Molecular Networking-Guided Annotation of Flavonoid Glycosides from Quercus mongolica Bee Pollen
Abstract
1. Introduction
2. Results
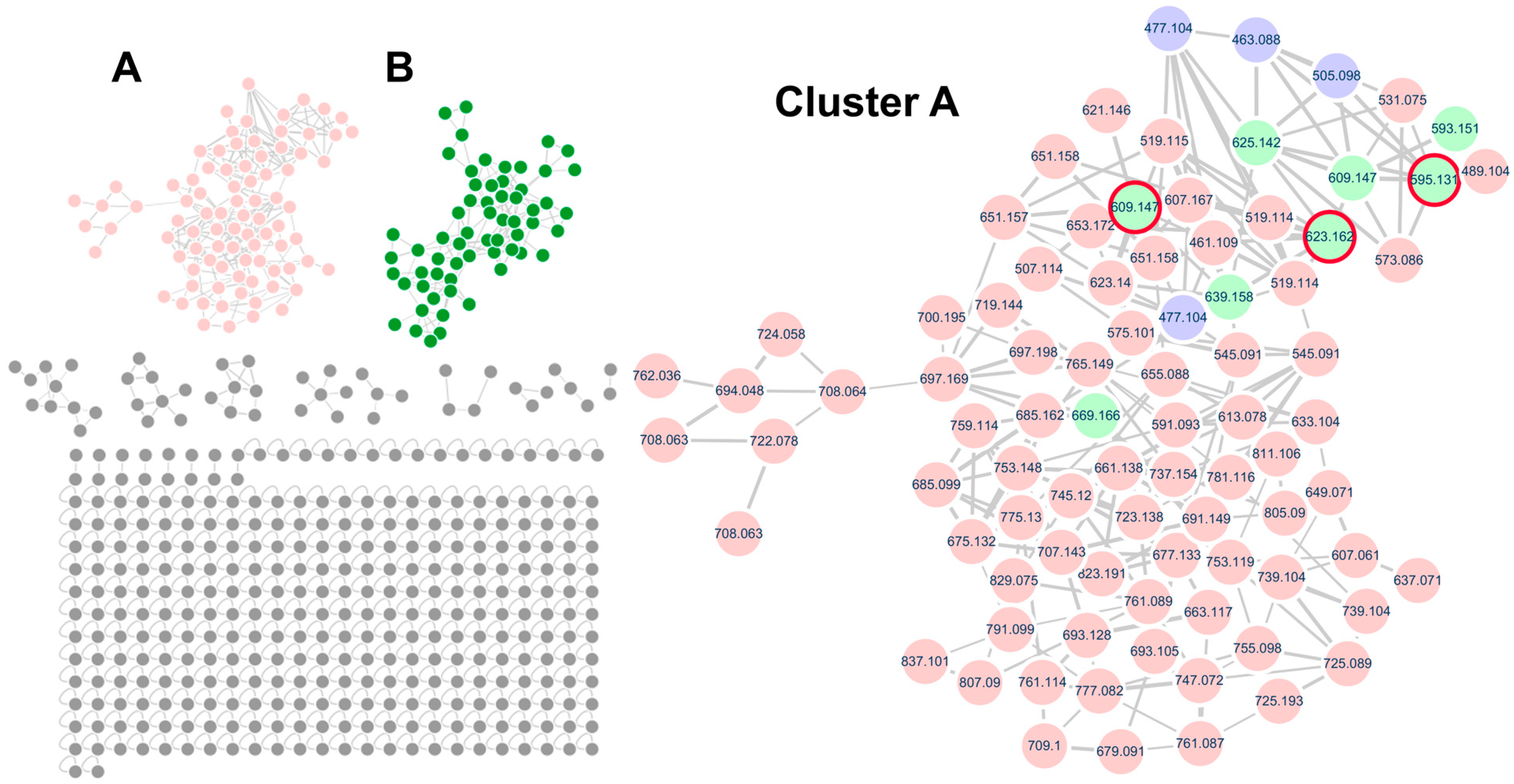
2.1. Flavonoid Glycoside Profiling of Q. mongolica Pollen Using Molecular Networking Based on UPLC–QTOF–MS/MS Analysis
2.2. Identification of Kaempferol Derivatives
| No. | RT (min) | [M−H]− (m/z) | Molecular Formula (Error in ppm) | Tentatively Identification | MS2 (m/z) | Ref |
|---|---|---|---|---|---|---|
| Kaempferol derivatives | ||||||
| 1 | 6.92 | 489.104 | C23H22O12 (−0.1) | Kaempferol-3-O-acetylglucoside | 429, 309, 285, 284, 255, 227 | [24] |
| 2 | 4.69 | 593.151 | C27H30O15 (−0.2) | Kaempferol-3-O-rutinoside a | 285, 284, 255, 227 | [24,35] |
| Quercetin derivatives | ||||||
| 3 | 4.95 | 463.088 | C21H20O12 (0.0) | Quercetin-3-O-glucoside a | 301, 300, 271, 255, 243 | [34,35] |
| 4 | 6.03 | 505.098 | C23H22O13 (−0.3) | Quercetin-3-O-acetylglucoside a | 301, 300, 271, 255, 243 | [24] |
| 5 | 6.27 | 573.086 | C33H18O10 (4.9) | Quercetin derivatives | 505, 301, 300, 271, 255 | |
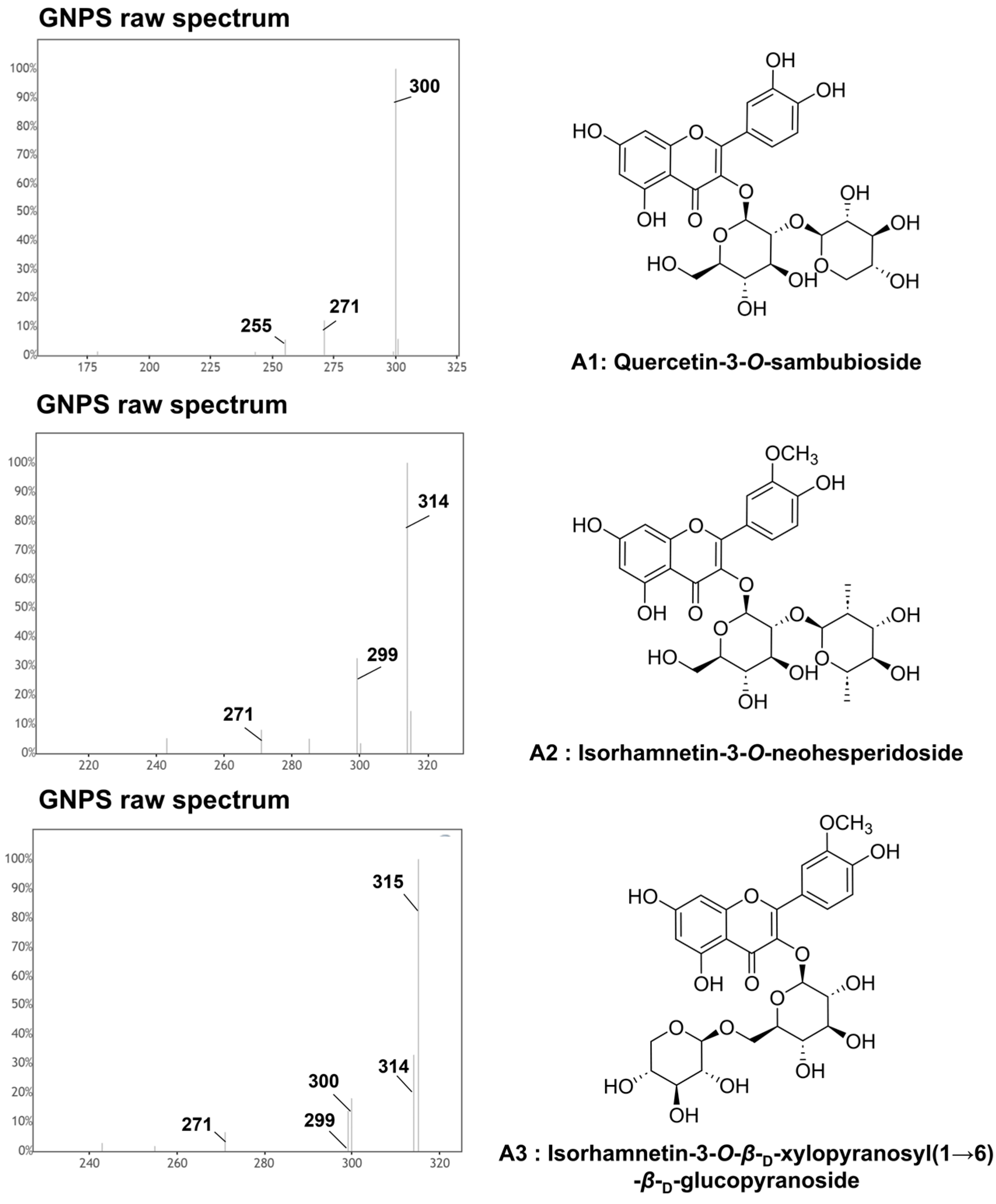
| 6 | 4.17 | 595.132 | C26H28O16 (1.2) | Quercetin-3-O-sambubioside a,b | 463, 445, 301, 300, 271, 255 | [36] |
| 7 | 4.13 | 609.147 | C27H30O16 (0.8) | Quercetin-3-O-rutinoside a | 463, 445, 301, 300, 271, 255 | [37] |
| 8 | 3.70 | 625.142 | C27H30O17 (1.1) | Quercetin-3-O-sophoroside a | 463, 445, 301, 300, 271, 255 | [36,37] |
| 9 | 4.17 | 663.117 | C29H28O18 (−4.1) | Quercetin derivatives | 595, 323, 301, 300 | |
| 10 | 4.19 | 679.091 | C32H24O17 (−5.1) | Quercetin derivatives | 611, 595, 301, 300, 271 | |
| 11 | 3.72 | 693.128 | C37H26O14 (4.2) | Quercetin derivatives | 647, 625, 323, 301, 300 | |
| 12 | 3.72 | 709.100 | C33H26O18 (−6.3) | Quercetin derivatives | 625, 399, 384, 301, 300 | |
| 13 | 4.27 | 725.193 | C29H26O22 (5.9) | Quercetin derivatives | 679, 633, 384, 301, 300, 284 | |
| 14 | 4.15 | 747.072 | C24H28O27 (−3.4) | Quercetin derivatives | 701, 655, 595, 523, 301, 300 | |
| 15 | 3.74 | 761.114 | C33H30O21 (−9.3) | Quercetin derivatives | 715, 647, 625, 323, 301, 300 | |
| 16 | 3.72 | 777.082 | C32H26O23 (3.6) | Quercetin derivatives | 731, 685, 625, 523, 301, 300 | |
| Isorhamnetin derivatives | ||||||
| 17 | 6.46 | 461.109 | C22H22O11 (−0.3) | Isorhamnetin-3-O-rhamnoside | 315, 314, 300, 299, 285, 271, 257, 243 | [24] |
