Genotype Structure Alterations in 5q SMA Patients as a Result of the Newborn Screening Program Implementation in the Russian Federation
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
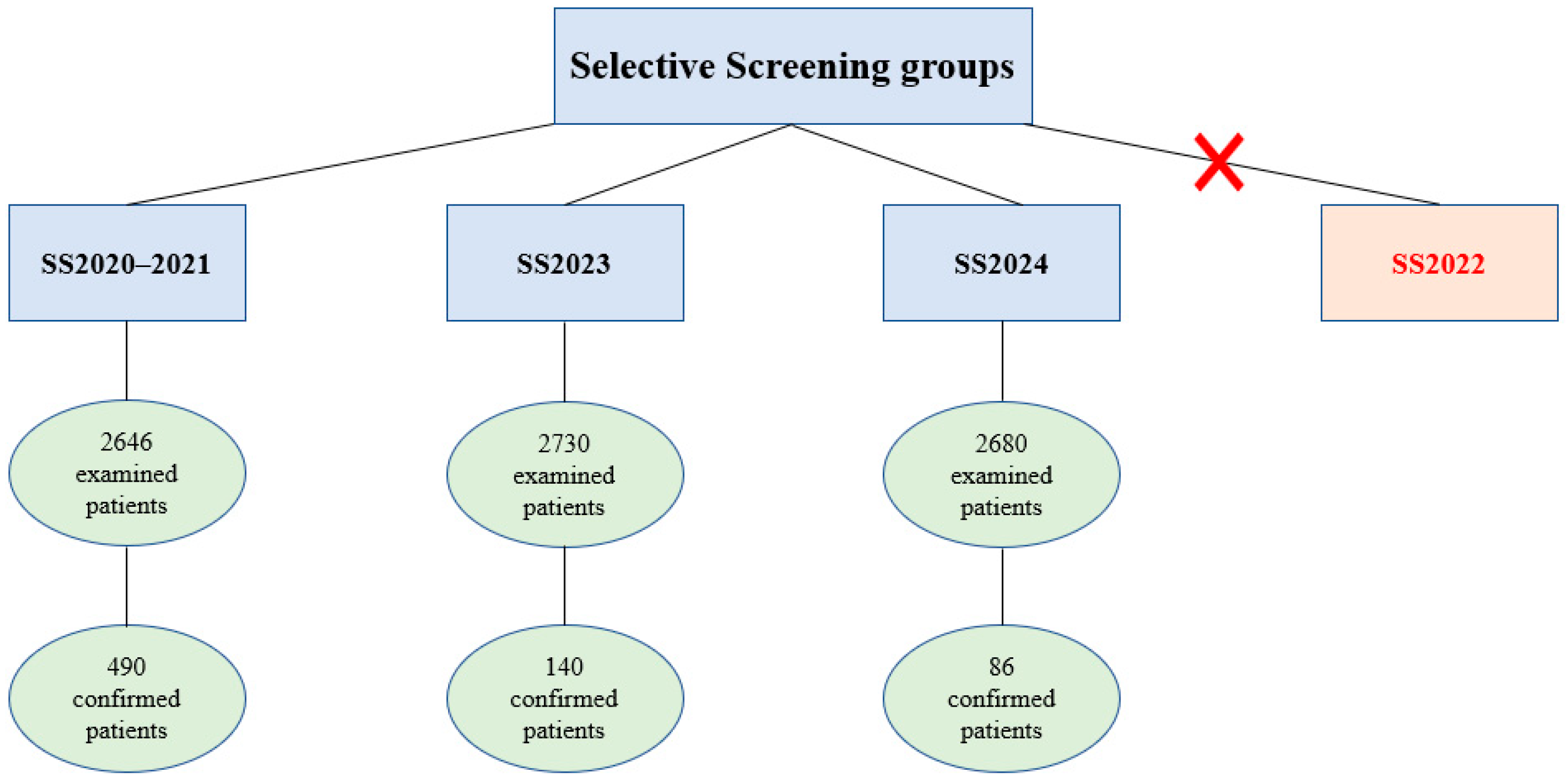
4.1. Selective 5q SMA Screening Program Design
- Group 1 (Selective Screening Prior to Nationwide Screening Projects, SS2020–2021): A total of 2646 patients who underwent selective screening for 5q SMA before the launch of pilot screening and newborn screening projects, from 6 March 2020, to 31 December 2021.
