Prime Editing Modification with FEN1 Improves F508del Variant Editing in the CFTR Gene in Airway Basal Cells
Abstract
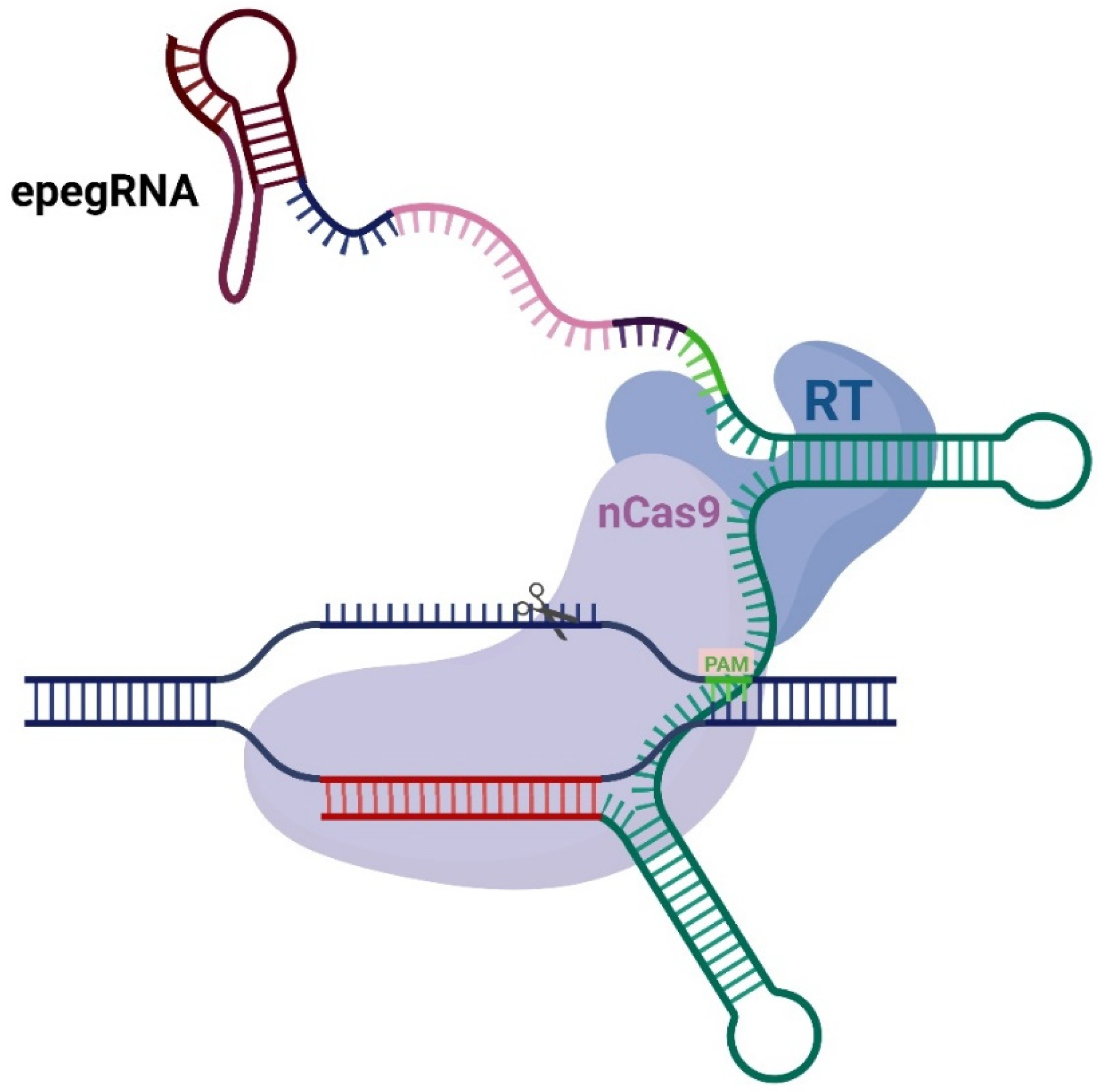
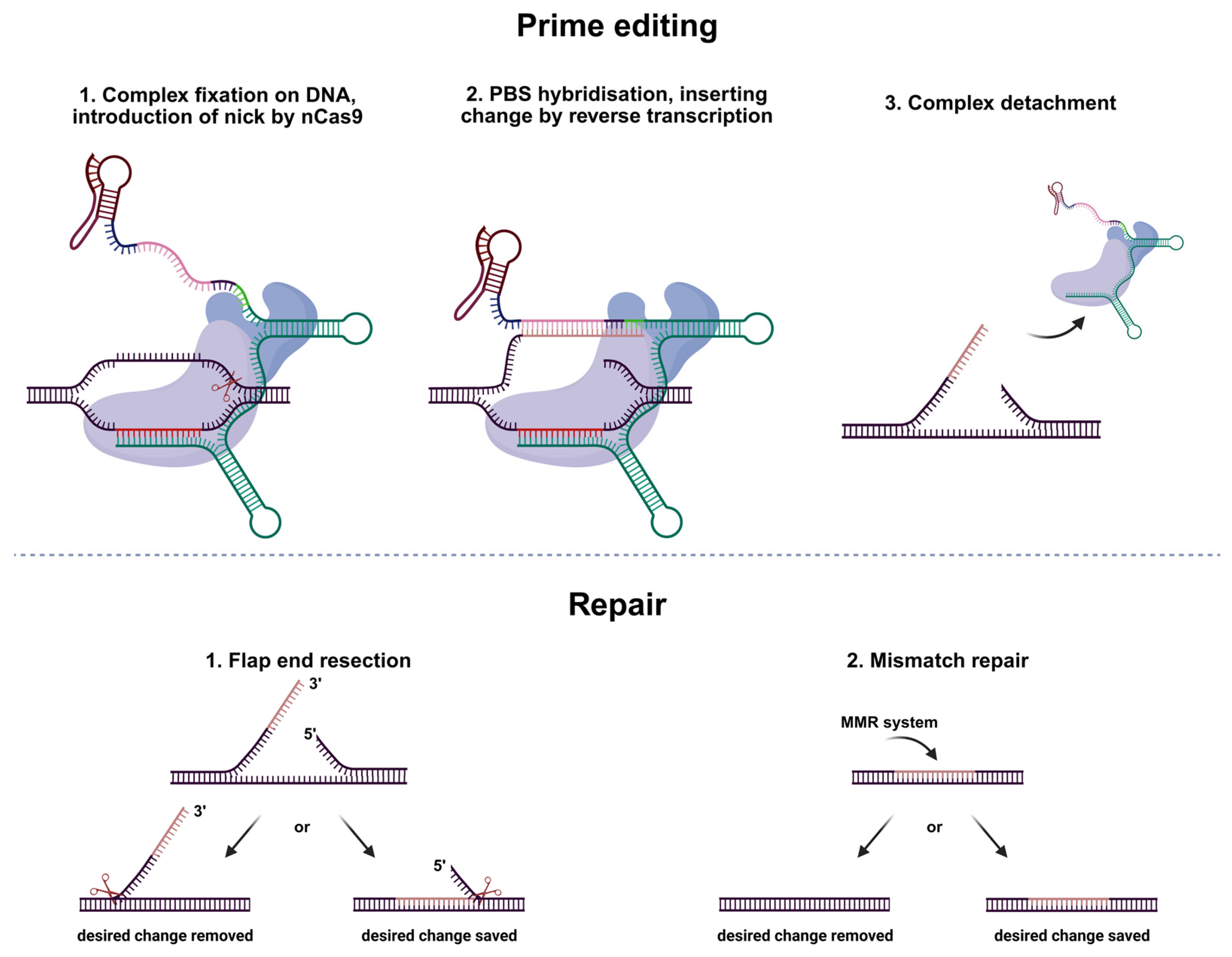
1. Introduction
2. Results
2.1. Primary Screening of All the Modifications
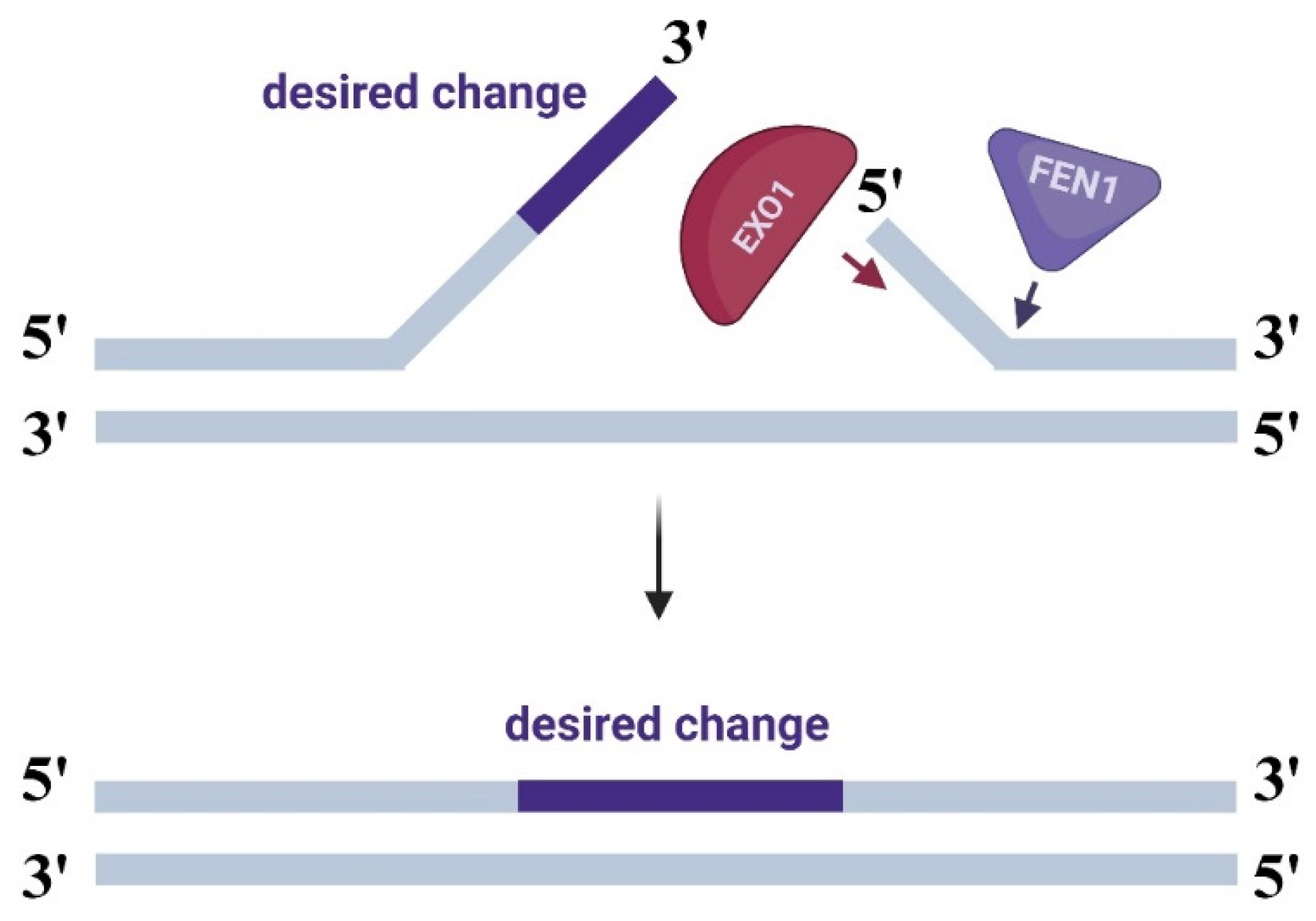
2.1.1. Modifications with FEN1 Nuclease
2.1.2. Modifications with EXO1 Nuclease
2.1.3. Modifications with the HEX-N2 Domain of EXO1 Nuclease
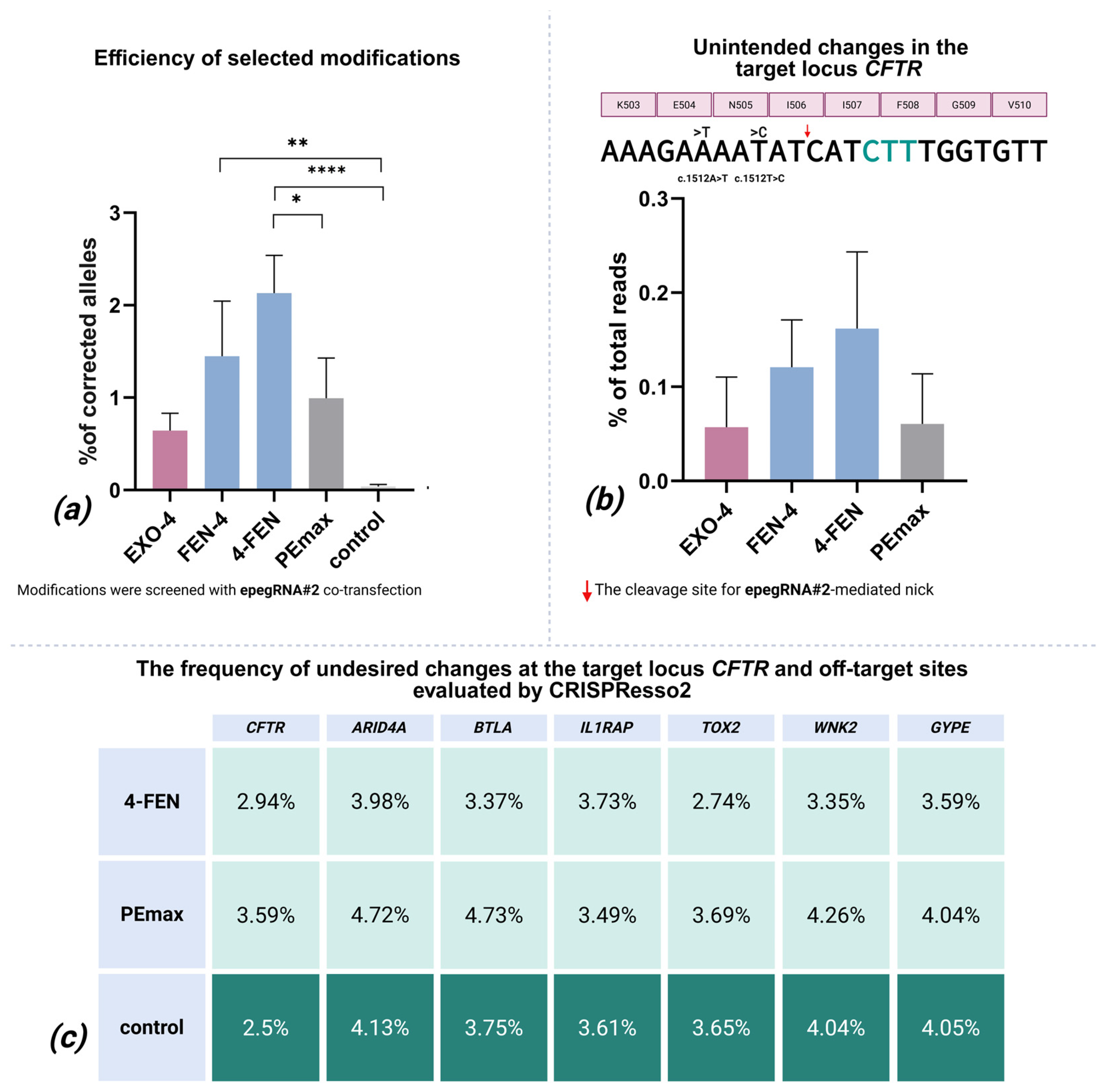
2.2. In-Depth Analysis of the Most Efficient Modifications
2.3. Indels and Mutagenesis in the Editing Locus
2.4. Undesired Changes in Edited Reads
2.5. Off-Target Effects
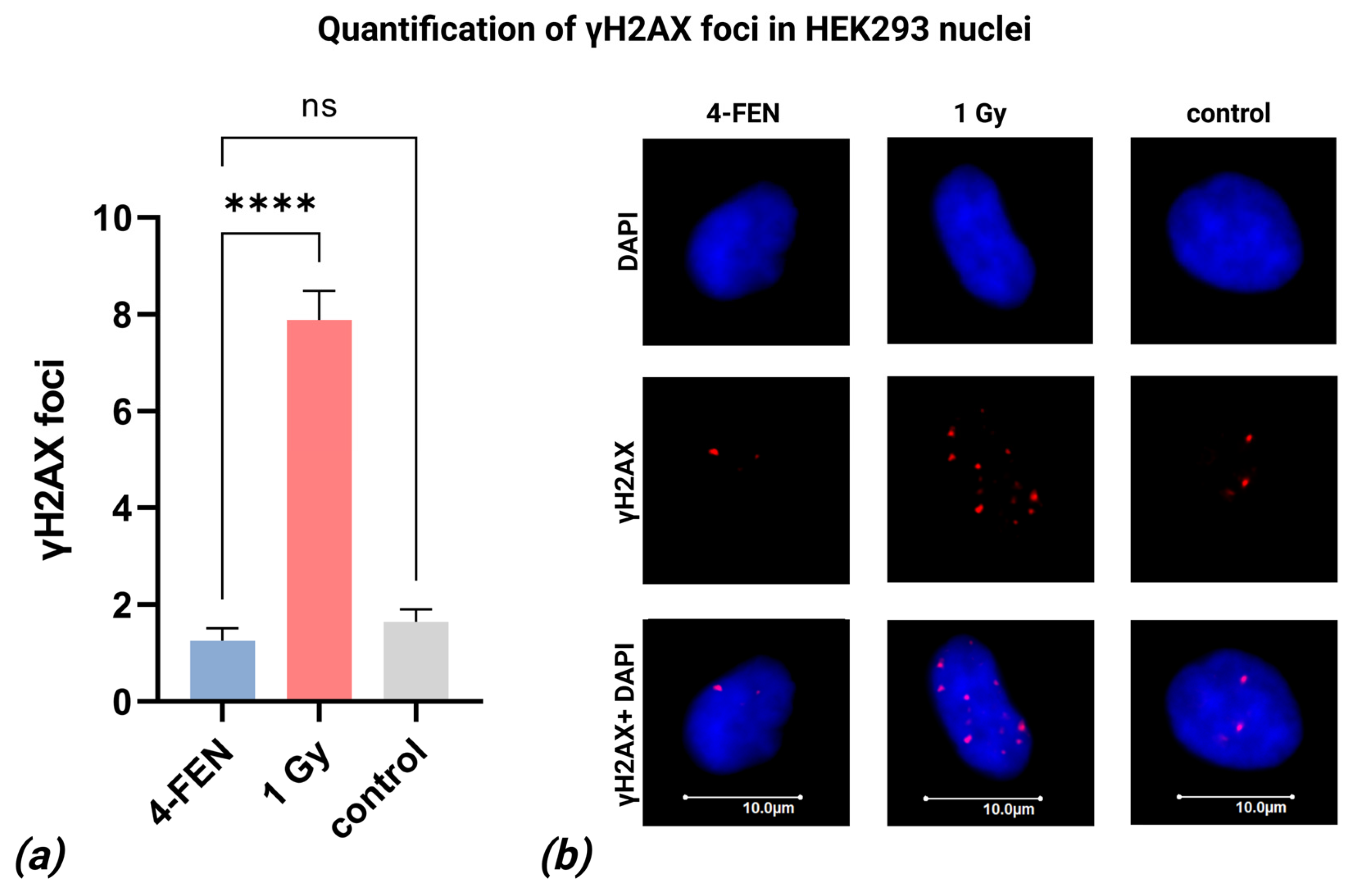
2.6. Quantification of γH2AX Foci
3. Discussion
4. Materials and Methods
4.1. Plasmid Assembly via Gibson Cloning
4.2. Derivation of Basal Cells from hiPSCs
4.3. Lipofection of Basal Cells
4.4. PegRNA Design
4.5. Editing Efficiency Analysis and Analysis of Unintended Changes in the Editing Site
4.6. Off-Target Evaluation
4.7. HEK293T Cultivation
4.8. Lipofection of HEK293T Cells
4.9. Irradiation
4.10. Immunofluorescence Staining
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| epegRNA | Engineered pegRNA |
| FEN1 | Flap endonuclease |
| EXO1 | Exonuclease 1 |
| RT | Reverse transcriptase |
| PE | Prime editing |
Appendix A
| Name | Direction | Sequence |
|---|---|---|
| 4-FEN insert | Forward | TTGAATTCCAGAACCACCAGAAGGGGAGGAGTTTTCAATCA |
| Reverse | TTTTGAGCCGCCAGATTTTCCCCTTTTAAACTTCCCTGC | |