| 18 | 5.79 | 477.104 | C22H22O12 (1.2) | Isorhamnetin-3-O-glucoside isomer a | 315, 314, 299, 285, 271, 257, 243 | [38] |
| 19 | 5.47 | 477.104 | C22H22O12 (1.4) | Isorhamnetin-3-O-glucoside isomer a | 315, 314, 300, 299, 271, 255 | [38] |
| 20 | 6.53 | 519.114 | C24H24O13 (−0.2) | Isorhamnetin-3-O-acetylglucoside isomer | 315, 314, 299, 285, 271, 257, 243 | |
| 21 | 6.79 | 519.114 | C24H24O13 (0) | Isorhamnetin-3-O-acetylglucoside isomer | 315, 314, 299, 285, 271, 257, 243 | |
| 22 | 7.11 | 519.114 | C24H24O13 (0.7) | Isorhamnetin-3-O-acetylglucoside isomer | 459, 315, 314, 300, 299, 285, 271 | |
| 23 | 5.45 | 545.091 | C25H22O14 (−3.3) | Isorhamnetin derivatives | 477, 315, 314, 300, 299, 271 | |
| 24 | 5.77 | 545.091 | C32H18O9 (5.3) | Isorhamnetin derivatives | 477, 315, 314, 299, 285, 271, | |
| 25 | 5.77 | 591.093 | C33H20O11 (−0.3) | Isorhamnetin derivatives | 477, 383, 315, 314, 285, 271 | |
| 26 | 6.58 | 607.167 | C28H32O15 (0.8) | Isorhamnetin derivatives | 315, 314, 299, 285, 271 | |
| 27 | 5.79 | 607.061 | C25H20O18 (5.8) | Isorhamnetin derivatives | 399, 383, 371, 315, 314, 299 | |
| 28 | 5.47 | 609.148 | C27H30O16 (1.5) | Isorhamnetin-3-O-β-D-xylopyranosyl(1→6)-β-D-glucopyranoside a,b | 315, 314, 300, 299, 271 | |
| 29 | 7.26 | 623.140 | C31H28O14 (0.1) | Isorhamnetin derivatives | 477, 315, 314, 300, 299, 271 | |
| 30 | 4.75 | 623.162 | C28H32O16 (1.2) | Isorhamnetin-3-O-neohesperidoside a,b | 315, 314, 300, 299, 285, 271 | [24,38] |
| 31 | 7.09 | 633.104 | C35H22O12 (0.4) | Isorhamnetin derivatives | 519, 383, 315, 314, 300, 299, 112 | |
| 32 | 4.08 | 639.158 | C28H32O17 (−0.6) | Isorhamnetin-3-O-sophoroside a | 459, 315, 314, 300, 299, 271 | [39] |
| 33 | 7.02 | 651.157 | C29H32O17 (0.5) | Isorhamnetin derivatives | 591, 315, 314, 299, 285, 243 | |
| 34 | 6.60 | 651.158 | C29H32O17 (0.5) | Isorhamnetin derivatives | 519, 383, 315, 314, 300, 299 | |
| 35 | 7.13 | 655.088 | C30H24O17 (−8.4) | Isorhamnetin derivatives | 519, 459, 315, 314, 300, 299 | |
| 36 | 4.73 | 669.166 | C29H34O18 (−1.1) | Isorhamnetin derivatives | 623, 315, 314, 299 | |
| 37 | 4.12 | 675.132 | C34H28O15 (−5.2) | Isorhamnetin derivatives | 639, 321, 315, 314, 300, 299 | |
| 38 | 5.47 | 677.133 | C30H30O18 (−5.2) | Isorhamnetin derivatives | 609, 315, 314, 300, 299 | |
| 39 | 4.15 | 685.099 | C29H34O19 (−1.0) | Isorhamnetin derivatives | 639, 315, 314, 300, 299 | |
