiol. 2019, 10, 577. [Google Scholar] [CrossRef]
- Mammalian Cry1 and Cry2 Are Essential for Maintenance of Circadian Rhythms | Nature. Available online: https://www.nature.com/articles/19323 (accessed on 1 July 2025).
- Parlak, G.C.; Baris, I.; Gul, S.; Kavakli, I.H. Functional Characterization of the CRY2 Circadian Clock Component Variant p.Ser420Phe Revealed a New Degradation Pathway for CRY2. J. Biol. Chem. 2023, 299, 105451. [Google Scholar] [CrossRef]
- Parnell, A.A.; De Nobrega, A.K.; Lyons, L.C. Translating around the Clock: Multi-Level Regulation of Post-Transcriptional Processes by the Circadian Clock. Cell Signal 2021, 80, 109904. [Google Scholar] [CrossRef]
- Anna, G.; Kannan, N.N. Post-Transcriptional Modulators and Mediators of the Circadian Clock. Chronobiol. Int. 2021, 38, 1244–1261. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-H.; Mohawk, J.A.; Siepka, S.M.; Shan, Y.; Huh, S.K.; Hong, H.-K.; Kornblum, I.; Kumar, V.; Koike, N.; Xu, M.; et al. Competing E3 Ubiquitin Ligases Govern Circadian Periodicity by Degradation of CRY in Nucleus and Cytoplasm. Cell 2013, 152, 1091–1105. [Google Scholar] [CrossRef]
- Takahashi, J.S. Transcriptional Architecture of the Mammalian Circadian Clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Hirano, A.; Yumimoto, K.; Tsunematsu, R.; Matsumoto, M.; Oyama, M.; Kozuka-Hata, H.; Nakagawa, T.; Lanjakornsiripan, D.; Nakayama, K.I.; Fukada, Y. FBXL21 Regulates Oscillation of the Circadian Clock through Ubiquitination and Stabilization of Cryptochromes. Cell 2013, 152, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Czarna, A.; Berndt, A.; Singh, H.R.; Grudziecki, A.; Ladurner, A.G.; Timinszky, G.; Kramer, A.; Wolf, E. Structures of Drosophila Cryptochrome and Mouse Cryptochrome1 Provide Insight into Circadian Function. Cell 2013, 153, 1394–1405. [Google Scholar] [CrossRef]
- Gul, S.; Aydin, C.; Ozcan, O.; Gurkan, B.; Surme, S.; Baris, I.; Kavakli, I.H. The Arg-293 of Cryptochrome1 Is Responsible for the Allosteric Regulation of CLOCK-CRY1 Binding in Circadian Rhythm. J. Biol. Chem. 2020, 295, 17187–17199. [Google Scholar] [CrossRef]
- Parico, G.C.G.; Perez, I.; Fribourgh, J.L.; Hernandez, B.N.; Lee, H.-W.; Partch, C.L. The Human CRY1 Tail Controls Circadian Timing by Regulating Its Association with CLOCK:BMAL1. Proc. Natl. Acad. Sci. USA 2020, 117, 27971–27979. [Google Scholar] [CrossRef]
- Gabriel, C.H.; del Olmo, M.; Rizki Widini, A.; Roshanbin, R.; Woyde, J.; Hamza, E.; Gutu, N.-N.; Zehtabian, A.; Ewers, H.; Granada, A.; et al. Circadian Period Is Compensated for Repressor Protein Turnover Rates in Single Cells. Proc. Natl. Acad. Sci. USA 2024, 121, e2404738121. [Google Scholar] [CrossRef] [PubMed]
- Gül, Z.M.; Aydoğan, S.; Surme, S.; Harputluoğlu Efendi, S.N.; Özcan, O.; Uyanık, E.; Baris, I.; Gul, S.; Kavakli, I.H. M54 Selectively Stabilizes the Circadian Clock Component of CRY1 and Enhances the Period of Circadian Rhythm at Cellular Level. J. Biol. Chem. 2025, 301, 110333. [Google Scholar] [CrossRef] [PubMed]
- Toledo, M.; Batista-Gonzalez, A.; Merheb, E.; Aoun, M.L.; Tarabra, E.; Feng, D.; Sarparanta, J.; Merlo, P.; Botrè, F.; Schwartz, G.J.; et al. Autophagy Regulates the Liver Clock and Glucose Metabolism by Degrading CRY1. Cell Metab. 2018, 28, 268–281.e4. [Google Scholar] [CrossRef]
- Xia, K.; Li, S.; Yang, Y.; Shi, X.; Zhao, B.; Lv, L.; Xin, Z.; Kang, J.; Ren, P.; Wu, H. Cryptochrome 2 Acetylation Attenuates Its Antiproliferative Effect in Breast Cancer. Cell Death Dis. 2023, 14, 250. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Jang, H.; Lee, G.; Jeon, Y.G.; Sohn, J.H.; Han, J.S.; Lee, W.T.; Park, J.; Huh, J.Y.; Nahmgoong, H.; et al. Hepatic GSK3β-Dependent CRY1 Degradation Contributes to Diabetic Hyperglycemia. Diabetes 2022, 71, 1373–1387. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Tian, H.; Yu, Z.; Zhao, H.; Li, W.; Sang, D.; Lin, K.; Cui, Y.; Liao, M.; Xu, Z.; et al. A Highland-Adaptation Mutation of the Epas1 Protein Increases Its Stability and Disrupts the Circadian Clock in the Plateau Pika. Cell Rep. 2022, 39, 110816. [Google Scholar] [CrossRef]
- Zhang, E.E.; Liu, A.C.; Hirota, T.; Miraglia, L.J.; Welch, G.; Pongsawakul, P.Y.; Liu, X.; Atwood, A.; Huss, J.W.; Janes, J.; et al. A Genome-Wide RNAi Screen for Modifiers of the Circadian Clock in Human Cells. Cell 2009, 139, 199–210. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, H.; Zhang, E.E.; Liu, N. Identification of PCBP1 as a Novel Modulator of Mammalian Circadian Clock. Front. Genet. 2021, 12, 656571. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, J.; Liu, N.; Ren, Y.; Wang, J.; Jin, Y.; Wang, X.; Wang, W.; Pan, J. CRY1 Lysine 151 Regulates Circadian Rhythms Through Ubiquitination-Independent Protein Interactions. Int. J. Mol. Sci. 2025, 26, 7962. https://doi.org/10.3390/ijms26167962
Peng J, Liu N, Ren Y, Wang J, Jin Y, Wang X, Wang W, Pan J. CRY1 Lysine 151 Regulates Circadian Rhythms Through Ubiquitination-Independent Protein Interactions. International Journal of Molecular Sciences. 2025; 26(16):7962. https://doi.org/10.3390/ijms26167962
Chicago/Turabian StylePeng, Jiawen, Na Liu, Yixuan Ren, Jiahui Wang, Yanxia Jin, Xianping Wang, Weidong Wang, and Jicheng Pan. 2025. "CRY1 Lysine 151 Regulates Circadian Rhythms Through Ubiquitination-Independent Protein Interactions" International Journal of Molecular Sciences 26, no. 16: 7962. https://doi.org/10.3390/ijms26167962
APA StylePeng, J., Liu, N., Ren, Y., Wang, J., Jin, Y., Wang, X., Wang, W., & Pan, J. (2025). CRY1 Lysine 151 Regulates Circadian Rhythms Through Ubiquitination-Independent Protein Interactions. International Journal of Molecular Sciences, 26(16), 7962. https://doi.org/10.3390/ijms26167962





