Multi-Omics Reveals Molecular and Genetic Mechanisms Underlying Egg Albumen Quality Decline in Aging Laying Hens
Abstract
1. Introduction
2. Results
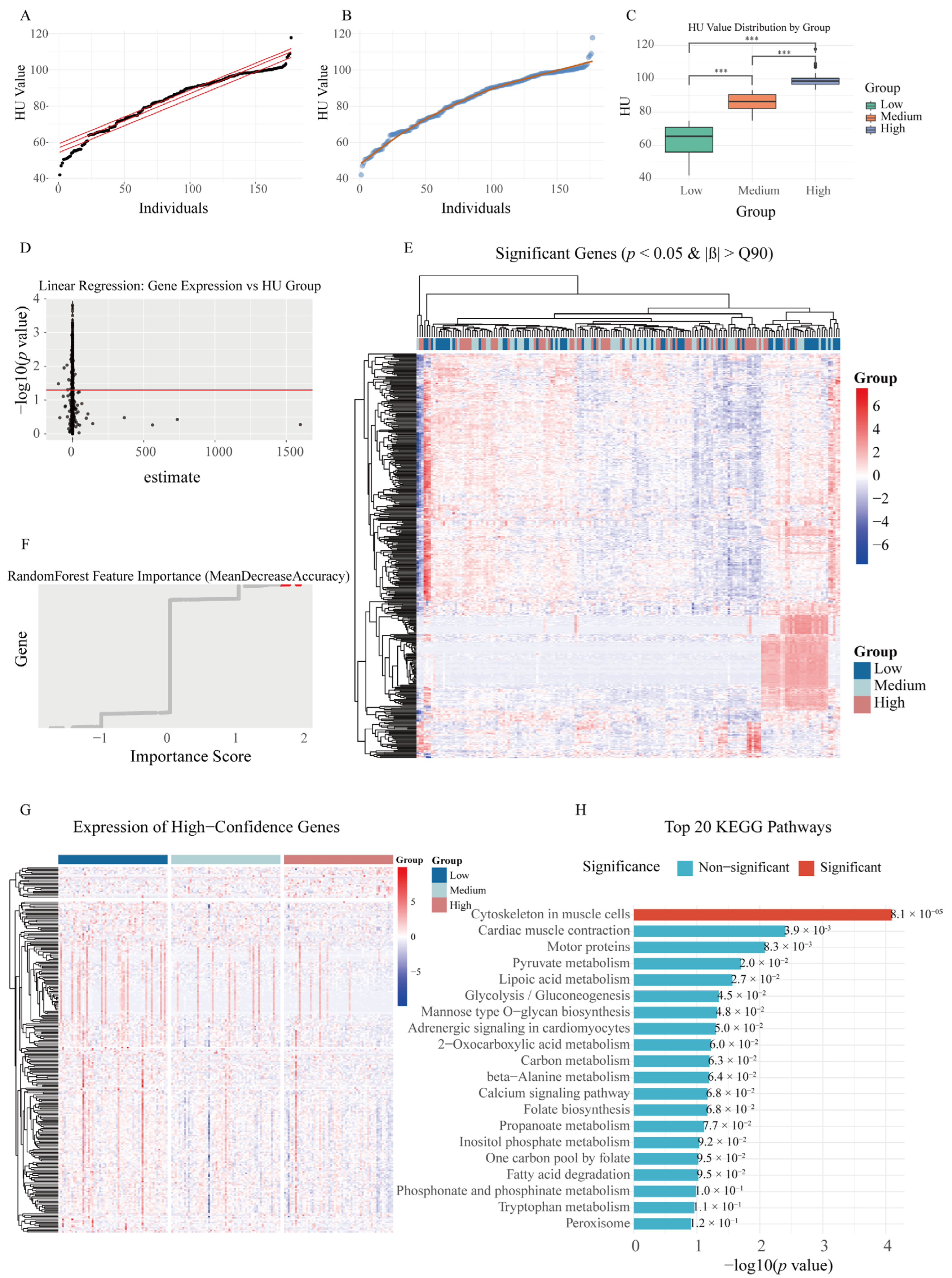
2.1. Integrated GWAS and eQTL Analysis Identifies Functional Loci for HU
2.2. Identification of Key Genes Related to HU in the Magnum Employing Linear Regression and Random Forest Analysis
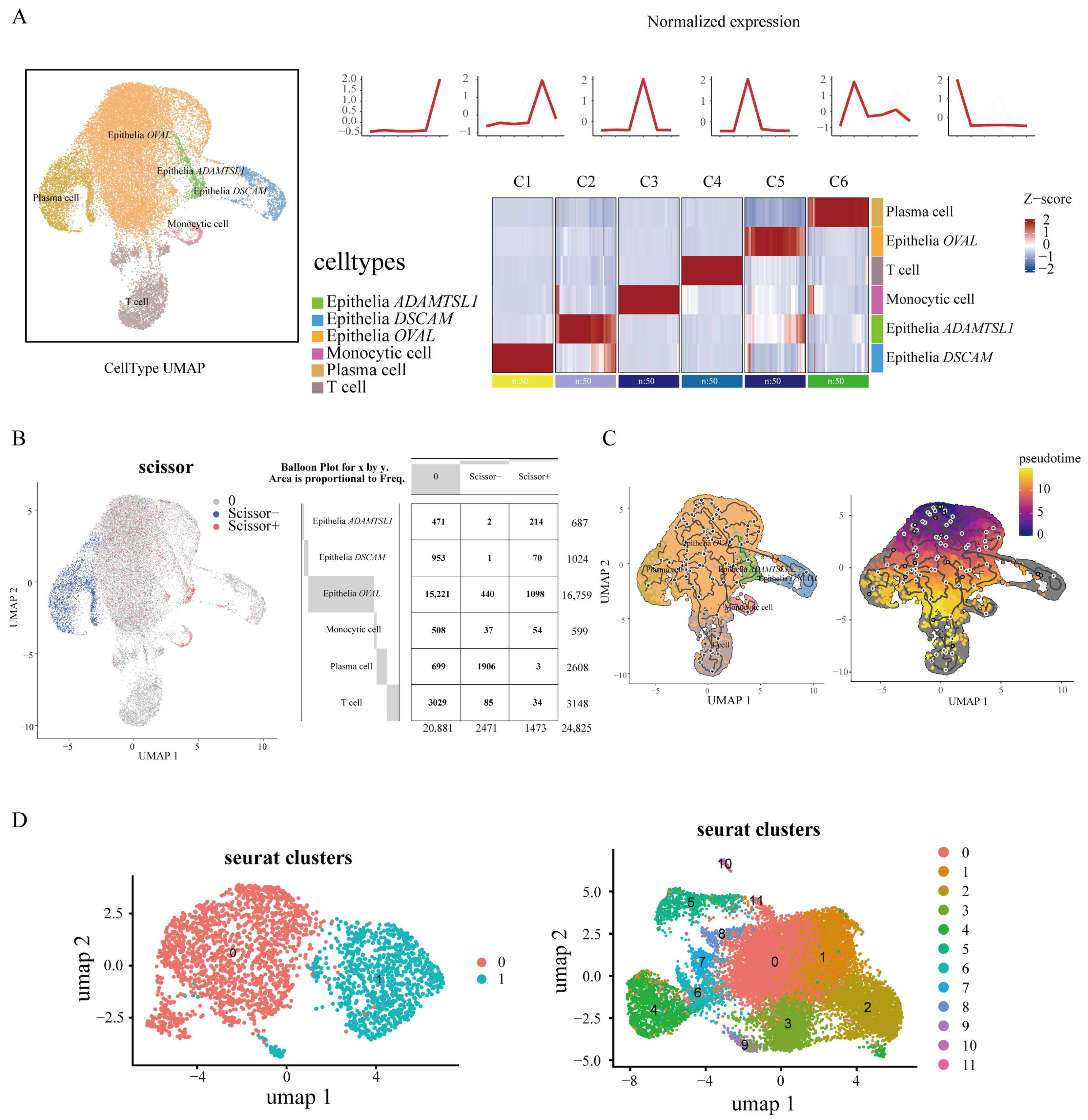
2.3. scRNA-Seq Uncovers Epithelial and Plasma Cell Contributions to HU
2.4. Subclustering of Plasma and Epithelial Cells Reveal Key Genes
2.5. Integrative Multi-Omics Analysis Identifies Key Genetic Regulators of HU
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Experiment Animals and Sample Collection
4.3. RNA Extraction, Quality Evaluation, and Sequencing
4.4. Transcriptome Data Processing
- Reads alignment. Reads from all samples were aligned to the chicken reference genome (Gallus gallus, GCA_016699485.1, https://mart.ensembl.org/Gallus_gallus/Info/Annotation# URL (accessed on 1 November 2024)) using HISAT2 (v 2.2.1). Prior to the alignment, low-quality reads were filtered based on the quality scores associated with each read. HISAT2 allows multiple alignments per read (up to 20 by default) and permits up to two mismatches during alignment. Additionally, HISAT2 constructs a database of potential splice junctions, enabling the mapping of reads that initially failed to align by comparing them against this junction database [51,52].