| 40 | 4.08 | 685.162 | C29H34O19 (−1.2) | Isorhamnetin derivatives | 639, 315, 314, 300, 299 | |
| 41 | 4.73 | 691.149 | C31H32O18 (−4.2) | Isorhamnetin derivatives | 623, 337, 315, 314 | |
| 42 | 5.47 | 693.105 | C33H26O17 (−6.5) | Isorhamnetin derivatives | 647, 609, 413, 315, 314 | |
| 43 | 7.07 | 697.169 | C23H26O17 (−0.5) | Isorhamnetin derivatives | 651, 605, 591, 315, 314, 300, 299 | |
| 44 | 7.15 | 697.198 | C31H38O18 (−3.2) | Isorhamnetin derivatives | 651, 605, 591, 315, 314, 299 | |
| 45 | 4.10 | 707.143 | C38H28O17 (−3.2) | Isorhamnetin derivatives | 639, 609, 315, 314, 300, 299 | |
| 46 | 7.00 | 719.144 | C32H32O19 (−3.6) | Isorhamnetin derivatives | 651, 591, 315, 314 | |
| 47 | 5.47 | 723.138 | C38H28O15 (3.9) | Isorhamnetin derivatives | 631, 609, 315, 314 | |
| 48 | 4.73 | 737.154 | C39H30O15 (3.8) | Isorhamnetin derivatives | 691, 645, 623, 315, 314 | |
| 49 | 4.92 | 739.104 | C23H32O27 (−3.5) | Isorhamnetin derivatives | 693, 609, 399, 399, 315, 314 | |
| 50 | 5.47 | 739.104 | C23H32O27 (−2.5) | Isorhamnetin derivatives | 693, 647, 609, 479, 413, 315 | |
| 51 | 5.47 | 745.120 | C33H30O20 (−8.1) | Isorhamnetin derivatives | 653, 609, 315, 314 | |
| 52 | 4.8 | 753.119 | C24H34O27 (−2.9) | Isorhamnetin derivatives | 707, 661, 623, 399, 315, 314 | |
| 53 | 4.1 | 753.148 | C39H30O16 (2.6) | Isorhamnetin derivatives | 707, 639, 315, 314, 300, 299 | |
| 54 | 5.49 | 761.089 | C25H30O27 (−2.1) | Isorhamnetin derivatives | 669, 663, 609, 435, 315, 314 | |
| 55 | 7.05 | 765.149 | C33H34O21 (−4.2) | Isorhamnetin derivatives | 701, 673, 651, 315, 314, 112 | |
| 56 | 4.08 | 775.130 | C34H32O21 (−8.4) | Isorhamnetin derivatives | 639, 609, 315, 314, 300, 299 | |
| 57 | 7.00 | 781.116 | C25H34O28 (−1.0) | Isorhamnetin derivatives | 689, 651, 629, 413, 315, 314 | |
| 58 | 4.12 | 791.099 | C26H32O28 (−2.4) | Isorhamnetin derivatives | 699, 639, 609, 315, 314, 301, 300, 299 | |
| 59 | 5.49 | 807.090 | C33H28O24 (−0.3) | Isorhamnetin derivatives | 715, 669, 609, 435, 315, 314 | |
| 60 | 4.75 | 811.106 | C29H32O27 (−0.1) | Isorhamnetin derivatives | 765, 623, 456, 315, 314 | |
| 61 | 4.34 | 823.191 | C36H40O22 (−3.7) | Isorhamnetin derivatives | 755, 315, 314 | |
| 62 | 4.08 | 837.101 | C34H30O25 (0.9) | Isorhamnetin derivatives | 723, 699, 639, 399, 315, 314 | |
| Quercetagetin-dimethyl derivatives | ||||||
| 63 | 5.64 | 507.114 | C23H24O13 (0.2) | Quercetagetin-dimethyl 3-O-hexoside | 345, 344, 330, 329, 314, 301, 286 | [40] |