- Group 2 (Selective Screening During NBS in 2023, SS2023): A total of 2730 patients referred to the RCMG during the implementation of the NBS program from 1 January 2023, to 31 December 2023.
- Group 3 (Selective Screening During NBS in 2024, SS2024): A total of 2680 patients referred to the RCMG during the implementation of the NBS program from 1 January 2024, to 31 December 2024.
4.2. Nationwide-Expanded Newborn Screening Program Design
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 5q SMA | Proximal spinal muscular atrophy 5q |
| SMN1 | Survival motor neuron 1 |
| SMN2 | Survival motor neuron 2 |
| RCMG | Research Centre for Medical Genetics |
| NBS | Newborn screening |
| RF | Russian Federation |
| SS2020–2021 | Selective screening in 2020–2021 |
| NBS2023–2024 | Newborn screening during 2023–2024 |
| SS2023 | Selective screening in 2023 |
| SS2024 | Selective screening in 2024 |
References
- Tisdale, S.; Pellizzoni, L. Disease mechanisms and therapeutic approaches in spinal muscular atrophy. J. Neurosci. 2015, 35, 8691–8700. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ogino, S.; Leonard, D.G.; Rennert, H.; Ewens, W.J.; Wilson, R.B. Genetic risk assessment in carrier testing for spinal muscular atrophy. Am. J. Med. Genet. 2002, 110, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Prior, T.W.; Snyder, P.J.; Rink, B.D.; Pearl, D.K.; Pyatt, R.E.; Mihal, D.C.; Conlan, T.; Schmalz, B.; Montgomery, L.; Ziegler, K.; et al. Newborn and carrier screening for spinal muscular atrophy. Am. J. Med. Genet. Part A 2010, 152, 1608–1616. [Google Scholar] [CrossRef] [PubMed]
