Bacteriophages, Antibiotics and Probiotics: Exploring the Microbial Battlefield of Colorectal Cancer
Abstract
1. Introduction
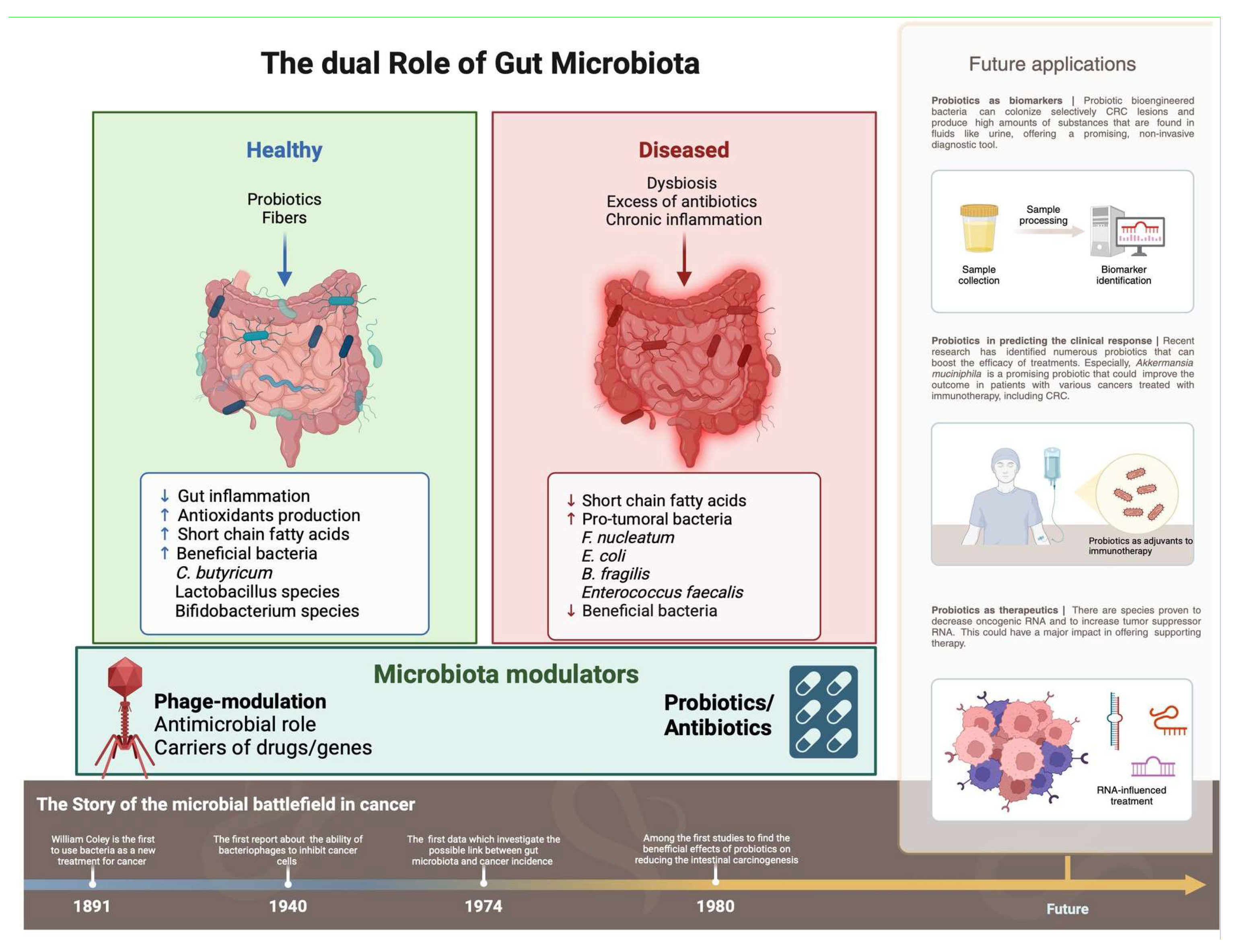
2. Gut Microbiota
2.1. Systemic Effects of the Gut Microbiota
2.2. Immunomodulation by Gut Microbiota
2.3. Role of Short-Chain Fatty Acids (SCFAs)
2.4. Mechanisms of GM in Carcinogenesis
3. Disruption of GM Homeostasis (Dysbiosis) and Gut Barrier Dysfunction
3.1. Dysbiosis and Chronic Inflammation
3.2. Macrophage-Mediated Chromosomal Instability
4. Key Microorganisms Implicated in Colorectal Cancer
4.1. Fusobacterium nucleatum (FN)
4.2. Escherichia coli
4.3. Enterotoxigenic Bacteroides fragilis (ETBF)
4.4. Additional Microbes of Interest
5. Bacteriophages: Emerging Modulators of the Gut Microbiome
5.1. Dynamics Across Lifespan and Impact of Diet
5.2. Synthetic Phage Engineering and CRISPR-Cas Applications
5.3. Phage Therapy and Immunomodulatory Properties
| In Vitro Studies | ||||
|---|---|---|---|---|
| Sample Type | Phage Modulation | Results | Novelty | References |
| BALB/c and C57BL/6 mice treated with CT26-CEA cells and MC38-CEA cells | Both local and systemic administration of M13 phage against CEA, an over-expressed tumor-associated antigen in CRC. | A significant decrease in tumor growth and extended survival in CRC mice models, as compared to the control group. The infiltration of macrophages and neutrophils into the tumor. The maturation of dendritic cells in the lymph nodes draining the tumor site. | The antitumor impact of M13-CEA is mediated through CD8+ T cells. | Murgas et al., [58] |
| NIH3T3 and HT-29 CRC cells lines | EGFR-targeted phage lambda (EGF-λ). | EGF-λ interfered with the initial formation and progression of cancer tissues. Moreover, these phages may be able to disturb the formation of metastasis. | λ phages exhibit the ability to traverse fibroblast and ECM layers and accumulate in CRC tissues without presenting cytotoxic effects. | Huh et al., [62] |
| Primary tumor cells (HCT116, SW480) and metastatic (SW620) CRC cells lines | The hybrid self-assembling nanotubes composed of the tail sheath protein gp053 derived from the phage vB_EcoM_FV3 and a label SNAP-tag protein, obtaining 053SNAP nanotubes. | HCT116 and SW480 efficiently accumulated nanotubes, in contrast with a low uptake in SW620 cells. Internalization into cancer cells escalated over time and did not result in direct cytotoxicity effects. Peritoneal macrophages actively took up 053SNAP, potentially posing a challenge to the utilization of these nanocarriers. | The self-assembling nanotubes generated from phage sheath proteins offer a promising foundation for the future development of innovative nanocarriers. | Gabrielaitis et al., [63] |
| CRC metastatic cells in liver, lung and lymph node | RNA nanoparticles, originating from the three-way junction of the phage phi29 motor pRNA, were employed to specifically target metastatic CRC cells. | The RNA NPs exhibited a tendency to home in on metastatic tumors without accumulating in the neighboring normal organ tissues. | pRNA-based NPs possess both specificity and advantageous pharmacokinetic characteristics. | Rychahou et al., [64] |
| HCT116 Human Colon Cancer Cell line co-cultured with E. faecalis | Phage EFA1 against EF. | Disrupted EF biofilms within just 2 h of treatment. Altered the growth-stimulatory effects of EF. Suppressed cell proliferation. Increased ROS production. | This study investigates the initial exploration of the impact of Enterococcus bacteriophages on colon cancer cells. | Kabwe et al., [65] |
| Orthotopic CRC mice and fecal samples of patients with CRC | Encapsulated irinotecan (a primary drug against CRC) within dextran nanoparticles were covalently linked to isolated phages from human saliva that selectively lyse FN. | Inhibition of pro-tumoral bacteria (FN). Promotion of anti-tumoral bacteria (Clostridium butyricum) that increase colonic SCFA. Elevated production of anti-tumoral butyrate. | Increase in the expression of anti-autophagy genes (Tspo, Poldip2, and Akt11), along with a decrease in the expression of pro-autophagy genes (Lrrk2, Adrb2, Zc3h12a, Dram1, and Dram2). | Zheng et al., [61] |
| Stool samples from FN-colonized CRC and orthotopic CT26 murine models | FN binding M13 phage linked to silver nanoparticles (M13@Ag). | Both in vivo and in vitro studies, M13@Ag demonstrated the ability to effectively remove FN from the gut and decrease the amplification of MDSCs in the tumor site, and to activate APCs, contributing to the stimulation of the host immune system. | The combination of M13@Ag with chemotherapy (FOLFIRI) and immune checkpoint inhibitors (α-PD1) resulted in a delayed tumorigenesis and significantly increased the lifespan of mice. | Dong et al., [66] |