- Transcript Assembly. Transcript abundance was estimated using StringTie (v 2.1.6) in combination with Ballgown. Gene and mRNA expression levels were quantified based on FPKM (Fragments Per Kilobase of transcript per Million mapped reads), which normalizes transcript counts for both the sequencing depth and transcript length [53,54,55].
4.5. Single-Cell RNA-Seq Sample Preparation and Data Analysis
4.6. GWAS
4.7. Linkage Disequilibrium (LD) Analysis
4.8. Expression Quantitative Trait Loci (eQTL) Analysis
4.9. Transcriptome-Wide Association Study (TWAS) Analysis
4.10. Summary-Based Mendelian Randomization (SMR) Analysis
4.11. SCISSOR Analysis
4.12. Monocle3-Based Pseudotime Analysis
4.13. Cell Subcluster Analysis
4.14. Enrichment Analysis
4.15. Linear Regression and Random Forest Analysis
4.16. PPI Network Construction and Network Integration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HU | Haugh unit |
| GWAS | Genome-wide association studies |
| eQTL mapping | Expression quantitative trait locus mapping |
| TWAS | Transcriptome-Wide Association Study |
| LD | Linear dichroism |
| SMR analyses | Summary-data-based Mendelian Randomization analyses |
| SNPs | single-nucleotide polymorphisms |
| scRNA-seq | single-cell RNA sequencing |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| DEGs | differentially expressed genes |
| PPI | protein–protein interaction |
| CISD1 | CDGSH Iron–Sulfur Domain-Containing Protein 1 |
| NQO2 | quinone oxidoreductase 2 |
| SLCs | Solute carriers |
| CMTM6 | CKLF-like MARVEL transmembrane domain-containing family member 6 |
References
- Sah, N.; Mishra, B. Regulation of egg formation in the oviduct of laying hen. World’s Poult. Sci. J. 2018, 74, 509–522. [Google Scholar] [CrossRef]
- Mann, K. The chicken egg white proteome. Proteomics 2007, 7, 3558–3568. [Google Scholar] [CrossRef]
- Stevens, L. Egg proteins: What are their functions? Sci. Prog. 1996, 79 Pt 1, 65–87. [Google Scholar]
- Edwards, N.A.; Luttrell, V. The protein content of the magnum of the domestic fowl in relation to the secretion and synthesis of albumin. Biochem. Soc. Trans. 1976, 4, 1103–1105. [Google Scholar] [CrossRef]
- Muramatsu, T.; Hiramoto, K.; Okumura, J. Changes in ovalbumin and protein synthesis in vivo in the magnum of laying hens during the egg formation cycle. Comp. Biochem. Physiol. B 1991, 99, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Cüneydioğlu, E.; Erdem, E.; Yalçın, S. Effects of the breeder age on the egg yield and egg quality traits of Landes geese (Anser anser). Trop. Anim. Health Prod. 2022, 54, 387. [Google Scholar] [CrossRef] [PubMed]
- Bain, M.M.; Nys, Y.; Dunn, I.C. Increasing persistency in lay and stabilising egg quality in longer laying cycles. What are the challenges? Br. Poult. Sci. 2016, 57, 330–338. [Google Scholar] [CrossRef]