| 4-FEN backbone 1 | Forward | GCAGGGAAGTTTAAAAGGGGAAAATCTGGCGGCTCAAAAAGAAC |
| Reverse | TTCTGGCCGCCAAGAACCTGTCCGACGCCATCCTGCTGAG | |
| 4-FEN backbone 2 | Forward | CTCAGCAGGATGGCGTCGGACAGGTTCTTGGCGGCCAGAA |
| Reverse | TTGAATTCCAGAACCACCAGAAGGGGAGGAGTTTTCAATCA | |
| FEN-4 insert | Forward | GTCACCAAAGAAGAAGCGGAAAGTCGGAATTCAAGGCCTGGCC |
| Reverse | CCAGGCCGATGCTGTACTTCTTGTCAGAACCACCAGATTTTCCCCTTTTAAACTTCCCTGC | |
| FEN-4 backbone 1 | Forward | GAAACCTGGGATATAGGGCATC |
| Reverse | GACTTTCCGCTTCTTCTTTGG | |
| FEN-4 backbone 2 | Forward | GACAAGAAGTACAGCATCGGC |
| Reverse | TGCTTCTGACAGATCTGGGC | |
| 16-FEN insert | Forward | TCAGGATCAGAAACTCCAGGTACTTCTGAATCTGCTACTCCTGAATCTGGAATTCAAGGCCTGGCCAAA |
| Reverse | TTTTGAGCCGCCAGATTTTCCCCTTTTAAACTTCCCTGC | |
| 16-FEN backbone 1 | Forward | GCAGGGAAGTTTAAAAGGGGAAAATCTGGCGGCTCAAAAAGAAC |
| Reverse | TTCTGGCCGCCAAGAACCTGTCCGACGCCATCCTGCTGAG | |
| 16-FEN backbone 2 | Forward | CTCAGCAGGATGGCGTCGGACAGGTTCTTGGCGGCCAGAA |
| Reverse | ACCTGGAGTTTCTGATCCTGAAGGGGAGGAGTTTTCAAT | |
| FEN-16 insert | Forward | GTCACCAAAGAAGAAGCGGAAAGTCGGAATTCAAGGCCTGGCC |
| Reverse | CAGATTCAGAAGTACCTGGAGTTTCTGATCCTGATTTTCCCCTTTTAAACTTCCCTGCT | |
| FEN-16 backbone 1 | Forward | GAAACCTGGGATATAGGGCATC |
| Reverse | GACTTTCCGCTTCTTCTTTGG | |
| FEN-16 backbone 2 | Forward | GGTACTTCTGAATCTGCTACTCCTGAATCTGACAAGAAGTACAGCATCGGC |
| Reverse | TGCTTCTGACAGATCTGGGC | |
| 4-EXO insert | Forward | ACTCCTCCCCTTCTGGTGGTTCTGGGATACAGGGATTGCTACA |
| Reverse | GGTTCTTTTTGAGCCGCCAGAGAATTTTTTAAATCCAAA | |
| 4-EXO backbone 1 | Forward | TTTGGATTTAAAAAATTCTCTGGCGGCTCAAAAAGAACC |