| HTC116 human colon adenocarcinoma and mouse MC38 colon cancer cells; mice with MC38 tumors induced | CEA targeting the expression of E gene (pCEA-E) specifically to colon cancer cells. | Substantial inhibition of cell growth in both human and mice colon cancer cells and a reduction in tumor volume. | The E gene expression presents antitumoral activity: formation of a toxic transmembrane pore, mitochondrial damage, and caspase protein expression. | Rama et al., [67] |
| CRC Mouse models | B. fragilis-targeting phage VA7. | Selective targeting of B. fragilis by phage VA7 restores chemosensitivity in CRC in mice. | A promising adjunct therapy for chemoresistant tumors. | Ding et al., [68] |
6. Antibiotics and Their Complex Relationship with CRC
7. Probiotics: Potential Adjuncts in CRC Management
| Study Design/ Sample Size | Probiotics | Results | Clinical Importance/Novelty | References |
|---|---|---|---|---|
| Randomized, single-blind, placebo-controlled prospective study/100 patients | B. infants, L. acidophilus, E. faecalis, and B. cereus | ↓ severity of chemotherapy-induced gastrointestinal complications; ↑ levels of SCFA; ↑ gut microbiota homeostasis. | Probiotics have the potential to be used as complementary approaches for chemoprevention. | Huang et al., [111] |
| Randomized, double-blind, placebo-controlled trial/52 patients | L. acidophilus, L. lactis, L. casei, B. longum, B. bifidum and B. infantis | ↓ IL-6, IL-10, IL-12, IL-17A, IL-17C, IL-22 and TNF-α. No difference in the levels of IFN-γ. | These strains of Lactobacillus and Bifidobacteria were shown to be safe and to have anti-inflammatory properties when consumed after surgery. | Zaharuddin et al., [98] |
| Randomized, double-blind, placebo-controlled trial/66 patients | L. rhamnosus, L. acidophilus | ↑ QoL with improved mental health status; ↓ cancer-related fatigue. | Probiotics could improve both physical and psychological health. | Lee et al., [112] |
| Randomized controlled trial/110 patients | L. acidophilus | ↓ inflammation by influencing CYLD protein. | The first study to investigate the link between L. acidophilus, CYLD expression and NF-κB/TNF-α signaling. | Zamani et al., [99] |
| Double-blind randomized trial/140 patients | L. acidophilus, L. casei, L. lactis, B. bifidum, B. longum, B. infantis + omega-3 fatty acid | ↑ QoL; ↓ IL-6; ↓ chemotherapy-related side effects. | This combination of probiotics and omega-3 might act as a synergic adjuvant to chemotherapy. | Golkhalkhali et al., [113] |
| Randomized prospective study/78 patients | L. acidophilus, L. casei, L. plantarum, L. rhamnosus, B. lactis, B. bifidum, B. breve, Streptococcus thermophilus | ↓ postoperative complications and hospitalization; ↓ post-surgery mortality within the first six months. | Incorporating probiotics into the postoperative care treatment for CRC patients could lead to improved outcomes. | Bajramagic et al., [114] |
| Randomized, prospective, double-blind, placebo-controlled study/73 patients | L. acidophilus, L. rhamnosus, L. casei, B. lactis + Fructose Oligosaccharide | Used preoperatively. ↓ inflammatory state; ↓ morbidity and hospitalization; ↓ duration of antibiotic usage; ↑ bowel functions. | Modulating the microbiota during the preoperative period in CRC patients could influence the incidence of postoperative complications. | Polakowski et al., [115] |
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Abs | Antibodies |
| Actinobacteria | (Bacterial phylum; generally not abbreviated) |
| A. muciniphila | Akkermansia muciniphila |
| bft | Bacteroides fragilis toxin |
| B. adolescentis | Bifidobacterium adolescentis |
| B. fragilis | Bacteroides fragilis |
| CD8+ T cells | Cluster of Differentiation 8 positive T lymphocytes |
| CRC | Colorectal Cancer |
| CRISPR-Cas | Clustered Regularly Interspaced Short Palindromic Repeats-Cas system |
| CTLA-4 | Cytotoxic T-Lymphocyte Antigen 4 |
| DCs | Dendritic Cells |
| DNA | Deoxyribonucleic Acid |
| Efa | Enterococcus faecalis |
| EMT | Epithelial–Mesenchymal Transition |
| ETBF | Enterotoxigenic Bacteroides fragilis |
| FadA | Fusobacterium adhesin A |
| FAP2 | Fibroblast Activation Protein-2 |
| FN | Fusobacterium nucleatum |
| Foxp3 | Forkhead box P3 (Regulatory T cell transcription factor) |
| GM | Gut Microbiota |
| GM-CSF | Granulocyte-Macrophage Colony-Stimulating Factor |
| GPCR | G Protein-Coupled Receptor |
| HCT116 | Human colorectal carcinoma cell line |
| IBD | Inflammatory Bowel Disease |
| ICIs | Immune Checkpoint Inhibitors |
| IgG | Immunoglobulin G |
| IgA | Immunoglobulin A |
| IL-12 | Interleukin 12 |
| IL-17A | Interleukin 17A |
| ITIM | Immunoreceptor Tyrosine-Based Inhibitory Motif |
| lncRNAs | Long non-coding RNAs |
| L. acidophilus | Lactobacillus acidophilus |
| MAPK | Mitogen-Activated Protein Kinase |
| METTL3 | Methyltransferase-like 3 |
| miRs | MicroRNAs |
| MMP-9 | Matrix Metalloprotease 9 |
| NK cells | Natural Killer Cells |
| NF-κB | Nuclear Factor kappa-light-chain-enhancer of activated B cells |