- Mikhalchuk, K.; Polyakov, A.; Zabnenkova, V.V.; Ismagilova, O.; Shchagina, O. Exploring the Prevalence of SMN1 Duplication and Deletion in Russia and Its Impact on Carrier Screening. Int. J. Mol. Sci. 2025, 26, 1984. [Google Scholar] [CrossRef] [PubMed]
- Zabnenkova, V.V.; Dadali, E.L.; Spiridonova, M.G.; Zinchenko, R.A.; Polyakov, A.V. Spinal muscular atrophy carrier frequency in Russian Federation. In Proceedings of the ASHG, Vancouver, BC, Canada, 18–22 October 2016; p. 2476. [Google Scholar] [CrossRef]
- Sugarman, E.A.; Nagan, N.; Zhu, H.; Akmaev, V.R.; Zhou, Z.; Rohlfs, E.M.; Flynn, K.; Hendrickson, B.C.; Scholl, T.; Sirko-Osadsa, D.A.; et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: Clinical laboratory analysis of >72,400 specimens. Eur. J. Hum. Genet. 2012, 20, 27–32. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mikhalchuk, K.; Zabnenkova, V.; Braslavskaya, S.; Chukhrova, A.; Ryadninskaya, N.; Dadaly, E.; Rudenskaya, G.; Sharkova, I.; Anisimova, I.; Bessonova, L.; et al. Rare Cause 5q SMA: Molecular Genetic and Clinical Analyses of Intragenic Subtle Variants in the SMN Locus. Clin. Genet. 2025, 108, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Mikhalchuk, K.; Shchagina, O.; Chukhrova, A.; Zabnenkova, V.; Chausova, P.; Ryadninskaya, N.; Vlodavets, D.; Kutsev, S.I.; Polyakov, A. Pilot Program of Newborn Screening for 5q Spinal Muscular Atrophy in the Russian Federation. Int. J. Neonatal. Screen. 