- Al-batshan, H.A.; Scheideler, S.E.; Black, B.L.; Garlich, J.D.; Anderson, K.E. Duodenal Calcium Uptake, Femur Ash, and Eggshell Quality Decline with Age and Increase Following Molt1. Poult. Sci. 1994, 73, 1590–1596. [Google Scholar] [CrossRef]
- Joyner, C.J.; Peddie, M.J.; Taylor, T.G. The effect of age on egg production in the domestic hen. Gen. Comp. Endocrinol. 1987, 65, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Molnár, A.; Maertens, L.; Ampe, B.; Buyse, J.; Kempen, I.; Zoons, J.; Delezie, E. Changes in egg quality traits during the last phase of production: Is there potential for an extended laying cycle? Br. Poult. Sci. 2016, 57, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Travel, A.; Nys, Y.; Bain, M. 13—Effect of hen age, moult, laying environment and egg storage on egg quality. In Improving the Safety and Quality of Eggs and Egg Products; Nys, Y., Bain, M., Van Immerseel, F., Eds.; Woodhead Publishing Series in Food Science, Technology and Nutrition; Woodhead Publishing: Cambridge, UK, 2011; pp. 300–329. [Google Scholar]
- Jing, M.; Munyaka, P.M.; Tactacan, G.B.; Rodriguez-Lecompte, J.C.; O, K.; House, J.D. Performance, serum biochemical responses, and gene expression of intestinal folate transporters of young and older laying hens in response to dietary folic acid supplementation and challenge with Escherichia coli lipopolysaccharide. Poult. Sci. 2014, 93, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Wang, B.; Zhang, H.; Qiu, K.; Wu, S. The change of albumen quality during the laying cycle and its potential physiological and molecular basis of laying hens. Poult. Sci. 2024, 103, 104004. [Google Scholar] [CrossRef]
- Wu, H.; Yuan, J.; Yin, H.; Jing, B.; Sun, C.; Nguepi Tsopmejio, I.S.; Jin, Z.; Song, H. Flammulina velutipes stem regulates oxidative damage and synthesis of yolk precursors in aging laying hens by regulating the liver–blood–ovary axis. Poult. Sci. 2023, 102, 102261. [Google Scholar] [CrossRef]
- González-Morán, M.G. Changes in progesterone receptor isoforms expression and in the morphology of the oviduct magnum of mature laying and aged nonlaying hens. Biochem. Biophys. Res. Commun. 2016, 478, 999–1005. [Google Scholar] [CrossRef]
- Kim, J.; Choi, Y.H. Differential abundance of egg white proteins in laying hens treated with corticosterone. J. Agric. Food Chem. 2014, 62, 12346–12359. [Google Scholar] [CrossRef]
- Schröder, H.C.; Messer, R.; Breter, H.J.; Müller, W.E. Evidence for age-dependent impairment of ovalbumin heterogeneous nuclear RNA (HnRNA) processing in hen oviduct. Mech. Ageing Dev. 1985, 30, 319–324. [Google Scholar] [CrossRef]
- Rattanawut, J.; Pimpa, O.; Yamauchi, K.E. Effects of dietary bamboo vinegar supplementation on performance, eggshell quality, ileal microflora composition, and intestinal villus morphology of laying hens in the late phase of production. Anim. Sci. J. 2018, 89, 1572–1580. [Google Scholar] [CrossRef]