| Reverse | TTCTGGCCGCCAAGAACCTGTCCGACGCCATCCTGCTGAG | |
| 4-EXO backbone 2 | Forward | CTCAGCAGGATGGCGTCGGACAGGTTCTTGGCGGCCAGAA |
| Reverse | TGTAGCAATCCCTGTATCCCAGAACCACCAGAAGGGGAGGAGT | |
| EXO-4 insert | Forward | GTCACCAAAGAAGAAGCGGAAAGTCGGGATACAGGGATTGCTACAAT |
| Reverse | AGGCCGATGCTGTACTTCTTGTCAGAACCACCAGAGAATTTTTTAAATCC | |
| EXO-4 backbone 1 | Forward | GAAACCTGGGATATAGGGCATC |
| Reverse | GACTTTCCGCTTCTTCTTTGG | |
| EXO-4 backbone 2 | Forward | TGCTTCTGACAGATCTGGGC |
| Reverse | TGCTTCTGACAGATCTGGGC | |
| 16-EXO insert | Forward 1 | ATTGAAAACTCCTCCCCTTCAGGATCAGAAACTCCAGGTACTTCTGAATCTGCTAC |
| Forward 2 | CAGGTACTTCTGAATCTGCTACTCCTGAATCTGGGATACAGGGATTGCTACAAT | |
| Reverse | GGTTCTTTTTGAGCCGCCAGAGAATTTTTTAAATCCAAA | |
| 16-EXO backbone 1 | Forward | TTTGGATTTAAAAAATTCTCTGGCGGCTCAAAAAGAACC |
| Reverse | TTCTGGCCGCCAAGAACCTGTCCGACGCCATCCTGCTGAG | |
| 16-EXO backbone 2 | Forward | CTCAGCAGGATGGCGTCGGACAGGTTCTTGGCGGCCAGAA |
| Reverse | ACCTGGAGTTTCTGATCCTGAAGGGGAGGAGTTTTCAAT | |
| EXO-16 insert | Forward | GTCACCAAAGAAGAAGCGGAAAGTCGGGATACAGGGATTGCTACAAT |
| Reverse | GGTACTTCTGAATCTGCTACTCCTGAATCTGACAAGAAGTACAGCATCGGC | |
| EXO-16 backbone 1 | Forward | GAAACCTGGGATATAGGGCATC |
| Reverse | GACTTTCCGCTTCTTCTTTGG | |
| EXO-16 backbone 2 | Forward | GACAAGAAGTACAGCATCGGC |
| Reverse | TGCTTCTGACAGATCTGGGC | |
| 4-HEX insert | Forward | ACTCCTCCCCTTCTGGTGGTTCTGGGATACAGGGATTGCTACA |
| Reverse | ACTTGGATTTTCCTTCAATTGTGG | |
| 4-HEX backbone 1 | Forward | ATTGAAGGAAAATCCAAGTTCTGGCGGCTCAAAAAGAACCGCCGA |
