Epidermal and Dermal T Cells Exhibit Distinct Proteomic Signatures
Abstract
1. Introduction
2. Results
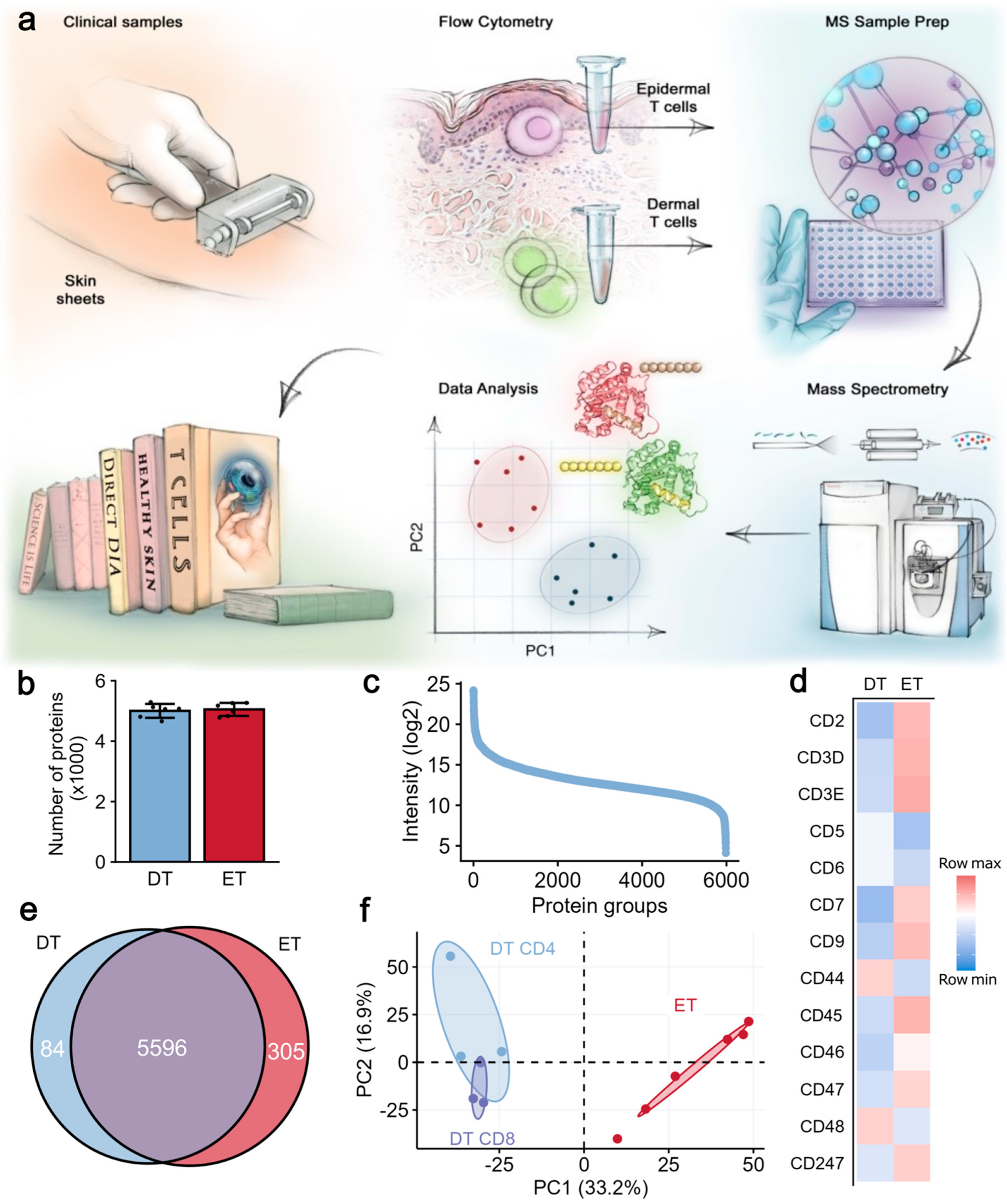
2.1. Proteomic Analysis Revealed Distinct Protein Signatures in Epidermal and Dermal T Cells
2.2. Pathway Enrichment Analysis Highlights Distinct Metabolic Profiles in Epidermal and Dermal T Cells
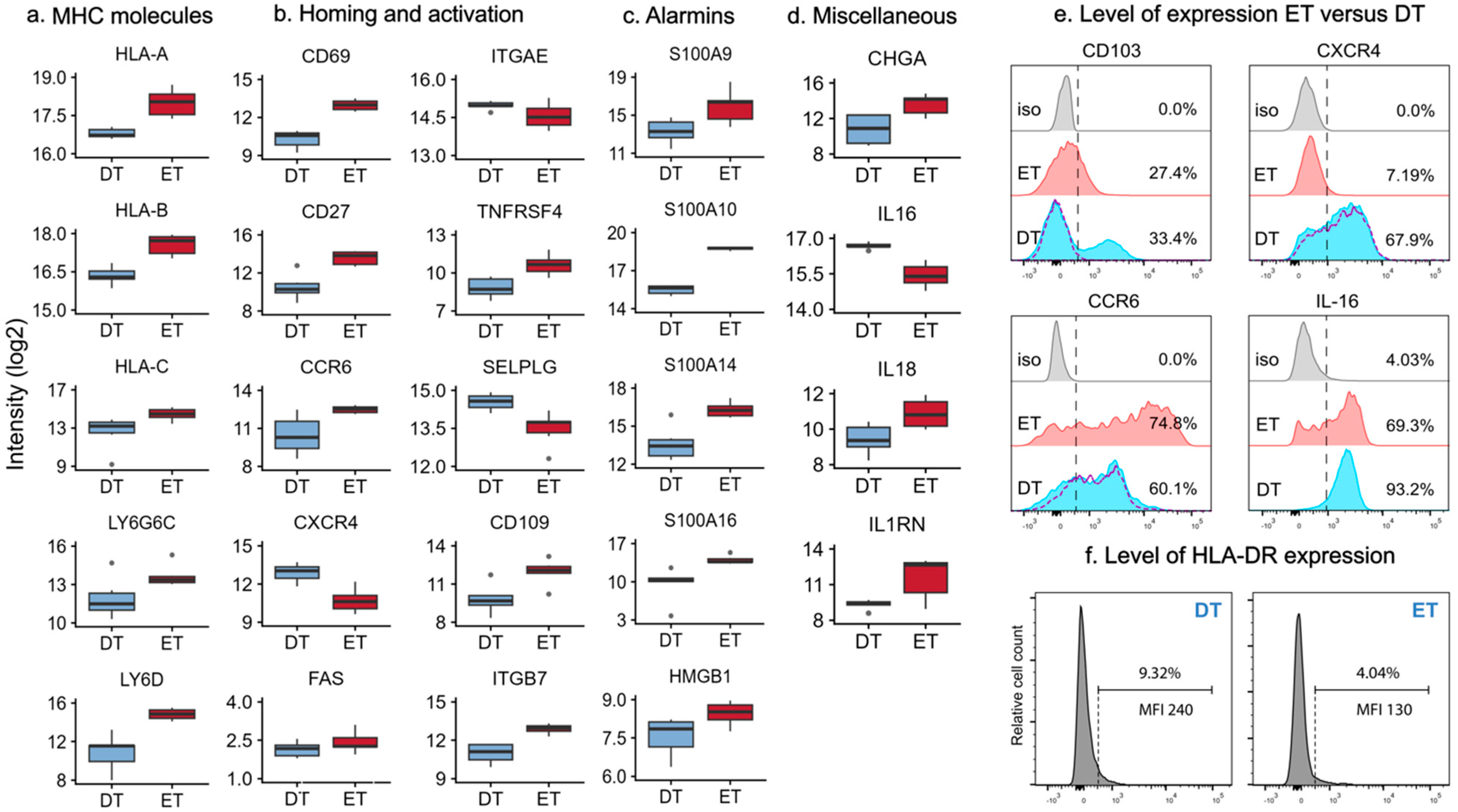
2.3. MHC Molecules and Lipid Antigen Receptors in Epidermal and Dermal T Cells
2.4. Differentially Expressed Proteins Involved in Homing and Activation
3. Discussion
4. Materials and Methods
4.1. Samples and Cell Isolation
4.2. Ethics Statement
4.3. LC-MS/MS and Data Analysis
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carbone, F.R. Unique Properties of Tissue-Resident Memory T Cells in the Lungs: Implications for COVID-19 and Other Respiratory Diseases. Nat. Rev. Immunol. 2023, 23, 329–335. [Google Scholar] [CrossRef]
- Clark, R.A.; Chong, B.; Mirchandani, N.; Brinster, N.K.; Yamanaka, K.-I.; Dowgiert, R.K.; Kupper, T.S. The Vast Majority of CLA+ T Cells Are Resident in Normal Skin. J. Immunol. Baltim. 2006, 176, 4431–4439. [Google Scholar] [CrossRef]
- Watanabe, R.; Gehad, A.; Yang, C.; Scott, L.L.; Teague, J.E.; Schlapbach, C.; Elco, C.P.; Huang, V.; Matos, T.R.; Kupper, T.S.; et al. Human Skin Is Protected by Four Functionally and Phenotypically Discrete Populations of Resident and Recirculating Memory T Cells. Sci. Transl. Med. 2015, 7, 279ra39. [Google Scholar] [CrossRef] [PubMed]
- Remedios, K.A.; Zirak, B.; Sandoval, P.M.; Lowe, M.M.; Boda, D.; Henley, E.; Bhattrai, S.; Scharschmidt, T.C.; Liao, W.; Naik, H.B.; et al. The TNFRSF Members CD27 and OX40 Coordinately Limit TH 17 Differentiation in Regulatory T Cells. Sci. Immunol. 2018, 3, eaau2042. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Hu, Q.; Li, Y.; Lu, L.; Xiang, Z.; Yin, Z.; Kabelitz, D.; Wu, Y. Γδ T Cells: Origin and Fate, Subsets, Diseases and Immunotherapy. Signal Transduct. Target. Ther. 2023, 8, 434. [Google Scholar] [CrossRef]
- Teunissen, M.B.M.; Yeremenko, N.G.; Baeten, D.L.P.; Chielie, S.; Spuls, P.I.; de Rie, M.A.; Lantz, O.; Res, P.C.M. The IL-17A-Producing CD8 + T-Cell Population in Psoriatic Lesional Skin Comprises Mucosa-Associated Invariant T Cells and Conventional T Cells. J. Investig. Dermatol. 2014, 134, 2898–2907. [Google Scholar] [CrossRef]