- Nasri, H.; van den Brand, H.; Najjar, T.; Bouzouaia, M. Egg storage and breeder age impact on egg quality and embryo development. J. Anim. Physiol. Anim. Nutr. 2020, 104, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Sacchetti, C.; Bai, Y.; Stanford, S.M.; Di Benedetto, P.; Cipriani, P.; Santelli, E.; Piera-Velazquez, S.; Chernitskiy, V.; Kiosses, W.B.; Ceponis, A.; et al. PTP4A1 promotes TGFβ signaling and fibrosis in systemic sclerosis. Nat. Commun. 2017, 8, 1060. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Hussein, S.; Yang, J.; Huang, J.; Zhang, F.; Hernandez-Anzaldo, S.; Fernandez-Patron, C.; Cao, Y.; Zeng, H.; Tang, J.; et al. A novel PKD2L1 C-terminal domain critical for trimerization and channel function. Sci. Rep. 2015, 5, 9460. [Google Scholar] [CrossRef]
- Rochat, B. Role of cytochrome P450 activity in the fate of anticancer agents and in drug resistance: Focus on tamoxifen, paclitaxel and imatinib metabolism. Clin. Pharmacokinet. 2005, 44, 349–366. [Google Scholar] [CrossRef]
- Ota, T.; Kamada, Y.; Hayashida, M.; Iwao-Koizumi, K.; Murata, S.; Kinoshita, K. Combination analysis in genetic polymorphisms of drug-metabolizing enzymes CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A5 in the Japanese population. Int. J. Med. Sci. 2015, 12, 78–82. [Google Scholar] [CrossRef]
- Sauer-Eriksson, A.E.; Hainzl, T. S-domain assembly of the signal recognition particle. Curr. Opin. Struct. Biol. 2003, 13, 64–70. [Google Scholar] [CrossRef]
- Lee, H.C.; Fu, C.Y.; Lin, C.Y.; Hu, J.R.; Huang, T.Y.; Lo, K.Y.; Tsai, H.-Y.; Sheu, J.-C.; Tsai, H.-J. Poly(U)-specific endoribonuclease ENDOU promotes translation of human CHOP mRNA by releasing uORF element-mediated inhibition. EMBO J. 2021, 40, e104123. [Google Scholar] [CrossRef]
- Bermann, M.; Legarra, A.; Hollifield, M.K.; Masuda, Y.; Lourenco, D.; Misztal, I. Validation of single-step GBLUP genomic predictions from threshold models using the linear regression method: An application in chicken mortality. J. Anim. Breed Genet. 2021, 138, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Küçüktopçu, E.; Cemek, B.; Yıldırım, D. Estimating Ross 308 Broiler Chicken Weight Through Integration of Random Forest Model and Metaheuristic Algorithms. Animals 2024, 14, 3082. [Google Scholar] [CrossRef] [PubMed]
- Polewko-Klim, A.; Lesiński, W.; Golińska, A.K.; Mnich, K.; Siwek, M.; Rudnicki, W.R. Sensitivity analysis based on the random forest machine learning algorithm identifies candidate genes for regulation of innate and adaptive immune response of chicken. Poult. Sci. 2020, 99, 6341–6354. [Google Scholar] [CrossRef]