| Reverse | TTCTGGCCGCCAAGAACCTGTCCGACGCCATCCTGCTGAG | |
| 4-HEX backbone 2 | Forward | CTCAGCAGGATGGCGTCGGACAGGTTCTTGGCGGCCAGAA |
| Reverse | TGTAGCAATCCCTGTATCCCAGAACCACCAGAAGGGGAGGAGT | |
| HEX-4 insert | Forward | GTCACCAAAGAAGAAGCGGAAAGTCGGGATACAGGGATTGCTACAAT |
| Reverse | GCTGTACTTCTTGTCACTACCGCCGGAACTTGGATTTTCCTTCAAT | |
| HEX-4 backbone 1 | Forward | GAAACCTGGGATATAGGGCATC |
| Reverse | GACTTTCCGCTTCTTCTTTGG | |
| HEX-4 backbone 2 | Forward | GACAAGAAGTACAGCATCGGC |
| Reverse | TGCTTCTGACAGATCTGGGC | |
| 16-HEX insert | Forward 1 | ATTGAAAACTCCTCCCCTTCAGGATCAGAAACTCCAGGTACTTCTGAATCTGCTAC |
| Forward 2 | CAGGTACTTCTGAATCTGCTACTCCTGAATCTGGGATACAGGGATTGCTACAAT | |
| Reverse | ACTTGGATTTTCCTTCAATTGTGG | |
| 16-HEX backbone 1 | Forward | ATTGAAGGAAAATCCAAGTTCTGGCGGCTCAAAAAGAACCGCCGA |
| Reverse | TTCTGGCCGCCAAGAACCTGTCCGACGCCATCCTGCTGAG | |
| 16-HEX backbone 2 | Forward | CTCAGCAGGATGGCGTCGGACAGGTTCTTGGCGGCCAGAA |
| Reverse | ACCTGGAGTTTCTGATCCTGAAGGGGAGGAGTTTTCAAT | |
| HEX-16 insert | Forward | GTCACCAAAGAAGAAGCGGAAAGTCGGGATACAGGGATTGCTACAAT |
| Reverse | AGATTCAGGAGTAGCAGATTCAGAAGTACCTGGAGTTTCTGATCCTGAACTTGGATTTTCCTTCAATTGTGG | |
| HEX-16 backbone 1 | Forward | GAAACCTGGGATATAGGGCATC |
| Reverse | GACTTTCCGCTTCTTCTTTGG | |
| HEX-16 backbone 2 | Forward | GGTACTTCTGAATCTGCTACTCCTGAATCTGACAAGAAGTACAGCATCGGC |
| Reverse | TGCTTCTGACAGATCTGGGC |
| Name | Direction | Sequence |
|---|---|---|
| X-FEN texting | Forward | ACACTCCGCCGAGGCAAG |
| Reverse | GCCGTTCTCCATCATGCGAAT | |
| FEN-X testing | Forward | AGGCGTGTACGGTGGGAG |
| Reverse | GCCGTTCTCCATCATGCGAAT | |