- Konjar, Š.; Veldhoen, M. Dynamic Metabolic State of Tissue Resident CD8 T Cells. Front. Immunol. 2019, 10, 1683. [Google Scholar] [CrossRef]
- Pan, Y.; Tian, T.; Park, C.O.; Lofftus, S.Y.; Mei, S.; Liu, X.; Luo, C.; O’Malley, J.T.; Gehad, A.; Teague, J.E.; et al. Survival of Tissue-Resident Memory T Cells Requires Exogenous Lipid Uptake and Metabolism. Nature 2017, 543, 252–256. [Google Scholar] [CrossRef]
- Dyring-Andersen, B.; Løvendorf, M.B.; Coscia, F.; Santos, A.; Møller, L.B.P.; Colaço, A.R.; Niu, L.; Bzorek, M.; Doll, S.; Andersen, J.L.; et al. Spatially and Cell-Type Resolved Quantitative Proteomic Atlas of Healthy Human Skin. Nat. Commun. 2020, 11, 5587. [Google Scholar] [CrossRef] [PubMed]
- Geiger, R.; Rieckmann, J.C.; Wolf, T.; Basso, C.; Feng, Y.; Fuhrer, T.; Kogadeeva, M.; Picotti, P.; Meissner, F.; Mann, M.; et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-Tumor Activity. Cell 2016, 167, 829–842.e13. [Google Scholar] [CrossRef]
- Rieckmann, J.C.; Geiger, R.; Hornburg, D.; Wolf, T.; Kveler, K.; Jarrossay, D.; Sallusto, F.; Shen-Orr, S.S.; Lanzavecchia, A.; Mann, M.; et al. Social Network Architecture of Human Immune Cells Unveiled by Quantitative Proteomics. Nat. Immunol. 2017, 18, 583–593. [Google Scholar] [CrossRef]
- Teunissen, M.B.M.; Pilgaard Møller, L.B.; Løvendorf, M.B.; Skov, L.; Bonefeld, C.M.; Bekkenk, M.W.; Clark, R.A.; Mann, M.; Dyring-Andersen, B. In-Depth Proteomic Map of Innate Lymphoid Cells from Healthy Human Skin and Blood. J. Investig. Dermatol. 2024, 144, 316–330.e3. [Google Scholar] [CrossRef]
- Slavov, N. Increasing Proteomics Throughput. Nat. Biotechnol. 2021, 39, 809–810. [Google Scholar] [CrossRef] [PubMed]
- Hotz, A.; Bourrat, E.; Küsel, J.; Oji, V.; Alter, S.; Hake, L.; Korbi, M.; Ott, H.; Hausser, I.; Zimmer, A.D.; et al. Mutation Update for CYP4F22 Variants Associated with Autosomal Recessive Congenital Ichthyosis. Hum. Mutat. 2018, 39, 1305–1313. [Google Scholar] [CrossRef]
- Hayashi, K.; Jutabha, P.; Endou, H.; Sagara, H.; Anzai, N. LAT1 Is a Critical Transporter of Essential Amino Acids for Immune Reactions in Activated Human T Cells. J. Immunol. 2013, 191, 4080–4085. [Google Scholar] [CrossRef]