| ROS | Reactive Oxygen Species |
| SCFAs | Short-Chain Fatty Acids (acetate, propionate, butyrate) |
| SOS response | DNA damage repair response in bacteria |
| TME | Tumor Microenvironment |
| TLR4 | Toll-Like Receptor 4 |
| Tregs | Regulatory T cells (Foxp3+ phenotype) |
References
- Shin, A.E.; Giancotti, F.G.; Rustgi, A.K. Metastatic colorectal cancer: Mechanisms and emerging therapeutics. Trends Pharmacol. Sci. 2023, 44, 222–236. [Google Scholar] [CrossRef]
- Liu, Y.; Lau, H.C.H.; Cheng, W.Y.; Yu, J. Gut Microbiome in Colorectal Cancer: Clinical Diagnosis and Treatment. Genom. Proteom. Bioinform. 2023, 21, 84–96. [Google Scholar] [CrossRef]
- Jerin, S.; Harvey, A.J.; Lewis, A. Therapeutic Potential of Protein Tyrosine Kinase 6 in Colorectal Cancer. Cancers 2023, 15, 3703. [Google Scholar] [CrossRef]
- Ni, J.J.; Li, X.S.; Zhang, H.; Xu, Q.; Wei, X.T.; Feng, G.J.; Zhao, M.; Zhang, Z.J.; Zhang, L.; Shen, G.H.; et al. Mendelian randomization study of causal link from gut microbiota to colorectal cancer. BMC Cancer 2022, 22, 1371. [Google Scholar] [CrossRef]
- Fong, W.; Li, Q.; Yu, J. Gut microbiota modulation: A novel strategy for prevention and treatment of colorectal cancer. Oncogene 2020, 39, 4925–4943. [Google Scholar] [CrossRef]
- Song, M.; Chan, A.T. Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clin. Gastroenterol. Hepatol. 2019, 17, 275–289. [Google Scholar] [CrossRef]
- Coker, O.O.; Liu, C.; Wu, W.K.K.; Wong, S.H.; Jia, W.; Sung, J.J.Y.; Yu, J. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome 2022, 10, 35. [Google Scholar] [CrossRef]
- Torres-Maravilla, E.; Boucard, A.-S.; Mohseni, A.H.; Taghinezhad-S, S.; Cortes-Perez, N.G.; Bermúdez-Humarán, L.G. Role of Gut Microbiota and Probiotics in Colorectal Cancer: Onset and Progression. Microorganisms 2021, 9, 1021. [Google Scholar] [CrossRef] [PubMed]
- Saus, E.; Iraola-Guzmán, S.; Willis, J.R.; Brunet-Vega, A.; Gabaldón, T. Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol. Asp. Med. 2019, 69, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Eslami, M.; Yousefi, B.; Kokhaei, P.; Hemati, M.; Nejad, Z.R.; Arabkari, V.; Namdar, A. Importance of probiotics in the prevention and treatment of colorectal cancer. J. Cell. Physiol. 2019, 234, 17127–17143. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef]
- Illiano, P.; Brambilla, R.; Parolini, C. The mutual interplay of gut microbiota, diet and human disease. FEBS J. 2020, 287, 833–855. [Google Scholar] [CrossRef]
- Pant, A.; Maiti, T.K.; Mahajan, D.; Das, B. Human Gut Microbiota and Drug Metabolism. Microb. Ecol. 2023, 86, 97–111. [Google Scholar] [CrossRef]
- Góralczyk-Bińkowska, A.; Szmajda-Krygier, D.; Kozłowska, E. The Microbiota–Gut–Brain Axis in Psychiatric Disorders. IJMS 2022, 23, 11245. [Google Scholar] [CrossRef]
- Quigley, E.M.M. Microbiota-Brain-Gut Axis and Neurodegenerative Diseases. Curr. Neurol. Neurosci. Rep. 2017, 17, 94. [Google Scholar] [CrossRef]
- Dang, A.T.; Marsland, B.J. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef]
- Huang, Y.; Xin, W.; Xiong, J.; Yao, M.; Zhang, B.; Zhao, J. The Intestinal Microbiota and Metabolites in the Gut-Kidney-Heart Axis of Chronic Kidney Disease. Front. Pharmacol. 2022, 13, 837500. [Google Scholar]
- Zhang, T.; Sun, P.; Geng, Q.; Fan, H.; Gong, Y.; Hu, Y.; Shan, L.; Sun, Y.; Shen, W.; Zhou, Y. Disrupted spermatogenesis in a metabolic syndrome model: The role of vitamin A metabolism in the gut–testis axis. Gut 2022, 71, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Mauro, D.; Nakamura, A.; Haroon, N.; Ciccia, F. The gut-enthesis axis and the pathogenesis of Spondyloarthritis. Semin. Immunol. 2021, 58, 101607. [Google Scholar] [CrossRef] [PubMed]
- Shi, N.; Li, N.; Duan, X.; Niu, H. Interaction between the gut microbiome and mucosal immune system. Mil. Med. Res. 2017, 4, 14. [Google Scholar] [CrossRef]
- Mangiola, F.; Nicoletti, A.; Gasbarrini, A.; Ponziani, F.R. Gut microbiota and aging. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7404–7413. [Google Scholar]
- Xu, J.Y.; Liu, M.T.; Tao, T.; Zhu, X.; Fei, F.Q. The role of gut microbiota in tumorigenesis and treatment. Biomed. Pharmacother. 2021, 138, 111444. [Google Scholar] [CrossRef]
- Yu, A.I.; Zhao, L.; Eaton, K.A.; Ho, S.; Chen, J.; Poe, S.; Becker, J.; Gonzalez, A.; McKinstry, D.; Hasso, M.; et al. Gut Microbiota Modulate CD8 T Cell Responses to Influence Colitis-Associated Tumorigenesis. Cell Rep. 2020, 31, 107471. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Xu, B.; Wang, X.; Wan, W.H.; Lu, J.; Kong, D.; Jin, Y.; You, W.; Sun, H.; Mu, X.; et al. Gut microbiota–derived short-chain fatty acids regulate group 3 innate lymphoid cells in HCC. Hepatology 2023, 77, 48–64. [Google Scholar] [CrossRef]