| X-EXO testing | Forward | ACACTCCGCCGAGGCAAG |
| Reverse | ATAGGCTTGATCCCGTGAGA | |
| EXO-X testing | Forward | AGGCGTGTACGGTGGGAG |
| Reverse | ATAGGCTTGATCCCGTGAGA | |
| X-HEX testing | Forward | ACACTCCGCCGAGGCAAG |
| Reverse | ATAGGCTTGATCCCGTGAGA | |
| HEX-X testing | Forward | AGGCGTGTACGGTGGGAG |
| Reverse | ATAGGCTTGATCCCGTGAGA | |
| Backbone testing 1 | Forward | GAGTGGAGGGACCCTGAGAT |
| Reverse | TGGGATGAACAGCCTGCAAA | |
| Backbone testing 2 | Forward | CCGAGGATGCCAAACTGCAGCT |
| Reverse | GCCGTTGTCGAAGGTCCG |
| epegRNA#1 | GACCATTAAAGAAAATATCATGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCACCAAAGATGATATTTTCTTTATACCAACTCGCGGTTCTATCTAGTTACGCGTTAAACCAACTAGAA |
| epegRNA#2 | GACCATTAAAGAAAATATCATGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCCACCAAAGATGATATTTTCTTTAACTACCCAGCGCGGTTCTATCTAGTTACGCGTTAAACCAACTAGAA |
| Gene | Forward | Reverse |
|---|---|---|
| IL1RAP | CTCGCAATGAGGTTTGGTGG | TCATTGACTAGAACACACATCAGAG |
| TOX2 | AGTCCTATCCTCAATGCGTGT | TGAGTACTTTTCTTCCTACAGGTCA |
| GYPE | GAAAGAGGCCAAAATCATGTTTATTTAAT | CAACTCTTACTGGCTTAGATGTCA |
| WNK2 | CGCTAGAAACAAATGAAACAGAAG | GGGCTGCACACATACTAACTG |
| ARID4A | TTTAAATGGCCTTCCCTAAGACT | TCAACTGTGATAACTATAGAAGTCCCT |
| BTLA | ATATCCCTTGTTAAGTCACTATAGGT | ATGTATAGAGAAAGGGGTTGCTCAAATG |
References
- Shishido, H.; Yoon, J.S.; Yang, Z.; Skach, W.R. CFTR trafficking mutations disrupt cotranslational protein folding by targeting biosynthetic intermediates. Nat. Commun. 2020, 11, 4258. [Google Scholar] [CrossRef]