- Kongsbak, M.; Levring, T.B.; Geisler, C.; von Essen, M.R. The Vitamin d Receptor and T Cell Function. Front. Immunol. 2013, 4, 148. [Google Scholar] [CrossRef] [PubMed]
- Líndez, A.-A.M.I.; Reith, W. Arginine-Dependent Immune Responses. Cell. Mol. Life Sci. 2021, 78, 5303–5324. [Google Scholar] [CrossRef]
- Aubatin, A.; Sako, N.; Decrouy, X.; Donnadieu, E.; Molinier-Frenkel, V.; Castellano, F. IL4-Induced Gene 1 Is Secreted at the Immune Synapse and Modulates TCR Activation Independently of Its Enzymatic Activity. Eur. J. Immunol. 2018, 48, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Revenfeld, A.L.S.; Steffensen, R.; Pugholm, L.H.; Jørgensen, M.M.; Stensballe, A.; Varming, K. Presence of HLA-DR Molecules and HLA-DRB1 mRNA in Circulating CD4(+) T Cells. Scand. J. Immunol. 2016, 84, 211–221. [Google Scholar] [CrossRef]
- Lannes-Costa, P.S.; Pimentel, B.A.d.S.; Nagao, P.E. Role of Caveolin-1 in Sepsis—A Mini-Review. Front. Immunol. 2022, 13, 902907. [Google Scholar] [CrossRef]
- Mallya, M.; Campbell, R.D.; Aguado, B. Characterization of the Five Novel Ly-6 Superfamily Members Encoded in the MHC, and Detection of Cells Expressing Their Potential Ligands. Protein Sci. 2006, 15, 2244–2256. [Google Scholar] [CrossRef]
- Clark, R.A.; Chong, B.F.; Mirchandani, N.; Yamanaka, K.-I.; Murphy, G.F.; Dowgiert, R.K.; Kupper, T.S. A Novel Method for the Isolation of Skin Resident T Cells from Normal and Diseased Human Skin. J. Investig. Dermatol. 2006, 126, 1059–1070. [Google Scholar] [CrossRef]
- Fuhlbrigge, R.C.; Kieffer, J.D.; Armerding, D.; Kupper, T.S. Cutaneous Lymphocyte Antigen Is a Specialized Form of PSGL-1 Expressed on Skin-Homing T Cells. Nature 1997, 389, 978–981. [Google Scholar] [CrossRef]
- Contento, R.L.; Molon, B.; Boularan, C.; Pozzan, T.; Manes, S.; Marullo, S.; Viola, A. CXCR4-CCR5: A Couple Modulating T Cell Functions. Proc. Natl. Acad. Sci. USA 2008, 105, 10101–10106. [Google Scholar] [CrossRef] [PubMed]
- Briolat, J.; Wu, S.D.; Mahata, S.K.; Gonthier, B.; Bagnard, D.; Chasserot-Golaz, S.; Helle, K.B.; Aunis, D.; Metz-Boutigue, M.H. New Antimicrobial Activity for the Catecholamine Release-Inhibitory Peptide from Chromogranin A. Cell. Mol. Life Sci. 2005, 62, 377–385. [Google Scholar] [CrossRef]