| 64 | 5.65 | 637.071 | C26H22O19 (5.1) | Quercetagetin-dimethyl derivatives | 429, 413, 386, 345, 329, 300 | |
| 65 | 4.50 | 653.172 | C29H34O17 (−0.1) | Quercetagetin-dimethyl derivatives | 345, 344, 330, 329, 301 | |
| Others | ||||||
| 66 | 5.62 | 575.101 | C26H24O15 (−4.8) | unknown | 507, 492, 344, 329, 301 | |
| 67 | 3.34 | 621.146 | C28H30O16 (−0.2) | unknown | 327, 326, 312, 311, 284, 283 | |
| 68 | 4.15 | 725.089 | C29H26O22 (5.9) | unknown | 679, 633, 595, 399, 384, 301 | |
| 69 | 3.74 | 755.098 | C30H28O23 (4.7) | unknown | 709, 663, 625, 399, 384, 301 | |
2.3. Identification of Quercetin Derivatives
2.4. Identification of Isorhamnetin Derivatives
2.5. Identification of Additional Flavonoid Glycosides
3. Discussion
4. Materials and Methods
4.1. Reagents and Materials
4.2. Plant Materials and Extract Preparation
4.3. UPLC–QTOF–MS/MS Analysis
4.4. Data Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kwak, J.-E.; Lee, J.-Y.; Baek, J.-Y.; Kim, S.W.; Ahn, M.-R. The Antioxidant and Anti-Inflammatory Properties of Bee Pollen from Acorn (Quercus acutissima Carr.) and Darae (Actinidia arguta). Antioxidants 2024, 13, 981. [Google Scholar] [CrossRef]
- Qiao, J.; Feng, Z.; Zhang, Y.; Xiao, X.; Dong, J.; Haubruge, E.; Zhang, H. Phenolamide and Flavonoid Glycoside Profiles of 20 Types of Monofloral Bee Pollen. Food Chem. 2023, 405, 134800. [Google Scholar] [CrossRef]
- El Ghouizi, A.; Bakour, M.; Laaroussi, H.; Ousaaid, D.; El Menyiy, N.; Hano, C.; Lyoussi, B. Bee Pollen as Functional Food: Insights into Its Composition and Therapeutic Properties. Antioxidants 2023, 12, 557. [Google Scholar] [CrossRef] [PubMed]
- Miłek, M.; Mołoń, M.; Kula-Maximenko, M.; Sidor, E.; Zaguła, G.; Dżugan, M. Chemical Composition and Bioactivity of Laboratory-Fermented Bee Pollen in Comparison with Natural Bee Bread. Biomolecules 2023, 13, 1025. [Google Scholar] [CrossRef]
- Bakour, M.; Laaroussi, H.; Ousaaid, D.; El Ghouizi, A.; Es-Safi, I.; Mechchate, H.; Lyoussi, B. Bee Bread as a Promising Source of Bioactive Molecules and Functional Properties: An up-to-Date Review. Antibiotics 2022, 11, 203. [Google Scholar] [CrossRef]