- Bobadilla, J.L.; Macek, M., Jr.; Fine, J.P.; Farrell, P.M. Cystic fibrosis: A worldwide analysis of CFTR mutations—Correlation with incidence data and application to screening. Hum. Mutat. 2002, 19, 575–606. [Google Scholar] [CrossRef]
- Petrova, N.; Balinova, N.; Marakhonov, A.; Vasilyeva, T.; Kashirskaya, N.; Galkina, V.; Ginter, E.; Kutsev, S.; Zinchenko, R. Ethnic Differences in the Frequency of CFTR Gene Mutations in Populations of the European and North Caucasian Part of the Russian Federation. Front. Genet. 2021, 12, 678374. [Google Scholar] [CrossRef]
- De Boeck, K. Cystic fibrosis in the year 2020: A disease with a new face. Acta Paediatr. 2020, 109, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Spoletini, G.; Gillgrass, L.; Pollard, K.; Shaw, N.; Williams, E.; Etherington, C.; Clifton, I.; Peckham, D. Dose adjustments of Elexacaftor/Tezacaftor/Ivacaftor in response to mental health side effects in adults with cystic fibrosis. J. Cyst. Fibros. 2022, 21, 1061–1065. [Google Scholar] [CrossRef]
- Papadakis, L.; Stander, T.; Mombourquette, J.; Richards, C.J.; Yonker, L.M.; Lawton, B.; Hardcastle, M.; Zweifach, J.; Sicilian, L.; Bringhurst, L.; et al. Heterogeneity in Reported Side Effects Following Initiation of Elexacaftor-Tezacaftor-Ivacaftor: Experiences at a Quaternary CF Care Center. Pediatr. Pulmonol. 2024, 60, e27382. [Google Scholar] [CrossRef] [PubMed]
- Safety and Possible Side Effects|TRIKAFTA® (Elexacaftor/Tezacaftor/Ivacaftor and Ivacaftor). (n.d.). Available online: https://www.trikafta.com/safety-side-effects (accessed on 10 March 2025).