2023, 9, 29. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kiselev, A.; Maretina, M.; Shtykalova, S.; Al-Hilal, H.; Maslyanyuk, N.; Plokhih, M.; Serebryakova, E.; Frolova, M.; Shved, N.; Krylova, N.; et al. Establishment of a Pilot Newborn Screening Program for Spinal Muscular Atrophy in Saint Petersburg. Int. J. Neonatal. Screen. 2024, 10, 9. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Efimova, I.Y.; Zinchenko, R.A.; Marakhonov, A.V.; Balinova, N.V.; Mikhalchuk, K.A.; Shchagina, O.A.; Polyakov, A.V.; Mudaeva, D.A.; Saydaeva, D.H.; Matulevich, S.A.; et al. Epidemiology of Spinal Muscular Atrophy Based on the Results of a Large-Scale Pilot Project on 202,908 Newborns. Pediatr. Neurol. 2024, 156, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Dangouloff, T.; Vrščaj, E.; Servais, L.; Osredkar, D.; Adoukonou, T.; Aryani, O.; Barisic, N.; Bashiri, F.; Bastaki, L.; Benitto, A.; et al. SMA NBS World Study Group. Newborn screening programs for spinal muscular atrophy worldwide: Where we stand and where to go. Neuromuscul. Disord. 2021, 31, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Oskoui, M.; Servais, L. Spinal Muscular Atrophy. Continuum (Minneap Minn). Lifelong Learn. Neurol. 2023, 29, 1564–1584. [Google Scholar] [CrossRef] [PubMed]
- Rudnik-Schöneborn, S.; Zerres, K. Emery and Rimoin’s Principles and Practice of Medical Genetics, 6th ed.; 2013. [Google Scholar]