- Kramer, M.F.; Mack, B.; Rasp, G. Immunohistological Expression of Interleukin 16 in Human Tonsils. Arch. Otolaryngol. Head Neck Surg. 2001, 127, 1120–1125. [Google Scholar] [CrossRef] [PubMed]
- Laberge, S.; Ghaffar, O.; Boguniewicz, M.; Center, D.M.; Leung, D.Y.; Hamid, Q. Association of Increased CD4+ T-Cell Infiltration with Increased IL-16 Gene Expression in Atopic Dermatitis. J. Allergy Clin. Immunol. 1998, 102, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Schärli, S.; Luther, F.; Domizio, J.D.; Hillig, C.; Radonjic-Hoesli, S.; Thormann, K.; Simon, D.; Rønnstad, A.T.M.; Ruge, I.F.; Fritz, B.G.; et al. IL-9 Sensitizes Human Th2 Cells to pro-Inflammatory IL-18 Signals in Atopic Dermatitis. J. Allergy Clin. Immunol. 2025, 155, 491–504.e9. [Google Scholar] [CrossRef]
- Saul, L.; Ilieva, K.M.; Bax, H.J.; Karagiannis, P.; Correa, I.; Rodriguez-Hernandez, I.; Josephs, D.H.; Tosi, I.; Egbuniwe, I.U.; Lombardi, S.; et al. IgG Subclass Switching and Clonal Expansion in Cutaneous Melanoma and Normal Skin. Sci. Rep. 2016, 6, 29736. [Google Scholar] [CrossRef]
- West, E.E.; Kunz, N.; Kemper, C. Complement and Human T Cell Metabolism: Location, Location, Location. Immunol. Rev. 2020, 295, 68–81. [Google Scholar] [CrossRef]
- West, E.E.; Kolev, M.; Kemper, C. Complement and the Regulation of T Cell Responses. Annu. Rev. Immunol. 2018, 36, 309–338. [Google Scholar] [CrossRef]
- Chapman, N.M.; Boothby, M.R.; Chi, H. Metabolic Coordination of T Cell Quiescence and Activation. Nat. Rev. Immunol. 2020, 20, 55–70. [Google Scholar] [CrossRef]
- van der Windt, G.J.W.; Everts, B.; Chang, C.-H.; Curtis, J.D.; Freitas, T.C.; Amiel, E.; Pearce, E.J.; Pearce, E.L. Mitochondrial Respiratory Capacity Is a Critical Regulator of CD8+ T Cell Memory Development. Immunity 2012, 36, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Feingold, K.R.; Elias, P.M. Role of Lipids in the Formation and Maintenance of the Cutaneous Permeability Barrier. Biochim. Biophys. Acta BBA-Mol. Cell Biol. Lipids 2014, 1841, 280–294. [Google Scholar] [CrossRef]
- Molnár, E.; Swamy, M.; Holzer, M.; Beck-García, K.; Worch, R.; Thiele, C.; Guigas, G.; Boye, K.; Luescher, I.F.; Schwille, P.; et al. Cholesterol and Sphingomyelin Drive Ligand-Independent T-Cell Antigen Receptor Nanoclustering. J. Biol. Chem. 2012, 287, 42664–42674. [Google Scholar] [CrossRef]