- Chen, P.J.; Hussmann, J.A.; Yan, J.; Knipping, F.; Ravisankar, P.; Chen, P.-F.; Chen, C.; Nelson, J.W.; Newby, G.A.; Sahin, M.; et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 2021, 184, 5635–5652.e29. [Google Scholar] [CrossRef]
- Scholefield, J.; Harrison, P.T. Prime editing—An update on the field. Gene Ther. 2021, 28, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.J.; Liu, D.R. Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 2022, 24, 161–177. [Google Scholar] [CrossRef]
- Nelson, J.W.; Randolph, P.B.; Shen, S.P.; Everette, K.A.; Chen, P.J.; Anzalone, A.V.; An, M.; Newby, G.A.; Chen, J.C.; Hsu, A.; et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 2021, 40, 402–410. [Google Scholar] [CrossRef]
- Westberg, I.; Carlsen, F.M.; Johansen, I.E.; Petersen, B.L. Cytosine base editors optimized for genome editing in potato protoplasts. Front. Genome Ed. 2023, 5, 1247702. [Google Scholar] [CrossRef] [PubMed]
- Kcam, M.C.V.; Tsong, A.J.; Chappell, J. Rational engineering of a modular bacterial CRISPR–Cas activation platform with expanded target range. Nucleic Acids Res. 2021, 49, 4793–4802. [Google Scholar] [CrossRef]
- FEN1 Flap Structure-Specific Endonuclease 1 [Homo Sapiens (Human)]—Gene—NCBI. (n.d.). Available online: https://www.ncbi.nlm.nih.gov/gene/2237 (accessed on 3 July 2025).
- Keijzers, G.; Bohr, V.A.; Rasmussen, L.J. Human exonuclease 1 (EXO1) activity characterization and its function on flap structures. Biosci. Rep. 2015, 35, e00206. [Google Scholar] [CrossRef]
- Bulcaen, M.; Kortleven, P.; Liu, R.B.; Maule, G.; Dreano, E.; Kelly, M.; Ensinck, M.M.; Thierie, S.; Smits, M.; Ciciani, M.; et al. Prime editing functionally corrects cystic fibrosis-causing CFTR mutations in human organoids and airway epithelial cells. Cell Rep. Med. 2024, 5, 101544. [Google Scholar] [CrossRef]
- Selection of Optimal pegRNAs to Enhance Efficiency of Prime Editing in AT-Rich Genome Regions|Biochemistry (Moscow). (n.d.). Available online: https://link.springer.com/article/10.1134/S0006297924604672#citeas (accessed on 3 July 2025).
- Ma, X.; Shao, Y.; Tian, L.; Flasch, D.A.; Mulder, H.L.; Edmonson, M.N.; Liu, Y.; Chen, X.; Newman, S.; Nakitandwe, J.; et al. Analysis of error profiles in deep next-generation sequencing data. Genome Biol. 2019, 20, 50. [Google Scholar] [CrossRef]
- Becker, J.R.; Gallo, D.; Leung, W.; Croissant, T.; Thu, Y.M.; Nguyen, H.D.; Starr, T.K.; Brown, G.W.; Bielinsky, A.-K. Flap endonuclease overexpression drives genome instability and DNA damage hypersensitivity in a PCNA-dependent manner. Nucleic Acids Res. 2018, 46, 5634–5650. [Google Scholar] [CrossRef]
- Xie, W.; Li, S.; Guo, H.; Zhang, J.; Tu, M.; Wang, R.; Lin, B.; Wu, Y.; Wang, X. Androgen receptor knockdown enhances prostate cancer chemosensitivity by down-regulating FEN1 through the ERK/ELK1 signalling pathway. Cancer Med. 2023, 12, 15317–15336. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wu, B.; Huang, X.; Wang, Z.; Zhu, M.; Zhu, Y.; Yu, L.; Liu, J. Inhibition of the FEN1-PBX1 axis elicits cellular senescence in breast cancer via the increased intracellular reactive oxygen species levels. J. Transl. Med. 2025, 23, 248. [Google Scholar] [CrossRef] [PubMed]
- Berfelde, J.; Hildebrand, L.S.; Kuhlmann, L.; Fietkau, R.; Distel, L.V. FEN1 Inhibition as a Potential Novel Targeted Therapy against Breast Cancer and the Prognostic Relevance of FEN1. Int. J. Mol. Sci. 2024, 25, 2110. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Tang, Z.; Yang, Z.; Zhang, L.; Deng, Q.; Zhang, X.; Yu, Y.; Liu, X.; Zhu, J. EXO1 overexpression is associated with poor prognosis of hepatocellular carcinoma patients. Cell Cycle 2018, 17, 2386–2397. [Google Scholar] [CrossRef]
- Doman, J.L.; Pandey, S.; Neugebauer, M.E.; An, M.; Davis, J.R.; Randolph, P.B.; McElroy, A.; Gao, X.D.; Raguram, A.; Richter, M.F.; et al. Phage-assisted evolution and protein engineering yield compact, efficient prime editors. Cell 2023, 186, 3983–4002.e26. [Google Scholar] [CrossRef]
- Yan, J.; Oyler-Castrillo, P.; Ravisankar, P.; Ward, C.C.; Levesque, S.; Jing, Y.; Simpson, D.; Zhao, A.; Li, H.; Yan, W.; et al. Improving prime editing with an endogenous small RNA-binding protein. Nature 2024, 628, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Lim, J.M.; Min, S.; Oh, J.-S.; Kim, D.Y.; Woo, J.-S.; Nishimasu, H.; Cho, S.-R.; Yoon, S.; Kim, H.H. Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain. Nat. Commun. 2021, 12, 5617. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-J.; Jeong, T.Y.; Shin, S.K.; Yoon, D.E.; Lim, S.-Y.; Kim, S.P.; Choi, J.; Lee, H.; Hong, J.-I.; Ahn, J.; et al. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome Biol. 2021, 22, 170. [Google Scholar] [CrossRef] [PubMed]