- Sen, N.B.; Vovk, I.; Kırmızıbekmez, H.; Guzelmeric, E. Phytochemical and Bioactivity Evaluation of Bee Pollen and Androecia of Castanea, Salix, and Quercus Species. Antioxidants 2024, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Rzepecka-Stojko, A.; Stojko, J.; Kurek-Górecka, A.; Górecki, M.; Kabała-Dzik, A.; Kubina, R.; Moździerz, A.; Buszman, E. Polyphenols from Bee Pollen: Structure, Absorption, Metabolism and Biological Activity. Molecules 2015, 20, 21732–21749, Erratum in: Molecules 2016, 21, 159. https://doi.org/10.3390/molecules21020159. [Google Scholar] [CrossRef] [PubMed]
- Miyata, R.; Hoshino, S.; Ahn, M.-R.; Kumazawa, S. Chemical Profiles of Korean Bee Pollens and Their Catechol-O-Methyltransferase Inhibitory Activities. J. Agric. Food Chem. 2022, 70, 1174–1181. [Google Scholar] [CrossRef]
- Rodríguez-Pólit, C.; Gonzalez-Pastor, R.; Heredia-Moya, J.; Carrera-Pacheco, S.E.; Castillo-Solis, F.; Vallejo-Imbaquingo, R.; Barba-Ostria, C.; Guamán, L.P. Chemical Properties and Biological Activity of Bee Pollen. Molecules 2023, 28, 7768. [Google Scholar] [CrossRef]
- Cornara, L.; Biagi, M.; Xiao, J.; Burlando, B. Therapeutic Properties of Bioactive Compounds from Different Honeybee Products. Front. Pharmacol. 2017, 8, 412. [Google Scholar] [CrossRef]
- Dias, M.C.; Pinto, D.C.G.A.; Silva, A.M.S. Plant Flavonoids: Chemical Characteristics and Biological Activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-Y.; Li, Q.; Bi, K.-S. Bioactive Flavonoids in Medicinal Plants: Structure, Activity and Biological Fate. Asian J. Pharm. Sci. 2018, 13, 12–23. [Google Scholar] [CrossRef]
- Joo, Y.; Seo, Y.H.; Lee, S.; Shin, E.; Yeon, S.W.; Kim, S.B.; Lee, M.K. Antioxidant and Tyrosinase-Inhibitory Activities and Biological Bioactivities of Flavonoid Derivatives from Quercus mongolica Pollen. Molecules 2025, 30, 794. [Google Scholar] [CrossRef]
- Serra Bonvehí, J.; Soliva Torrentó, M.; Centelles Lorente, E. Evaluation of Polyphenolic and Flavonoid Compounds in Honeybee-Collected Pollen Produced in Spain. J. Agric. Food Chem. 2001, 49, 1848–1853. [Google Scholar] [CrossRef]
- Hu, L.; Luo, Y.; Yang, J.; Cheng, C. Botanical Flavonoids: Efficacy, Absorption, Metabolism and Advanced Pharmaceutical Technology for Improving Bioavailability. Molecules 2025, 30, 1184. [Google Scholar] [CrossRef]
- Sajid, M.; Channakesavula, C.N.; Stone, S.R.; Kaur, P. Synthetic Biology towards Improved Flavonoid Pharmacokinetics. Biomolecules 2021, 11, 754. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lin, C.; Ma, X.; Tan, Y.; Wang, J.; Zeng, M. Functional Characterization of a Flavonoid Glycosyltransferase in Sweet Orange (Citrus sinensis). Front. Plant Sci. 2018, 9, 166. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J. Dietary Flavonoid Aglycones and Their Glycosides: Which Show Better Biological Significance? Crit. Rev. Food Sci. Nutr. 2017, 57, 1874–1905. [Google Scholar] [CrossRef]