- De Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Zhang, K.; Wei, J.; Ding, Y.; Wang, X.; Hou, H.; Wu, J.; Liu, T.; Wang, B.; Cao, H. Gut microbiota-derived short-chain fatty acids regulate gastrointestinal tumor immunity: A novel therapeutic strategy? Front. Immunol. 2023, 14, 1158200. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, R.; Afaghi, A.; Babakhani, S.; Sohrabi, M.R.; Hosseini-Fard, S.R.; Babolhavaeji, K.; Khani Ali Akbari, S.; Yousefimashouf, R.; Karampoor, S. Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed. Pharmacother. 2021, 139, 111619. [Google Scholar] [CrossRef]
- Binienda, A.; Owczarek, K.; Sałaga, M.; Fichna, J. Synthetic free fatty acid receptor (FFAR) 2 agonist 4-CMTB and FFAR4 agonist GSK13764 inhibit colon cancer cell growth and migration and regulate FFARs expression in in vitro and in vivo models of colorectal cancer. Pharmacol. Rep. 2024, 76, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Domínguez, A.; Pastor, N.; Martínez-López, L.; Colón-Pérez, J.; Bermúdez, B.; Orta, M.L. The Role of DNA Damage Response in Dysbiosis-Induced Colorectal Cancer. Cells 2021, 10, 1934. [Google Scholar] [CrossRef]
- Artemev, A.; Naik, S.; Pougno, A.; Honnavar, P.; Shanbhag, N.M. The Association of Microbiome Dysbiosis with Colorectal Cancer. Cureus 2022, 14, e22156. Available online: https://www.cureus.com/articles/86171-the-association-of-microbiome-dysbiosis-with-colorectal-cancer (accessed on 10 January 2025). [CrossRef]
- Yu, L.C.H. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: Exploring a common ground hypothesis. J. Biomed. Sci. 2018, 25, 79. [Google Scholar] [CrossRef]
- Wang, X.; Huycke, M.M. Colorectal cancer: Role of commensal bacteria and bystander effects. Gut Microbes 2015, 6, 370–376. [Google Scholar] [CrossRef]
- Clay, S.L.; Fonseca-Pereira, D.; Garrett, W.S. Colorectal cancer: The facts in the case of the microbiota. J. Clin. Investig. 2022, 132, e155101. [Google Scholar] [CrossRef]
- Debelius, J.W.; Engstrand, L.; Matussek, A.; Brusselaers, N.; Morton, J.T.; Stenmarker, M.; Olsen, R.S. The Local Tumor Microbiome Is Associated with Survival in Late-Stage Colorectal Cancer Patients. Xu ZZ, editor. Microbiol. Spectr. 2023, 11, e05066-22. [Google Scholar] [CrossRef]
- Karpiński, T.M.; Ożarowski, M.; Stasiewicz, M. Carcinogenic microbiota and its role in colorectal cancer development. Semin. Cancer Biol. 2022, 86, 420–430. [Google Scholar] [CrossRef]
- Tran, H.N.H.; Thu, T.N.H.; Nguyen, P.H.; Vo, C.N.; Doan, K.V.; Nguyen Ngoc Minh, C.; Nguyen, N.T.; Ta, V.N.D.; Vu, K.A.; Hua, T.D.; et al. Tumour microbiomes and Fusobacterium genomics in Vietnamese colorectal cancer patients. npj Biofilms Microbiomes 2022, 8, 87. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, C.G.; Kim, W.K.; Kim, K.A.; Yoo, J.; Min, B.S.; Paik, S.; Shin, S.J.; Lee, H.; Lee, K.; et al. Fusobacterium nucleatum induces a tumor microenvironment with diminished adaptive immunity against colorectal cancers. Front. Cell. Infect. Microbiol. 2023, 13, 1101291. [Google Scholar] [CrossRef] [PubMed]
- Padma, S.; Patra, R.; Sen Gupta, P.S.; Panda, S.K.; Rana, M.K.; Mukherjee, S. Cell Surface Fibroblast Activation Protein-2 (Fap2) of Fusobacterium nucleatum as a Vaccine Candidate for Therapeutic Intervention of Human Colorectal Cancer: An Immunoinformatics Approach. Vaccines 2023, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Sun, T.; Zhou, J.; Zhi, M.; Shen, S.; Wang, Y.; Gu, X.; Li, Z.; Gao, H.; Wang, P.; et al. Pangenomic Study of Fusobacterium nucleatum Reveals the Distribution of Pathogenic Genes and Functional Clusters at the Subspecies and Strain Levels. Microbiol. Spectr. 2023, 11, e05184-22. [Google Scholar] [CrossRef] [PubMed]
- Baik, J.E.; Li, L.; Shah, M.A.; Freedberg, D.E.; Jin, Z.; Wang, T.C.; Han, Y.W. Circulating IgA Antibodies Against Fusobacterium nucleatum Amyloid Adhesin FadA are a Potential Biomarker for Colorectal Neoplasia. Cancer Res. Commun. 2022, 2, 1497–1503. [Google Scholar] [CrossRef]
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef]
- Fan, X.; Jin, Y.; Chen, G.; Ma, X.; Zhang, L. Gut Microbiota Dysbiosis Drives the Development of Colorectal Cancer. Digestion 2021, 102, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H. Full-length recombinant antibodies from Escherichia coli: Production, characterization, effector function (Fc) engineering, and clinical evaluation. mAbs 2022, 14, 2111748. [Google Scholar] [CrossRef]
- Meng, C.; Bai, C.; Brown, T.D.; Hood, L.E.; Tian, Q. Human Gut Microbiota and Gastrointestinal Cancer. Genom. Proteom. Bioinform. 2018, 16, 33–49. [Google Scholar] [CrossRef]
- Purcell, R.V.; Permain, J.; Keenan, J.I. Enterotoxigenic Bacteroides fragilis activates IL-8 expression through Stat3 in colorectal cancer cells. Gut Pathog. 2022, 14, 16. [Google Scholar] [CrossRef]
- Butt, J.; Jenab, M.; Werner, J.; Fedirko, V.; Weiderpass, E.; Dahm, C.C.; Tjønneland, A.; Olsen, A.; Boutron-Ruault, M.C.; Rothwell, J.A.; et al. Association of Pre-diagnostic Antibody Responses to Escherichia coli and Bacteroides fragilis Toxin Proteins with Colorectal Cancer in a European Cohort. Gut Microbes 2021, 13, 1903825. [Google Scholar] [CrossRef]