- Velimirovic, M.; Zanetti, L.C.; Shen, M.W.; Fife, J.D.; Lin, L.; Cha, M.; Akinci, E.; Barnum, D.; Yu, T.; Sherwood, R.I. Peptide fusion improves prime editing efficiency. Nat. Commun. 2022, 13, 3512. [Google Scholar] [CrossRef]
- Truong, D.-J.J.; Geilenkeuser, J.; Wendel, S.V.; Wilming, J.C.H.; Armbrust, N.; Binder, E.M.H.; Santl, T.H.; Siebenhaar, A.; Gruber, C.; Phlairaharn, T.; et al. Exonuclease-enhanced prime editors. Nat. Methods 2024, 21, 455–464. [Google Scholar] [CrossRef]
- Johnson, L.G.; Olsen, J.C.; Sarkadi, B.; Moore, K.L.; Swanstrom, R.; Boucher, R.C. Efficiency of gene transfer for restoration of normal airway epithelial function in cystic fibrosis. Nat. Genet. 1992, 2, 21–25. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Gröber, C.; Blank, M.; Händler, K.; Beyer, M.; Schultze, J.L.; Mayer, G. Systematic evaluation of error rates and causes in short samples in next-generation sequencing. Sci. Rep. 2018, 8, 10950. [Google Scholar] [CrossRef]
- Kondrateva, E.; Adilgereeva, E.; Amelina, E.; Tabakov, V.; Demchenko, A.; Ustinov, K.; Yasinovsky, M.; Voronina, E.; Lavrov, A.; Smirnikhina, S. Generation of induced pluripotent stem cell line (RCMGi001-A) from human skin fibroblasts of a cystic fibrosis patient with p.F508del mutation. Stem Cell Res. 2020, 48, 101933. [Google Scholar] [CrossRef]
- Panchuk, I.; Kondrateva, E.; Demchenko, A.; Grigorieva, O.; Erofeeva, A.; Amelina, E.; Tabakov, V.; Orlova, M.; Voronina, E.; Pozhitnova, V.; et al. Generation of two induced pluripotent stem cell lines (RCMGi005-A/B) from human skin fibroblasts of a cystic fibrosis patient with homozygous F508del mutation in CFTR gene. Stem Cell Res. 2022, 64, 102896. [Google Scholar] [CrossRef]
- Kondrateva, E.; Demchenko, A.; Slesarenko, Y.; Yasinovsky, M.; Amelina, E.; Tabakov, V.; Voronina, E.; Lavrov, A.; Smirnikhina, S. Derivation of iPSC line (RCMGi002-A) from dermal fibroblasts of a cystic fibrosis female patient with homozygous F508del mutation. Stem Cell Res. 2021, 53, 102251. [Google Scholar] [CrossRef] [PubMed]
- Demchenko, A.; Belova, L.; Balyasin, M.; Kochergin-Nikitsky, K.; Kondrateva, E.; Voronina, E.; Pozhitnova, V.; Tabakov, V.; Salikhova, D.; Bukharova, T.; et al. Airway basal cells from human-induced pluripotent stem cells: A new frontier in cystic fibrosis research. Front. Cell Dev. Biol. 2024, 12, 1336392. [Google Scholar] [CrossRef] [PubMed]
- Clement, K.; Rees, H.; Canver, M.C.; Gehrke, J.M.; Farouni, R.; Hsu, J.Y.; Cole, M.A.; Liu, D.R.; Joung, J.K.; Bauer, D.E.; et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019, 37, 224–226. [Google Scholar] [CrossRef]
- Bae, S.; Park, J.; Kim, J.S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volodina, O.V.; Demchenko, A.G.; Anuchina, A.A.; Ryzhkova, O.P.; Kovalskaya, V.A.; Kondrateva, E.V.; Artemova, E.V.; Tabakov, V.Y.; Ignatov, M.A.; Vorobyeva, N.Y.; et al. Prime Editing Modification with FEN1 Improves F508del Variant Editing in the CFTR Gene in Airway Basal Cells. Int. J. Mol. Sci. 2025, 26, 7943. https://doi.org/10.3390/ijms26167943
Volodina OV, Demchenko AG, Anuchina AA, Ryzhkova OP, Kovalskaya VA, Kondrateva EV, Artemova EV, Tabakov VY, Ignatov MA, Vorobyeva NY, et al. Prime Editing Modification with FEN1 Improves F508del Variant Editing in the CFTR Gene in Airway Basal Cells. International Journal of Molecular Sciences. 2025; 26(16):7943. https://doi.org/10.3390/ijms26167943
Chicago/Turabian StyleVolodina, Olga V., Anna G. Demchenko, Arina A. Anuchina, Oxana P. Ryzhkova, Valeriia A. Kovalskaya, Ekaterina V. Kondrateva, Ekaterina V. Artemova, Vyacheslav Y. Tabakov, Maxim A. Ignatov, Natalia Y. Vorobyeva, and et al. 2025. "Prime Editing Modification with FEN1 Improves F508del Variant Editing in the CFTR Gene in Airway Basal Cells" International Journal of Molecular Sciences 26, no. 16: 7943. https://doi.org/10.3390/ijms26167943
APA StyleVolodina, O. V., Demchenko, A. G., Anuchina, A. A., Ryzhkova, O. P., Kovalskaya, V. A., Kondrateva, E. V., Artemova, E. V., Tabakov, V. Y., Ignatov, M. A., Vorobyeva, N. Y., Osipov, A. N., Lavrov, A. V., & Smirnikhina, S. A. (2025). Prime Editing Modification with FEN1 Improves F508del Variant Editing in the CFTR Gene in Airway Basal Cells. International Journal of Molecular Sciences, 26(16), 7943. https://doi.org/10.3390/ijms26167943