- Maaiden, E.E.; Ullah, N.; Ezzariai, A.; Mazar, A.; Boukcim, H.; Hirich, A.; Nasser, B.; Qarah, N.; Kouisni, L.; Kharrassi, Y.E. Comparing Antioxidant and Cytoprotective Effects: Quercetin Glycoside vs. Aglycone from Ephedra alata. Phytomed. Plus 2024, 4, 100603. [Google Scholar] [CrossRef]
- Burlacu, E.; Nisca, A.; Tanase, C. A Comprehensive Review of Phytochemistry and Biological Activities of Quercus Species. Forests 2020, 11, 904. [Google Scholar] [CrossRef]
- Morales, D. Oak Trees (Quercus Spp.) as a Source of Extracts with Biological Activities: A Narrative Review. Trends Food Sci. Technol. 2021, 109, 116–125. [Google Scholar] [CrossRef]
- Di Marco, G.; D’Agostino, A.; Braglia, R.; Redi, E.L.; Iacobelli, S.; Gismondi, A.; Canini, A. Pollen Variability in Quercus L. Species and Relative Systematic Implications. Plant Physiol. Biochem. 2023, 204, 108079. [Google Scholar] [CrossRef]
- Lee, S.H.; Park, Y.D. Carbon Stock and Analysis on Factors Influencing in Quercus mongolica Stand at Mt. Gariwang, South Korea. Korean J. Agric. Sci. 2024, 51, 413–427. [Google Scholar] [CrossRef]
- Marzouk, M.M.; Hegazi, N.M.; El Shabrawy, M.O.A.; Farid, M.M.; Kawashty, S.A.; Hussein, S.R.; Saleh, N.A.M. Discriminative Metabolomics Analysis and Cytotoxic Evaluation of Flowers, Leaves, and Roots Extracts of Matthiola longipetala Subsp. Livida. Metabolites 2023, 13, 909. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Zhang, M.; Otsuki, K.; Li, W. Molecular Networking as a Natural Products Discovery Strategy. Acta Mater. Medica 2023, 2, 126–141. [Google Scholar] [CrossRef]
- Araújo, J.S.; Chambó, E.D.; de Carvalho Costa, M.A.P.; da Silva, S.M.P.C.; de Carvalho, C.A.L.; Estevinho, L.M. Chemical Composition and Biological Activities of Mono- and Heterofloral Bee Pollen of Different Geographical Origins. Int. J. Mol. Sci. 2017, 18, 921. [Google Scholar] [CrossRef] [PubMed]
- Prđun, S.; Svečnjak, L.; Valentić, M.; Marijanović, Z.; Jerković, I. Characterization of Bee Pollen: Physico-Chemical Properties, Headspace Composition and FTIR Spectral Profiles. Foods 2021, 10, 2103. [Google Scholar] [CrossRef]