- McDonough, A.; Urquia, L.; Boulis, N. Chapter 5-An Introduction to the Natural History, Genetic Mapping, and Clinical Spectrum of Spinal Muscular Atrophy. In Anthony Donsante, Molecular and Cellular Therapies for Motor Neuron Diseases; Boulis, N., O’Connor, D., Eds.; Academic Press: Amsterdam, The Netherlands, 2017; pp. 101–120. ISBN 9780128022573. [Google Scholar] [CrossRef]
- Oskoui, M.; Darras, B.T.; DeVivo, D.C. Chapter 1-Spinal Muscular Atrophy: 125 Years Later and on the Verge of a Cure. In Spinal Muscular Atrophy; Sumner, C.J., Paushkin, S., Ko, C.P., Eds.; Academic Press: Amsterdam, The Netherlands, 2017; pp. 3–19. ISBN 780128036853. [Google Scholar] [CrossRef]
- Seliverstov, Y.A.; Klyushnikov, S.A.; Illarioshkin, S.N. Spinal muscular atrophy: The concept, differential diagnosis, treatment prospects. Nerv. Dis. 2015, 3, 9–17. [Google Scholar]
- Order of the Ministry of Health of the Russian Federation No. 274n dated April 21, 2022. In On Approval of the Procedure for Providing Medical Care to Patients with Congenital and (or) Hereditary Diseases. Moscow, Russia, 2022; 21 April.
- Voronin, S.V.; Zakharova, E.Y.; Baydakova, G.V.; Marakhonov, A.V.; Shchagina, O.A.; Ryzhkova, O.P.; Shilova, N.V.; Rumyantsev, A.G.; Shcherbina, A.Y.; Mukhina, A.A.; et al. Advanced neonatal screening for hereditary diseases in Russia: First results and future prospects. Pediatr. Na GN Speransky 2024, 103, 16–29. [Google Scholar] [CrossRef]