- De Almeida, C.V.; Taddei, A.; Amedei, A. The controversial role of Enterococcus faecalis in colorectal cancer. Ther. Adv. Gastroenterol. 2018, 11, 1756284818783606. [Google Scholar] [CrossRef] [PubMed]
- Koliarakis, I.; Messaritakis, I.; Nikolouzakis, T.K.; Hamilos, G.; Souglakos, J.; Tsiaoussis, J. Oral Bacteria and Intestinal Dysbiosis in Colorectal Cancer. IJMS 2019, 20, 4146. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Mendes, B.G.; Alves, B.S.; Duan, Y. Phage therapy in gut microbiome. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2023; pp. 93–118. Available online: https://linkinghub.elsevier.com/retrieve/pii/S1877117323001138 (accessed on 10 January 2025).
- Han, S.; Ding, K. Intestinal phages interact with bacteria and are involved in human diseases. Gut Microbes 2022, 14, 2113717. [Google Scholar] [CrossRef]
- Mushegian, A.R. Are There 1031 Virus Particles on Earth, or More, or Fewer? J. Bacteriol. 2020, 202, e00052-20. [Google Scholar] [CrossRef]
- Cao, Z.; Sugimura, N.; Burgermeister, E.; Ebert, M.P.; Zuo, T.; Lan, P. The gut virome: A new microbiome component in health and disease. eBioMedicine 2022, 81, 104113. [Google Scholar] [CrossRef]
- Tobin, C.A.; Hill, C.; Shkoporov, A.N. Factors Affecting Variation of the Human Gut Phageome. Annu. Rev. Microbiol. 2023, 77, 363–379. [Google Scholar] [CrossRef]
- Liu, S.; Lu, H.; Zhang, S.; Shi, Y.; Chen, Q. Phages against Pathogenic Bacterial Biofilms and Biofilm-Based Infections: A Review. Pharmaceutics 2022, 14, 427. [Google Scholar] [CrossRef]
- El Haddad, L.; Mendoza, J.F.; Jobin, C. Bacteriophage-mediated manipulations of microbiota in gastrointestinal diseases. Front. Microbiol. 2022, 13, 1055427. [Google Scholar] [CrossRef] [PubMed]
- Marongiu, L.; Burkard, M.; Venturelli, S.; Allgayer, H. Dietary Modulation of Bacteriophages as an Additional Player in Inflammation and Cancer. Cancers 2021, 13, 2036. [Google Scholar] [CrossRef]
- Murgas, P.; Bustamante, N.; Araya, N.; Cruz-Gómez, S.; Durán, E.; Gaete, D.; Oyarce, C.; López, E.; Herrada, A.A.; Ferreira, N.; et al. A filamentous bacteriophage targeted to carcinoembryonic antigen induces tumor regression in mouse models of colorectal cancer. Cancer Immunol. Immunother. 2018, 67, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Ru, J.; Mirzaei, M.K.; Xue, J.; Peng, X.; Ralser, A.; Mejías-Luque, R.; Gerhard, M.; Deng, L. Gut virome profiling identifies an association between temperate phages and colorectal cancer promoted by Helicobacter pylori infection. Gut Microbes 2023, 15, 2257291. [Google Scholar] [CrossRef]
- Marongiu, L.; Allgayer, H. Viruses in colorectal cancer. Mol. Oncol. 2022, 16, 1423–1450. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.W.; Dong, X.; Pan, P.; Chen, K.W.; Fan, J.X.; Cheng, S.X.; Zhang, X.Z. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 2019, 3, 717–728. [Google Scholar] [CrossRef]
- Dong, X.; Pan, P.; Zheng, D.W.; Bao, P.; Zeng, X.; Zhang, X.Z. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci. Adv. 2020, 6, eaba1590. [Google Scholar] [CrossRef]
- Huh, H.; Chen, D.W.; Foldvari, M.; Slavcev, R.; Blay, J. EGFR-targeted bacteriophage lambda penetrates model stromal and colorectal carcinoma tissues, is taken up into carcinoma cells, and interferes with 3-dimensional tumor formation. Front. Immunol. 2022, 13, 957233. [Google Scholar] [CrossRef] [PubMed]
- Gabrielaitis, D.; Zitkute, V.; Saveikyte, L.; Labutyte, G.; Skapas, M.; Meskys, R.; Casaite, V.; Sasnauskiene, A.; Neniskyte, U. Nanotubes from bacteriophage tail sheath proteins: Internalisation by cancer cells and macrophages. Nanoscale Adv. 2023, 5, 3705–3716. [Google Scholar] [CrossRef]
- Rychahou, P.; Haque, F.; Shu, Y.; Zaytseva, Y.; Weiss, H.L.; Lee, E.Y.; Mustain, W.; Valentino, J.; Guo, P.; Evers, B.M. Delivery of RNA Nanoparticles into Colorectal Cancer Metastases Following Systemic Administration. ACS Nano 2015, 9, 1108–1116. [Google Scholar] [CrossRef]
- Kabwe, M.; Meehan-Andrews, T.; Ku, H.; Petrovski, S.; Batinovic, S.; Chan, H.T.; Tucci, J. Lytic Bacteriophage EFA1 Modulates HCT116 Colon Cancer Cell Growth and Upregulates ROS Production in an Enterococcus faecalis Co-culture System. Front. Microbiol. 2021, 12, 650849. [Google Scholar] [CrossRef]
- Rama, A.R.; Hernandez, R.; Perazzoli, G.; Burgos, M.; Melguizo, C.; Vélez, C.; Prados, J. Specific Colon Cancer Cell Cytotoxicity Induced by Bacteriophage E Gene Expression under Transcriptional Control of Carcinoembryonic Antigen Promoter. IJMS 2015, 16, 12601–12615. [Google Scholar] [CrossRef]
- Prados, E Phage gene transfection enhances sensitivity of lung and colon cancer cells to chemotherapeutic agents. Int. J. Oncol. 2010, 37, 1503–1514. Available online: http://www.spandidos-publications.com/ijo/37/6/1503 (accessed on 10 January 2025).