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, A.P.N.; Du, J.; Hatfield, J.S.; Levi, E.; Adsay, V.; Schmelz, E.M.; Nagothu, K.K.; Jaszewski, R.; Kucuk, O.; Sarkar, F.H. Expression of EGF-Receptor Related Protein (ERRP) Decreases in Gastric Mucosa During Aging and Carcinogenesis. Dig. Dis. Sci. 2003, 48, 856–864. [Google Scholar] [CrossRef]
- Kasper, G.; Mao, L.; Geissler, S.; Draycheva, A.; Trippens, J.; Kühnisch, J.; Tschirschmann, M.; Kaspar, K.; Perka, C.; Duda, G.N.; et al. Insights into Mesenchymal Stem Cell Aging: Involvement of Antioxidant Defense and Actin Cytoskeleton. Stem Cells 2009, 27, 1288–1297. [Google Scholar] [CrossRef]
- Deindl, E.; Boengler, K.; van Royen, N.; Schaper, W. Differential expression of GAPDH and β-actin in growing collateral arteries. Mol. Cell. Biochem. 2002, 236, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Akbari, M.; Kirkwood, T.B.L.; Bohr, V.A. Mitochondria in the signaling pathways that control longevity and health span. Ageing Res. Rev. 2019, 54, 100940. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef]
- Yang, L.; Li, S.; Mo, C.; Zhou, B.; Fan, S.; Shi, F.; Wei, X.; Zhao, Q.; Yang, G.; Li, S.; et al. Transcriptome analysis and identification of age-associated fertility decreased genes in hen uterovaginal junction. Poult. Sci. 2021, 100, 100892. [Google Scholar] [CrossRef]
- Barua, A.; Yoshimura, Y.; Tamura, T. Effects of ageing and oestrogen on the localization of immunoglobulin-containing cells in the chicken ovary. J. Reprod. Fertil. 1998, 114, 11–16. [Google Scholar] [CrossRef]
- Wasti, S.; Sah, N.; Kuehu, D.L.; Kim, Y.S.; Jha, R.; Mishra, B. Expression of follistatin is associated with egg formation in the oviduct of laying hens. Anim. Sci. J. 2020, 91, e13396. [Google Scholar] [CrossRef]
- Mai, J.; Lu, M.; Gao, Q.; Zeng, J.; Xiao, J. Transcriptome-wide association studies: Recent advances in methods, applications and available databases. Commun. Biol. 2023, 6, 899. [Google Scholar] [CrossRef]
- Chen, S.; Wu, K.; Knox, R. Structure-function studies of DT-diaphorase (NQO1) and NRH: Quinone oxidoreductase (NQO2). Free Radic. Biol. Med. 2000, 29, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, C.; Zhang, W.; Gao, R.; Wang, Q.; Chen, H.; Zhang, S.; Mao, X.; Leblanc, M.; Behensky, A.; et al. Identification of the Potential Key Long Non-coding RNAs in Aged Mice With Postoperative Cognitive Dysfunction. Front. Aging Neurosci. 2019, 11, 181. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xie, H.Q.; Zhao, Y.; Zhang, W.; Zhang, Y. DNA methylation-mediated down-regulation of TMEM130 promotes cell migration in breast cancer. Acta Histochem. 2021, 123, 151814. [Google Scholar] [CrossRef]
- Wiley, S.E.; Murphy, A.N.; Ross, S.A.; van der Geer, P.; Dixon, J.E. MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc. Natl. Acad. Sci. USA 2007, 104, 5318–5323. [Google Scholar] [CrossRef]