- O’Sullivan, D.; vanderWindt, G.W.J.; Huang, S.C.C.; Curtis, J.D.; Chang, C.H.; Buck, M.D.L.; Qiu, J.; Smith, A.M.; Lam, W.Y.; DiPlato, L.M.; et al. Memory CD8+ T Cells Use Cell-Intrinsic Lipolysis to Support the Metabolic Programming Necessary for Development. Immunity 2014, 41, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Kidani, Y.; Bensinger, S.J. Liver X Receptor and Peroxisome Proliferator-activated Receptor as Integrators of Lipid Homeostasis and Immunity. Immunol. Rev. 2012, 249, 72–83. [Google Scholar] [CrossRef]
- Gebhardt, T.; Whitney, P.G.; Zaid, A.; MacKay, L.K.; Brooks, A.G.; Heath, W.R.; Carbone, F.R.; Mueller, S.N. Different Patterns of Peripheral Migration by Memory CD4+ and CD8+ T Cells. Nature 2011, 477, 216–219. [Google Scholar] [CrossRef]
- Sinclair, L.V.; Rolf, J.; Emslie, E.; Shi, Y.-B.; Taylor, P.M.; Cantrell, D.A. Control of Amino-Acid Transport by Antigen Receptors Coordinates the Metabolic Reprogramming Essential for T Cell Differentiation. Nat. Immunol. 2013, 14, 500–508. [Google Scholar] [CrossRef]
- Cibrian, D.; Saiz, M.L.; de la Fuente, H.; Sánchez-Díaz, R.; Moreno-Gonzalo, O.; Jorge, I.; Ferrarini, A.; Vázquez, J.; Punzón, C.; Fresno, M.; et al. CD69 Controls the Uptake of L-Tryptophan through LAT1-CD98 and AhR-Dependent Secretion of IL-22 in Psoriasis. Nat. Immunol. 2016, 17, 985–996, Erratum in Nat. Immunol. 2016, 17, 1235. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, M.; Ramirez, M.E.; Sierra, R.A.; Raber, P.; Thevenot, P.; Al-Khami, A.A.; Sanchez-Pino, D.; Hernez, C.; Wyczechowska, D.D.; Ochoa, A.C.; et al. L-Arginine Depletion Blunts Antitumor T-Cell Responses by Inducing Myeloid-Derived Suppressor Cells. Cancer Res. 2015, 75, 275–283. [Google Scholar] [CrossRef]
- Vu, T.T.; Koguchi-Yoshioka, H.; Watanabe, R. Skin-Resident Memory T Cells: Pathogenesis and Implication for the Treatment of Psoriasis. J. Clin. Med. 2021, 10, 3822. [Google Scholar] [CrossRef]
- Kumar, B.V.; Ma, W.; Miron, M.; Granot, T.; Guyer, R.S.; Carpenter, D.J.; Senda, T.; Sun, X.; Ho, S.-H.; Lerner, H.; et al. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 2017, 20, 2921–2934. [Google Scholar] [CrossRef] [PubMed]