- Dil, A.V.; Nazarov, V.D.; Sidorenko, D.V.; Lapin, S.V.; Emanuel, V.L. Characteristics of genetic changes in the SMN1 gene in spinal muscular atrophy 5q. Neuromuscul. Dis. 2022, 12, 36–44. (In Russian) [Google Scholar] [CrossRef]
- Niba, E.T.E.; Nishio, H.; Wijaya, Y.O.S.; San Lai, P.; Tozawa, T.; Chiyonobu, T.; Yamadera, M.; Okamoto, K.; Awano, H.; Takeshima, Y.; et al. Clinical phenotypes of spinal muscular atrophy patients with hybrid SMN gene. Brain Dev. 2021, 43, 294–302. [Google Scholar] [CrossRef] [PubMed]
| Samples | SS2020–2021 | NBS2023–2024 | SS2023 | SS2024 | ||||
|---|---|---|---|---|---|---|---|---|
| number | % | number | % | number | % | number | % | |
| Samples analyzed | 2646 | 100 | 505 | 100 | 2730 | 100 | 2680 | 100 |
| Deletions detected | 490 | 18.5 | 288 | 57 | 140 | 5.1 | 86 | 3.2 |
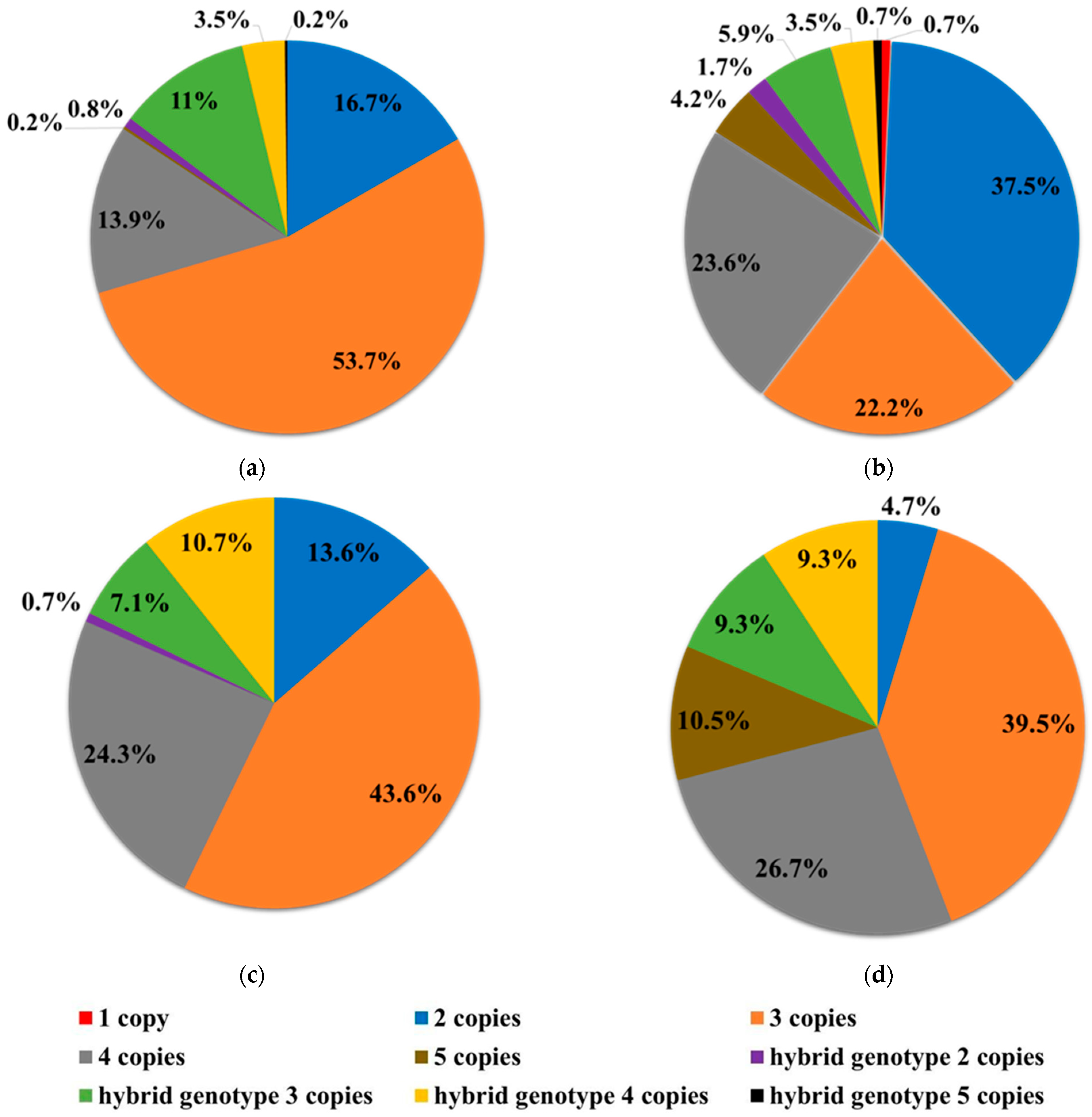
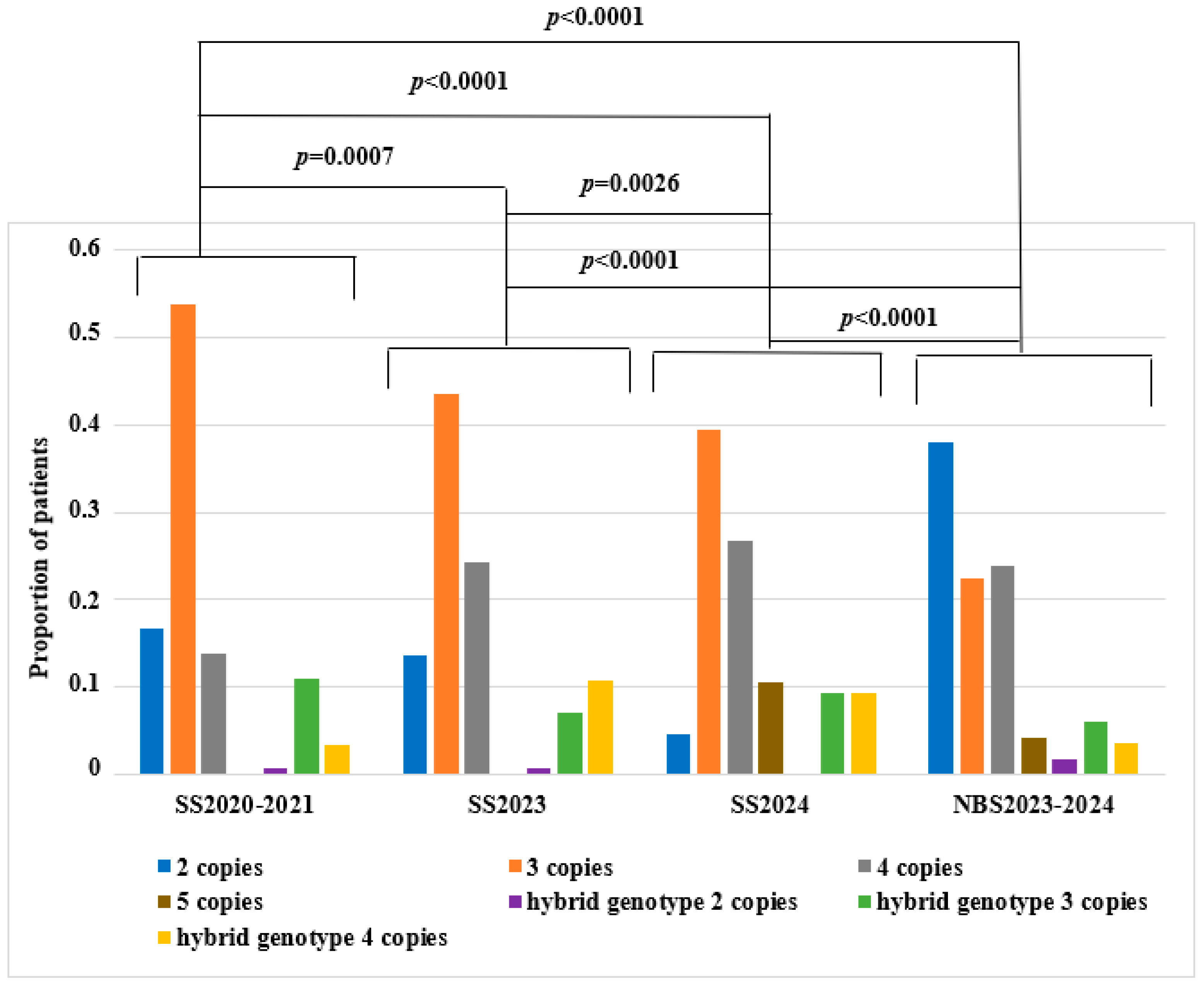
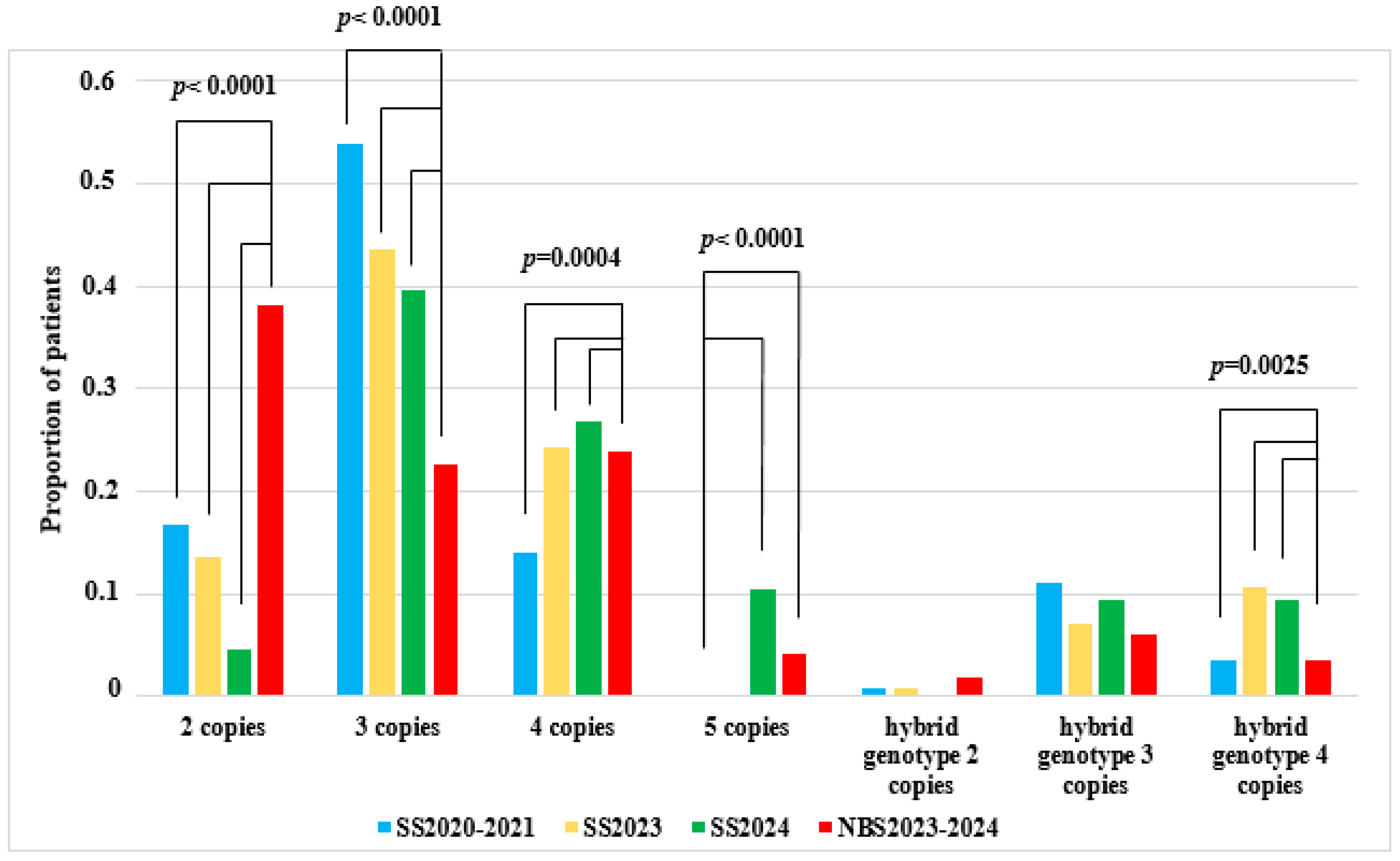
| Genotype | SS2020–2021 | NBS2023–2024 | SS2023 | SS2024 | ||||
|---|---|---|---|---|---|---|---|---|
| number | % | number | % | number | % | number | % | |
| 1 SMN2 copy | 0 | 0 | 2 | 0.7 | 0 | 0 | 0 | 0 |
| 2 SMN2 copies | 82 | 16.7 | 108 | 37.5 | 19 | 13.6 | 4 | 4.7 |
| 3 SMN2 copies | 263 | 53.7 | 64 | 22.2 | 61 | 43.6 | 34 | 39.5 |
| 4 SMN2 copies | 68 | 13.9 | 68 | 23.6 | 34 | 24.3 | 23 | 26.7 |
| 5 SMN2 copies | 1 | 0.2 | 12 | 4.2 | 0 | 0 | 9 | 10.5 |
| hybrid gene 2 SMN2 copies | 4 | 0.8 | 5 | 1.7 | 1 | 0.7 | 0 | 0 |
| hybrid gene 3 SMN2 copies | 54 | 11 | 17 | 5.9 | 10 | 7.1 | 8 | 9.3 |