- Kwak, W.; Song, K.D.; Oh, J.D.; Heo, K.N.; Lee, J.H.; Lee, W.K.; Yoon, S.H.; Kim, H.; Cho, S.; Lee, H.-K. Uncovering genomic features and maternal origin of korean native chicken by whole genome sequencing. PLoS ONE 2014, 9, e114763. [Google Scholar] [CrossRef]
- Sah, N.; Kuehu, D.L.; Khadka, V.S.; Deng, Y.; Jha, R.; Wasti, S.; Mishra, B. RNA sequencing-based analysis of the magnum tissues revealed the novel genes and biological pathways involved in the egg-white formation in the laying hen. BMC Genom. 2021, 22, 318. [Google Scholar] [CrossRef]
- Bhutia, Y.D.; Babu, E.; Ramachandran, S.; Yang, S.; Thangaraju, M.; Ganapathy, V. SLC transporters as a novel class of tumour suppressors: Identity, function and molecular mechanisms. Biochem. J. 2016, 473, 1113–1124. [Google Scholar] [CrossRef]
- Colas, C.; Ung, P.M.U.; Schlessinger, A. SLC Transporters: Structure, Function, and Drug Discovery. MedChemComm 2016, 7, 1069–1081. [Google Scholar] [CrossRef]
- Schlessinger, A.; Zatorski, N.; Hutchinson, K.; Colas, C. Targeting SLC transporters: Small molecules as modulators and therapeutic opportunities. Trends Biochem. Sci. 2023, 48, 801–814. [Google Scholar] [CrossRef]
- Chang, X.Y.; Uchechukwu Edna, O.; Wang, J.; Zhang, H.J.; Zhou, J.M.; Qiu, K.; Wu, S.-G. Histological and molecular difference in albumen quality between post-adolescent hens and aged hens. Poult. Sci. 2024, 103, 103618. [Google Scholar] [CrossRef]
- Yousefzadeh, M.J.; Zhu, Y.; McGowan, S.J.; Angelini, L.; Fuhrmann-Stroissnigg, H.; Xu, M.; Linga, Y.Y.; Melosa, K.I.; Pirtskhalavab, T.; Inman, C.L.; et al. Fisetin is a senotherapeutic that extends health and lifespan. EBioMedicine 2018, 36, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Thompson, O.; von Meyenn, F.; Hewitt, Z.; Alexander, J.; Wood, A.; Weightman, R.; Gregory, S.; Krueger, F.; Andrews, S.; Barbaric, I.; et al. Low rates of mutation in clinical grade human pluripotent stem cells under different culture conditions. Nat. Commun. 2020, 11, 1528. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Sun, D.; Guan, X.; Moran, A.E.; Wu, L.Y.; Qian, D.Z.; Schedin, P.; Dai, M.-S.; Danilov, A.V.; Alumkal, J.J.; Adey, A.C.; et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat. Biotechnol. 2022, 40, 527–538. [Google Scholar] [CrossRef]
- Binns, D.; Dimmer, E.; Huntley, R.; Barrell, D.; O’Donovan, C.; Apweiler, R. QuickGO: A web-based tool for Gene Ontology searching. Bioinformatics 2009, 25, 3045–3046. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsuet, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [PubMed]

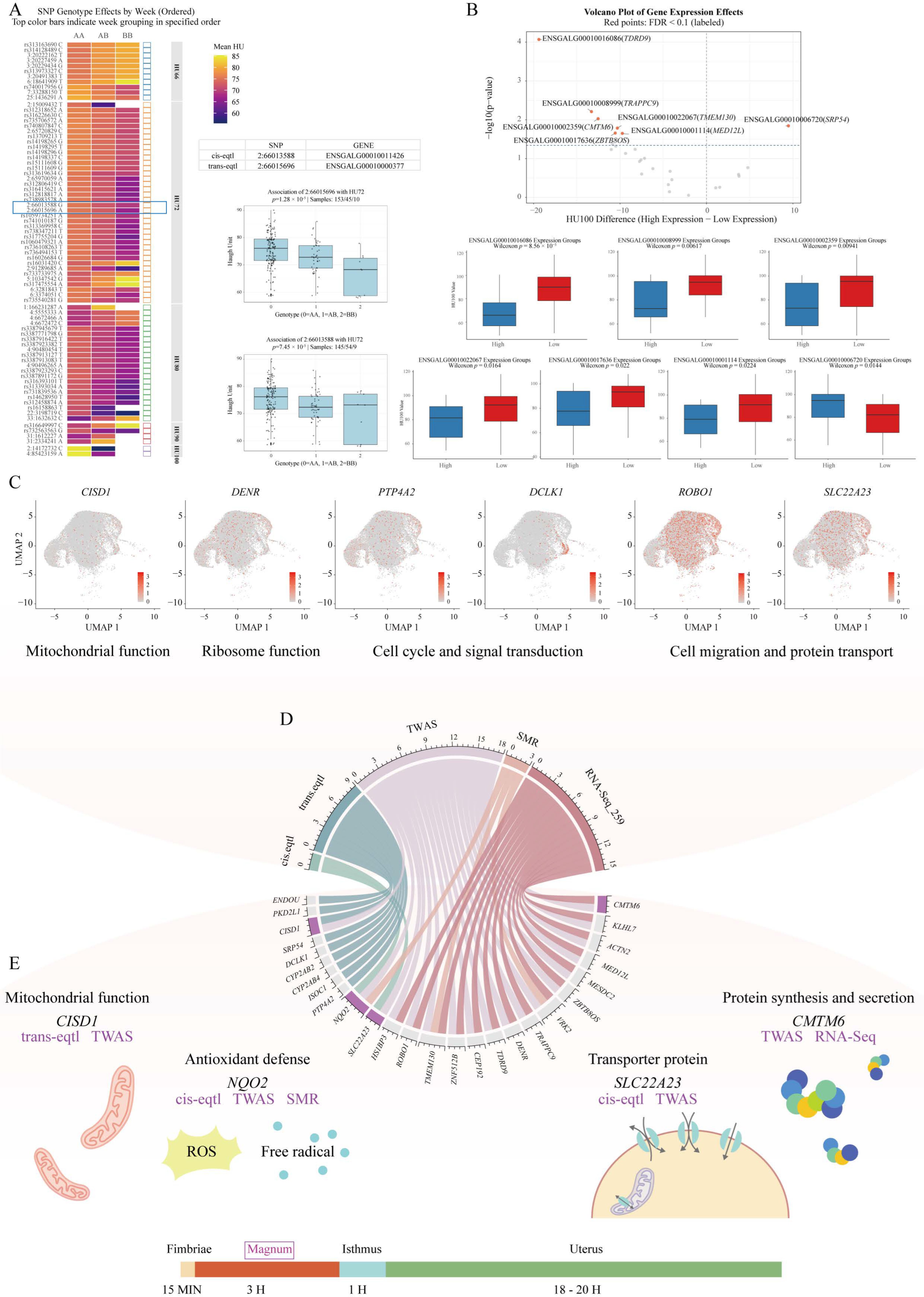
| Trait | Gene | Gene Symbol | Zscore | Effect Size | p Value |
|---|---|---|---|---|---|
| HU66 | ENSGALG00010009119 | CISD1 | −3.364072985 | −7.744721041 | 0.000768012 |
| ENSGALG00010002359 | CMTM6 | 2.334779743 | 3.195354674 | 0.01955493 | |