- Zhang, W.; Zhang, J.; Liu, T.; Xing, J.; Zhang, H.; Wang, D.; Tang, D. Bidirectional effects of intestinal microbiota and antibiotics: A new strategy for colorectal cancer treatment and prevention. J. Cancer Res. Clin. Oncol. 2022, 148, 2387–2404. [Google Scholar] [CrossRef]
- Qu, G.; Sun, C.; Sharma, M.; Uy, J.P.; Song, E.J.; Bhan, C.; Shu, L. Is antibiotics use really associated with increased risk of colorectal cancer? An updated systematic review and meta-analysis of observational studies. Int. J. Color. Dis. 2020, 35, 1397–1412. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.; Jin, F.; Shi, J.; Qiu, Z.; Chen, L.; Li, Q.; He, C.; Cheng, Z. Antibiotics use and risk of colorectal neoplasia: An updated meta-analysis. Int. J. Color. Dis. 2022, 37, 2291–2301. [Google Scholar] [CrossRef] [PubMed]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Stott, K.J.; Phillips, B.; Parry, L.; May, S. Recent advancements in the exploitation of the gut microbiome in the diagnosis and treatment of colorectal cancer. Biosci. Rep. 2021, 41, BSR20204113. [Google Scholar] [CrossRef]
- Lu, S.S.M.; Mohammed, Z.; Häggström, C.; Myte, R.; Lindquist, E.; Gylfe, Å.; Van Guelpen, B.; Harlid, S. Antibiotics Use and Subsequent Risk of Colorectal Cancer: A Swedish Nationwide Population-Based Study. JNCI J. Natl. Cancer Inst. 2022, 114, 38–46. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Derosa, L.; Routy, B.; Desilets, A.; Daillère, R.; Terrisse, S.; Kroemer, G.; Zitvogel, L. Microbiota-Centered Interventions: The Next Breakthrough in Immuno-Oncology? Cancer Discov. 2021, 11, 2396–2412. [Google Scholar] [CrossRef] [PubMed]
- RoRoberti, M.P.; Yonekura, S.; Duong, C.P.M.; Picard, M.; Ferrere, G.; Tidjani Alou, M.; Rauber, C.; Iebba, V.; Lehmann, C.H.K.; Amon, L.; et al. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nat. Med. 2020, 26, 919–931. [Google Scholar] [CrossRef]
- Bhatt, A.P.; Redinbo, M.R.; Bultman, S.J. The role of the microbiome in cancer development and therapy. CA A Cancer J. Clin. 2017, 67, 326–344. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Deng, Y.; Wang, J.; Yan, Z.; Wei, Q.; Ye, J.; Zhang, J.; He, T.C.; Qiao, M. Effect of antibiotic monensin on cell proliferation and IGF1R signaling pathway in human colorectal cancer cells. Ann. Med. 2023, 55, 954–964. [Google Scholar] [CrossRef]
- Seçme, M.; Kocoglu, S.S. Investigation of the TLR4 and IRF3 signaling pathway-mediated effects of monensin in colorectal cancer cells. Med. Oncol. 2023, 40, 187. [Google Scholar] [CrossRef]
- Liu, X.M.; Zhu, W.T.; Jia, M.L.; Li, Y.T.; Hong, Y.; Liu, Z.Q.; Yan, P.K. Rapamycin Liposomes Combined with 5-Fluorouracil Inhibits Angiogenesis and Tumor Growth of APC (Min/+) Mice and AOM/DSS-Induced Colorectal Cancer Mice. Int. J. Nanomed. 2022, 17, 5049–5061. [Google Scholar] [CrossRef] [PubMed]
- Klose, J.; Trefz, S.; Wagner, T.; Steffen, L.; Preißendörfer Charrier, A.; Radhakrishnan, P.; Volz, C.; Schmidt, T.; Ulrich, A.; Dieter, S.M.; et al. Salinomycin: Anti-tumor activity in a pre-clinical colorectal cancer model. PLoS ONE 2019, 14, e0211916. [Google Scholar] [CrossRef] [PubMed]
- Klose, J.; Kattner, S.; Borgström, B.; Volz, C.; Schmidt, T.; Schneider, M.; Oredsson, S.; Strand, D.; Ulrich, A. Semi-synthetic salinomycin analogs exert cytotoxic activity against human colorectal cancer stem cells. Biochem. Biophys. Res. Commun. 2018, 495, 53–59. [Google Scholar] [CrossRef]
- Liu, F.; Lv, R.B.; Liu, Y.; Hao, Q.; Liu, S.J.; Zheng, Y.Y.; Li, C.; Zhu, C.; Wang, M. Salinomycin and Sulforaphane Exerted Synergistic Antiproliferative and Proapoptotic Effects on Colorectal Cancer Cells by Inhibiting the PI3K/Akt Signaling Pathway in vitro and in vivo. OncoTargets Ther. 2020, 13, 4957–4969. [Google Scholar] [CrossRef]
- Liu, F.; Li, W.; Hua, S.; Han, Y.; Xu, Z.; Wan, D.; Wang, Y.; Chen, W.; Kuang, Y.; Shi, J.; et al. Nigericin Exerts Anticancer Effects on Human Colorectal Cancer Cells by Inhibiting Wnt/β-catenin Signaling Pathway. Mol. Cancer Ther. 2018, 17, 952–965. [Google Scholar] [CrossRef]