- Fabre, N.; Rustan, I.; Hoffmann, E.; Quetin-Leclercq, J. Determination of Flavone, Flavonol, and Flavanone Aglycones by Negative Ion Liquid Chromatography Electrospray Ion Trap Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2001, 12, 707–715. [Google Scholar] [CrossRef]
- Cuyckens, F.; Claeys, M. Mass Spectrometry in the Structural Analysis of Flavonoids. J. Mass Spectrom. 2004, 39, 1–15. [Google Scholar] [CrossRef]
- Kachlicki, P.; Piasecka, A.; Stobiecki, M.; Marczak, Ł. Structural Characterization of Flavonoid Glycoconjugates and Their Derivatives with Mass Spectrometric Techniques. Molecules 2016, 21, 1494. [Google Scholar] [CrossRef]
- Li, Z.-H.; Guo, H.; Xu, W.-B.; Ge, J.; Li, X.; Alimu, M.; He, D.-J. Rapid Identification of Flavonoid Constituents Directly from PTP1B Inhibitive Extract of Raspberry (Rubus idaeus L.) Leaves by HPLC-ESI-QTOF-MS-MS. J. Chromatogr. Sci. 2016, 54, 805–810. [Google Scholar] [CrossRef]
- Cuyckens, F.; Claeys, M. Determination of the Glycosylation Site in Flavonoid Mono-O-Glycosides by Collision-Induced Dissociation of Electrospray-Generated Deprotonated and Sodiated Molecules. J. Mass Spectrom. 2005, 40, 364–372. [Google Scholar] [CrossRef]
- Abdallah, R.H.; Hassan, W.H.B.; Al-Massarani, S.M.; Abdel-Mageed, W.M.; Eldahmy, S.I.; Basudan, O.A.; Parveen, M.; El Senosy, E.; Abdelaziz, S. UPLC-ESI-MS/MS Profiling of Secondary Metabolites from Methanol Extracts of in Vivo and in Vitro Tissues of Daucus capillifolius Gilli (A Comparative Study). Molecules 2024, 29, 2694. [Google Scholar] [CrossRef]
- Avula, B.; Bae, J.-Y.; Wang, Y.-H.; Wang, M.; Osman, A.G.; Smith, K.; Yuk, J.; Ali, Z.; Plumb, R.; Isaac, G.; et al. Chemical Profiling and Characterization of Phenolic Acids, Flavonoids, Terpene Glycosides from Vangueria agrestis Using Ultra-High-Performance Liquid Chromatography/Ion Mobility Quadrupole Time-of-Flight Mass Spectrometry and Metabolomics Approach. Biomed. Chromatogr. 2020, 34, e4840. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Yang, Y.; Duan, Y.; Li, C.; Gao, H.; Liu, H.; Cui, Q.; Guo, Z.; Liu, X.; Wang, Z. Quality Marker Discovery and Quality Evaluation of Eucommia ulmoides Pollen Using UPLC-QTOF-MS Combined with a DPPH-HPLC Antioxidant Activity Screening Method. Molecules 2023, 28, 5288. [Google Scholar] [CrossRef] [PubMed]