| hybrid gene 4 SMN2 copies | 17 | 3.5 | 10 | 3.5 | 15 | 10.7 | 8 | 9.3 |
| hybrid gene 5 SMN2 copies | 1 | 0.2 | 2 | 0.7 | 0 | 0 | 0 | 0 |
| Total | 490 | 100 | 288 | 100 | 140 | 100 | 86 | 100 |
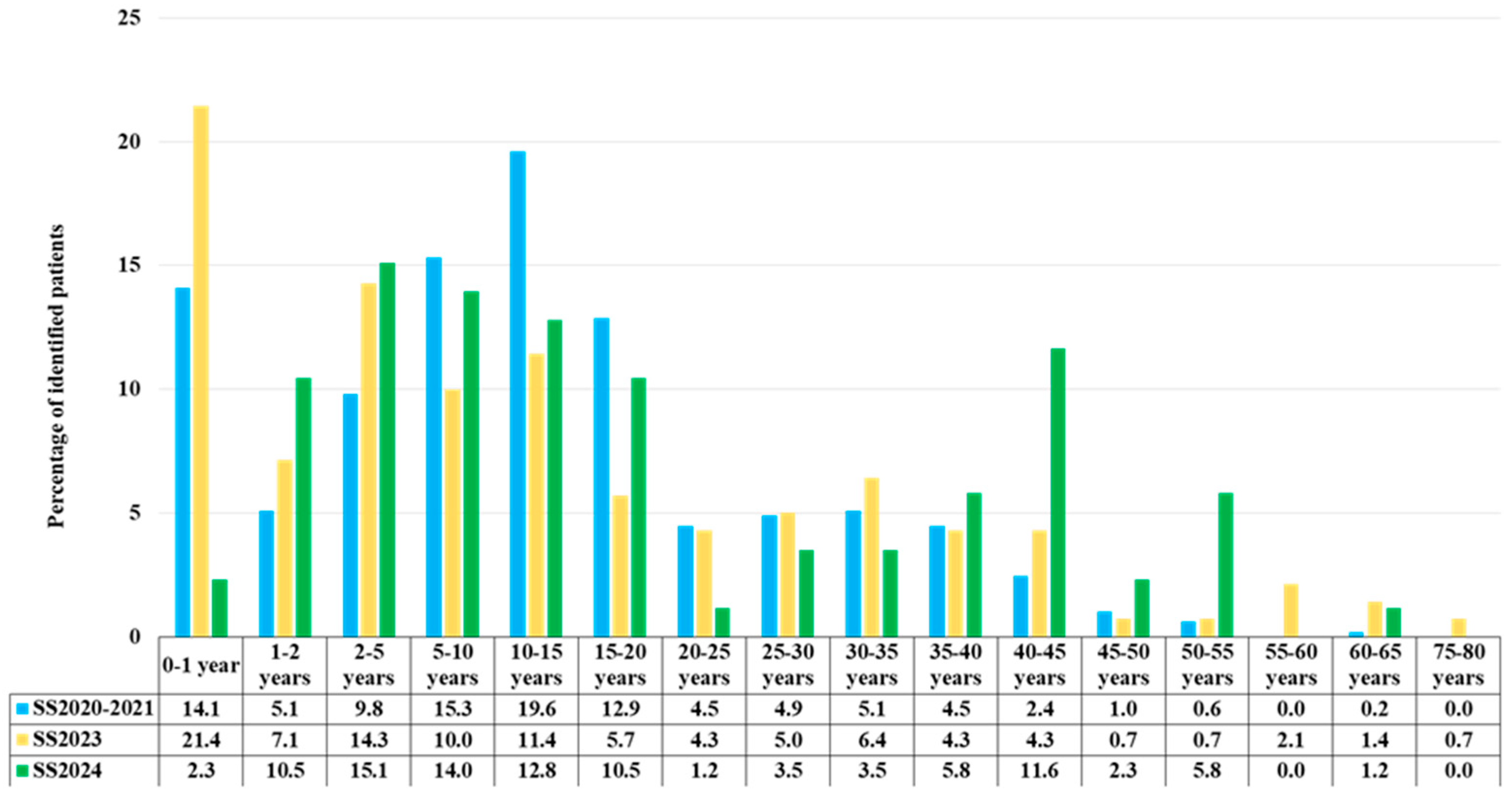
| Age, Years | SS2020–2021 | SS2023 | SS2024 | |||
|---|---|---|---|---|---|---|
| number | % | number | % | number | % | |
| 0–1 | 69 | 14.1 | 30 | 21.4 | 2 | 2.3 |
| 1–2 | 25 | 5.1 | 10 | 7.1 | 9 | 10.5 |
| 2–5 | 48 | 9.8 | 20 | 14.3 | 13 | 15.1 |
| 5–10 | 75 | 15.3 | 14 | 10.0 | 12 | 14.0 |
| 10–15 | 96 | 19.6 | 16 | 11.4 | 11 | 12.8 |
| 15–20 | 63 | 12.9 | 8 | 5.7 | 9 | 10.5 |
| 20–25 | 22 | 4.5 | 6 | 4.3 | 1 | 1.2 |
| 25–30 | 24 | 4.9 | 7 | 5.0 | 3 | 3.5 |
| 30–35 | 25 | 5.1 | 9 | 6.4 | 3 | 3.5 |
| 35–40 | 22 | 4.5 | 6 | 4.3 | 5 | 5.8 |
| 40–45 | 12 | 2.4 | 6 | 4.3 | 10 | 11.6 |
| 45–50 | 5 | 1.0 | 1 | 0.7 | 2 | 2.3 |
| 50–55 | 3 | 0.6 | 1 | 0.7 | 5 | 5.8 |
| 55–60 | 0 | 0 | 3 | 2.1 | 0 | 0 |
| 60–65 | 1 | 0.2 | 2 | 1.4 | 1 | 1.2 |
| 75–80 | 0 | 0 | 1 | 0.7 | 0 | 0 |
| Total | 490 | 100 | 140 | 100 | 86 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akhkiamova, M.A.; Marakhonov, A.V.; Zabnenkova, V.V.; Mikhalchuk, K.A.; Voronin, S.V.; Kutsev, S.I.; Musatova, V.V.; Kekeeva, T.N.; Ryadninskaya, N.V.; Chukhrova, A.L.; et al. Genotype Structure Alterations in 5q SMA Patients as a Result of the Newborn Screening Program Implementation in the Russian Federation. Int. J. Mol. Sci. 2025, 26, 7891. https://doi.org/10.3390/ijms26167891