- Garcia-Princival, I.M.R.; Princival, J.L.; Dias da Silva, E.; de Arruda Lima, S.M.; Carregosa, J.C.; Wisniewski, A., Jr.; de Lucena, C.C.O.; Halwass, F.; Alves Franca, J.A.; Ferreira, L.F.G.R.; et al. Streptomyces hygroscopicus UFPEDA 3370: A valuable source of the potent cytotoxic agent nigericin and its evaluation against human colorectal cancer cells. Chem. -Biol. Interact. 2021, 333, 109316. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Malagón, A.J.; Hidalgo-García, L.; Rodríguez-Sojo, M.J.; Molina-Tijeras, J.A.; García, F.; Diez-Echave, P.; Vezza, T.; Becerra, P.; Marchal, J.A.; Redondo-Cerezo, E.; et al. Tigecycline reduces tumorigenesis in colorectal cancer via inhibition of cell proliferation and modulation of immune response. Biomed. Pharmacother. 2023, 163, 114760. [Google Scholar] [CrossRef]
- Yang, L.; Yang, J.; Liu, H.; Lan, J.; Xu, Y.; Wu, X.; Mao, Y.; Wu, D.; Pan, K.; Zhang, T. Minocycline binds and inhibits LYN activity to prevent STAT3-meditated metastasis of colorectal cancer. Int. J. Biol. Sci. 2022, 18, 2540–2552. [Google Scholar] [CrossRef]
- Kabir, S.R.; Islam, T. Antimycin A induced apoptosis in HCT-116 colorectal cancer cells through the up- and downregulation of multiple signaling pathways. Med. Oncol. 2022, 40, 51. [Google Scholar] [CrossRef]
- Petroni, G.; Bagni, G.; Iorio, J.; Duranti, C.; Lottini, T.; Stefanini, M.; Kragol, G.; Becchetti, A.; Arcangeli, A. Clarithromycin inhibits autophagy in colorectal cancer by regulating the hERG1 potassium channel interaction with PI3K. Cell Death Dis. 2020, 11, 161. [Google Scholar] [CrossRef] [PubMed]
- Kvakova, M.; Kamlarova, A.; Stofilova, J.; Benetinova, V.; Bertkova, I. Probiotics and postbiotics in colorectal cancer: Prevention and complementary therapy. World J. Gastroenterol. 2022, 28, 3370–3382. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, P.; Maiolini, M.; Alnafoosi, O.; Hussein, S.; Alnafoosi, H.; Umbela, S.; Richardson, T.; Alla, N.; Lamichhane, N.; Subhadra, B.; et al. Colorectal Cancer and Probiotics: Are Bugs Really Drugs? Cancers 2020, 12, 1162. [Google Scholar] [CrossRef]
- Tang, G.; Zhang, L. Update on Strategies of Probiotics for the Prevention and Treatment of Colorectal Cancer. Nutr. Cancer 2022, 74, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liao, Y.; Wei, C.; Ma, Y.; Wang, F.; Chen, Y.; Zhao, B.; Ji, H.; Wang, D.; Tang, D. Potential Ability of Probiotics in the Prevention and Treatment of Colorectal Cancer. Clin. Med. Insights Oncol. 2023, 17, 11795549231188225. [Google Scholar] [CrossRef]
- Zhao, T.; Wang, H.; Liu, Z.; Liu, Y.; DeJi; Li, B.; Huang, X. Recent Perspective of Lactobacillus in Reducing Oxidative Stress to Prevent Disease. Antioxidants 2023, 12, 769. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.; Jiang, X.; Wang, H.; Chen, S.; Wang, X.; Liu, Y.; Guo, S.; Li, D.; Yu, W.; Zhao, Z.; et al. The inhibitory effects of probiotics on colon cancer cells: In vitro and in vivo studies. J. Gastrointest. Oncol. 2020, 11, 1224–1232. [Google Scholar] [CrossRef]
- Tripathy, A.; Dash, J.; Kancharla, S.; Kolli, P.; Mahajan, D.; Senapati, S.; Jena, M.K. Probiotics: A Promising Candidate for Management of Colorectal Cancer. Cancers 2021, 13, 3178. [Google Scholar] [CrossRef]
- Zaharuddin, L.; Mokhtar, N.M.; Muhammad Nawawi, K.N.; Raja Ali, R.A. A randomized double-blind placebo-controlled trial of probiotics in post-surgical colorectal cancer. BMC Gastroenterol. 2019, 19, 131. [Google Scholar] [CrossRef]
- Zamani, F.; Khalighfard, S.; Kalhori, M.R.; Poorkhani, A.; Amiriani, T.; Hosseinzadeh, P.; Esmati, E.; Alemrajabi, M.; Nikoofar, A.; Safarnezhad Tameshkel, F.; et al. Expanding CYLD protein in NF-κβ/TNF-α signaling pathway in response to Lactobacillus acidophilus in non-metastatic rectal cancer patients. Med. Oncol. 2023, 40, 302. [Google Scholar] [CrossRef]
- Khodaii, Z.; Mehrabani Natanzi, M.; Khalighfard, S.; Ghandian Zanjan, M.; Gharghi, M.; Khori, V.; Amiriani, T.; Rahimkhani, M.; Alizadeh, A.M. Novel targets in rectal cancer by considering lncRNA–miRNA–mRNA network in response to Lactobacillus acidophilus consumption: A randomized clinical trial. Sci. Rep. 2022, 12, 9168. [Google Scholar] [CrossRef]
- Ha, S.; Zhang, X.; Yu, J. Probiotics intervention in colorectal cancer: From traditional approaches to novel strategies. Chin. Med. J. 2024, 137, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Stoeva, M.K.; Garcia-So, J.; Justice, N.; Myers, J.; Tyagi, S.; Nemchek, M.; McMurdie, P.J.; Kolterman, O.; Eid, J. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 2021, 13, 1907272. [Google Scholar] [CrossRef]
- Zhang, K.; Dong, Y.; Li, M.; Zhang, W.; Ding, Y.; Wang, X.; Chen, D.; Liu, T.; Wang, B.; Cao, H.; et al. Clostridium butyricum inhibits epithelial–mesenchymal transition of intestinal carcinogenesis through downregulating METTL3. Cancer Sci. 2023, 114, 3114–3127. [Google Scholar] [CrossRef]