- Hefny Gad, M.; Tuenter, E.; El-Sawi, N.; Younes, S.; El-Ghadban, E.; Demeyer, K.; Pieters, L.; Vander Heyden, Y.; Mangelings, D. Identification of Some Bioactive Metabolites in a Fractionated Methanol Extract from Ipomoea aquatica (Aerial Parts) through TLC, HPLC, UPLC-ESI-QTOF-MS and LC-SPE-NMR Fingerprints Analyses. Phytochem. Anal. 2018, 29, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yu, H.; Wu, H.; Pan, Y.; Wang, K.; Jin, Y.; Zhang, C. Characterization and Quantification by LC–MS/MS of the Chemical Components of the Heating Products of the Flavonoids Extract in Pollen Typhae for Transformation Rule Exploration. Molecules 2015, 20, 18352–18366. [Google Scholar] [CrossRef]
- Liu, M.; Dong, J.; Lin, Z.; Niu, Y.; Zhang, X.; Jiang, H.; Guo, N.; Li, W.; Wang, H.; Chen, S. Rapid Screening of Transferrin-Binders in the Flowers of Bauhinia Blakeana Dunn by on-Line High-Performance Liquid Chromatography–Diode-Array Detector–Electrospray Ionization–Ion-Trap–Time-of-Flight–Mass Spectrometry–Transferrin–Fluorescence Detection System. J. Chromatogr. A 2016, 1450, 17–28. [Google Scholar] [CrossRef]
- Parejo, I.; Jáuregui, O.; Viladomat, F.; Bastida, J.; Codina, C. Characterization of Acylated Flavonoid-O-Glycosides and Methoxylated Flavonoids from Tagetes Maxima by Liquid Chromatography Coupled to Electrospray Ionization Tandem Mass Spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 2801–2810. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joo, Y.; Shin, E.; Kim, H.; Lee, M.K.; Kim, S.B. Molecular Networking-Guided Annotation of Flavonoid Glycosides from Quercus mongolica Bee Pollen. Int. J. Mol. Sci. 2025, 26, 7930. https://doi.org/10.3390/ijms26167930
Joo Y, Shin E, Kim H, Lee MK, Kim SB. Molecular Networking-Guided Annotation of Flavonoid Glycosides from Quercus mongolica Bee Pollen. International Journal of Molecular Sciences. 2025; 26(16):7930. https://doi.org/10.3390/ijms26167930
Chicago/Turabian StyleJoo, Yerim, Eunbeen Shin, Hyunwoo Kim, Mi Kyeong Lee, and Seon Beom Kim. 2025. "Molecular Networking-Guided Annotation of Flavonoid Glycosides from Quercus mongolica Bee Pollen" International Journal of Molecular Sciences 26, no. 16: 7930. https://doi.org/10.3390/ijms26167930
APA StyleJoo, Y., Shin, E., Kim, H., Lee, M. K., & Kim, S. B. (2025). Molecular Networking-Guided Annotation of Flavonoid Glycosides from Quercus mongolica Bee Pollen. International Journal of Molecular Sciences, 26(16), 7930. https://doi.org/10.3390/ijms26167930