- MacKay, L.K.; Rahimpour, A.; Ma, J.Z.; Collins, N.; Stock, A.T.; Hafon, M.L.; Vega-Ramos, J.; Lauzurica, P.; Mueller, S.N.; Stefanovic, T.; et al. The Developmental Pathway for CD103+ CD8+ Tissue-Resident Memory T Cells of Skin. Nat. Immunol. 2013, 14, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Nateghi-Rostami, M.; Sohrabi, Y. Memory T Cells: Promising Biomarkers for Evaluating Protection and Vaccine Efficacy against Leishmaniasis. Front. Immunol. 2024, 15, 1304696. [Google Scholar] [CrossRef]
- Poon, M.M.L.; Caron, D.P.; Wang, Z.; Wells, S.B.; Chen, D.; Meng, W.; Szabo, P.A.; Lam, N.; Kubota, M.; Matsumoto, R.; et al. Tissue Adaptation and Clonal Segregation of Human Memory T Cells in Barrier Sites. Nat. Immunol. 2023, 24, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Tyanova, S.; Cox, J. Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research. Methods Mol. Biol. 2018, 1711, 133–148. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arvesen, A.; Teunissen, M.B.M.; Agerbæk, S.; Kromann, B.; Bruun Pilgaard Møller, L.; Gehad, A.; Clark, R.A.; Løvendorf, M.B.; Dyring-Andersen, B. Epidermal and Dermal T Cells Exhibit Distinct Proteomic Signatures. Int. J. Mol. Sci. 2025, 26, 7942. https://doi.org/10.3390/ijms26167942
Arvesen A, Teunissen MBM, Agerbæk S, Kromann B, Bruun Pilgaard Møller L, Gehad A, Clark RA, Løvendorf MB, Dyring-Andersen B. Epidermal and Dermal T Cells Exhibit Distinct Proteomic Signatures. International Journal of Molecular Sciences. 2025; 26(16):7942. https://doi.org/10.3390/ijms26167942
Chicago/Turabian StyleArvesen, Amalie, Marcel B. M. Teunissen, Sofie Agerbæk, Bjørn Kromann, Line Bruun Pilgaard Møller, Ahmed Gehad, Rachael A. Clark, Marianne Bengtson Løvendorf, and Beatrice Dyring-Andersen. 2025. "Epidermal and Dermal T Cells Exhibit Distinct Proteomic Signatures" International Journal of Molecular Sciences 26, no. 16: 7942. https://doi.org/10.3390/ijms26167942
APA StyleArvesen, A., Teunissen, M. B. M., Agerbæk, S., Kromann, B., Bruun Pilgaard Møller, L., Gehad, A., Clark, R. A., Løvendorf, M. B., & Dyring-Andersen, B. (2025). Epidermal and Dermal T Cells Exhibit Distinct Proteomic Signatures. International Journal of Molecular Sciences, 26(16), 7942. https://doi.org/10.3390/ijms26167942