Akhkiamova MA, Marakhonov AV, Zabnenkova VV, Mikhalchuk KA, Voronin SV, Kutsev SI, Musatova VV, Kekeeva TN, Ryadninskaya NV, Chukhrova AL, et al. Genotype Structure Alterations in 5q SMA Patients as a Result of the Newborn Screening Program Implementation in the Russian Federation. International Journal of Molecular Sciences. 2025; 26(16):7891. https://doi.org/10.3390/ijms26167891
Chicago/Turabian StyleAkhkiamova, Maria A., Andrey V. Marakhonov, Victoria V. Zabnenkova, Kristina A. Mikhalchuk, Sergei V. Voronin, Sergey I. Kutsev, Victoria V. Musatova, Tatyana N. Kekeeva, Nina V. Ryadninskaya, Alena L. Chukhrova, and et al. 2025. "Genotype Structure Alterations in 5q SMA Patients as a Result of the Newborn Screening Program Implementation in the Russian Federation" International Journal of Molecular Sciences 26, no. 16: 7891. https://doi.org/10.3390/ijms26167891
APA StyleAkhkiamova, M. A., Marakhonov, A. V., Zabnenkova, V. V., Mikhalchuk, K. A., Voronin, S. V., Kutsev, S. I., Musatova, V. V., Kekeeva, T. N., Ryadninskaya, N. V., Chukhrova, A. L., Braslavskaya, S. I., Chausova, P. A., Beskorovainaya, T. S., Polyakov, A. V., Savostyanov, K. V., Zhanin, I. S., Bilalov, F. S., Koroteev, A. L., Trofimov, D. Y., ... Shchagina, O. A. (2025). Genotype Structure Alterations in 5q SMA Patients as a Result of the Newborn Screening Program Implementation in the Russian Federation. International Journal of Molecular Sciences, 26(16), 7891. https://doi.org/10.3390/ijms26167891