| ENSGALG00010014250 | - | 2.246253145 | 5.884450865 | 0.024687798 | |
| ENSGALG00010001397 | KLHL7 | −2.081680939 | −2.798993813 | 0.037371623 | |
| ENSGALG00010009118 | ACTN2 | 2.027599228 | 5.276012722 | 0.042601168 | |
| ENSGALG00010001114 | MED12L | −2.005956946 | −2.705734531 | 0.044860842 | |
| ENSGALG00010019300 | MESDC2 | −1.990618634 | −4.600845455 | 0.04652283 | |
| ENSGALG00010017636 | ZBTB8OS | −1.968763637 | −2.268956978 | 0.04898024 | |
| HU72 | ENSGALG00010009119 | CISD1 | −3.64988896 | −8.863673597 | 0.000262354 |
| ENSGALG00010011426 | NQO2 | −3.332953216 | −3.121989042 | 0.000859294 | |
| ENSGALG00010017065 | VRK2 | 2.480160034 | 2.156703244 | 0.013132343 | |
| ENSGALG00010008999 | TRAPPC9 | −2.468575317 | −6.050582223 | 0.01356521 | |
| ENSGALG00010011348 | SLC22A23 | 2.189596504 | 4.01736676 | 0.028553513 | |
| HU80 | ENSGALG00010008999 | TRAPPC9 | −2.679306244 | −7.693929247 | 0.007377489 |
| ENSGALG00010009118 | ACTN2 | 2.457223144 | 7.795617395 | 0.014001569 | |
| ENSGALG00010025026 | DENR | −2.124010969 | −3.587129831 | 0.033669226 | |
| ENSGALG00010016086 | TDRD9 | 1.990443239 | 4.307971134 | 0.046542131 | |
| ENSGALG00010001205 | CEP192 | −1.963226047 | −2.906827479 | 0.049619914 | |
| HU90 | ENSGALG00010001205 | CEP192 | −2.316489697 | −4.126937371 | 0.020531546 |
| ENSGALG00010022578 | ZNF512B | 2.024957407 | 4.407960469 | 0.042871738 | |
| HU100 | ENSGALG00010016086 | TDRD9 | −2.393592171 | −12.11901395 | 0.016684289 |
| ENSGALG00010022067 | TMEM130 | −2.277763246 | −6.529320887 | 0.022740687 | |
| ENSGALG00010007360 | ROBO1 | −2.19131752 | −6.630930423 | 0.028428823 | |
| ENSGALG00010007956 | HS1BP3 | −1.969423368 | −13.89364606 | 0.048904495 | |
| ENSGALG00010017065 | VRK2 | −1.9624531 | −4.634019521 | 0.049709756 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, M.; Zhang, J.; Yang, N.; Sun, C. Multi-Omics Reveals Molecular and Genetic Mechanisms Underlying Egg Albumen Quality Decline in Aging Laying Hens. Int. J. Mol. Sci. 2025, 26, 7876. https://doi.org/10.3390/ijms26167876
Gao M, Zhang J, Yang N, Sun C. Multi-Omics Reveals Molecular and Genetic Mechanisms Underlying Egg Albumen Quality Decline in Aging Laying Hens. International Journal of Molecular Sciences. 2025; 26(16):7876. https://doi.org/10.3390/ijms26167876
Chicago/Turabian StyleGao, Mingyue, Junnan Zhang, Ning Yang, and Congjiao Sun. 2025. "Multi-Omics Reveals Molecular and Genetic Mechanisms Underlying Egg Albumen Quality Decline in Aging Laying Hens" International Journal of Molecular Sciences 26, no. 16: 7876. https://doi.org/10.3390/ijms26167876
APA StyleGao, M., Zhang, J., Yang, N., & Sun, C. (2025). Multi-Omics Reveals Molecular and Genetic Mechanisms Underlying Egg Albumen Quality Decline in Aging Laying Hens. International Journal of Molecular Sciences, 26(16), 7876. https://doi.org/10.3390/ijms26167876