- Dikeocha, I.J.; Al-Kabsi, A.M.; Chiu, H.T.; Alshawsh, M.A. Faecalibacterium prausnitzii Ameliorates Colorectal Tumorigenesis and Suppresses Proliferation of HCT116 Colorectal Cancer Cells. Biomedicines 2022, 10, 1128. [Google Scholar] [CrossRef]
- Guo, J.; Meng, F.; Hu, R.; Chen, L.; Chang, J.; Zhao, K.; Ren, H.; Liu, Z.; Hu, P.; Wang, G.; et al. Inhibition of the NF-κB/HIF-1α signaling pathway in colorectal cancer by tyrosol: A gut microbiota-derived metabolite. J. Immunother. Cancer 2024, 12, e008831. [Google Scholar] [CrossRef]
- Faghfuri, E.; Gholizadeh, P. The role of Akkermansia muciniphila in colorectal cancer: A double-edged sword of treatment or disease progression? Biomed. Pharmacother. 2024, 173, 116416. [Google Scholar] [CrossRef]
- Derosa, L.; Routy, B.; Thomas, A.M.; Iebba, V.; Zalcman, G.; Friard, S.; Mazieres, J.; Audigier-Valette, C.; Moro-Sibilot, D.; Goldwasser, F.; et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat. Med. 2022, 28, 315–324. [Google Scholar] [CrossRef]
- Terrisse, S.; Goubet, A.G.; Ueda, K.; Thomas, A.M.; Quiniou, V.; Thelemaque, C.; Dunsmore, G.; Clave, E.; Gamat-Huber, M.; Yonekura, S.; et al. Immune system and intestinal microbiota determine efficacy of androgen deprivation therapy against prostate cancer. J. Immunother. Cancer 2022, 10, e004191. [Google Scholar] [CrossRef]
- Derosa, L.; Routy, B.; Fidelle, M.; Iebba, V.; Alla, L.; Pasolli, E.; Segata, N.; Desnoyer, A.; Pietrantonio, F.; Ferrere, G.; et al. Gut Bacteria Composition Drives Primary Resistance to Cancer Immunotherapy in Renal Cell Carcinoma Patients. Eur. Urol. 2020, 78, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Gurbatri, C.R.; Radford, G.A.; Vrbanac, L.; Im, J.; Thomas, E.M.; Coker, C.; Taylor, S.R.; Jang, Y.; Sivan, A.; Rhee, K.; et al. Engineering tumor-colonizing E. coli Nissle 1917 for detection and treatment of colorectal neoplasia. Nat. Commun. 2024, 15, 646. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Li, S.; Chen, W.; Han, Y.; Yao, Y.; Yang, L.; Li, Q.; Xiao, Q.; Wei, J.; Liu, Z.; et al. Postoperative Probiotics Administration Attenuates Gastrointestinal Complications and Gut Microbiota Dysbiosis Caused by Chemotherapy in Colorectal Cancer Patients. Nutrients 2023, 15, 356. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Chu, S.H.; Jeon, J.Y.; Lee, M.K.; Park, J.H.; Lee, D.C.; Lee, J.W.; Kim, N.K. Effects of 12 weeks of probiotic supplementation on quality of life in colorectal cancer survivors: A double-blind, randomized, placebo-controlled trial. Dig. Liver Dis. 2014, 46, 1126–1132. [Google Scholar] [CrossRef]
- Golkhalkhali, B.; Rajandram, R.; Paliany, A.S.; Ho, G.F.; Wan Ishak, W.Z.; Johari, C.S.; Chin, K.D. Strain-specific probiotic (microbial cell preparation) and omega-3 fatty acid in modulating quality of life and inflammatory markers in colorectal cancer patients: A randomized controlled trial. Asia-Pac. J. Clin. Oncol. 2018, 14, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Bajramagic, S.; Hodzic, E.; Mulabdic, A.; Holjan, S.; Smajlovic, S.; Rovcanin, A. Usage of Probiotics and its Clinical Significance at Surgically Treated Patients Sufferig from Colorectal Carcinoma. Med. Arch. 2019, 73, 316. [Google Scholar] [CrossRef] [PubMed]
- Polakowski, C.B.; Kato, M.; Preti, V.B.; Schieferdecker, M.E.M.; Ligocki Campos, A.C. Impact of the preoperative use of synbiotics in colorectal cancer patients: A prospective, randomized, double-blind, placebo-controlled study. Nutrition 2019, 58, 40–48. [Google Scholar] [CrossRef] [PubMed]
| Antibiotics | Sample Type | Results | Novelty | References |
|---|---|---|---|---|
| Metronidazole | Human primary CRC mice xenografts | ↓ FN load; ↓ Cancer cell proliferation; ↓ Tumor growth. | Further antimicrobial interventions are needed for FN-associated CRC patients. | Bullman et al., [72] |
| Monensin | Human CRC cell lines RKO and HCT-116 | Induction of G1 arrest and apoptosis. ⊝ cell migration and proliferation. ⊝ IGF1R expression. | This study highlights the importance of IGF1R signaling pathway. | Zhou et al., [79] |
| HCT-116, HT29 | Modulation of TLR4/IRF3 signaling pathway. Anti-inflammatory effect. | The first evidence establishing a connection between monensin and TLRs. | Seçme et al., [80] | |
| Rapamycin liposomes and 5-Fluorouracil | Azoxymethane/dextran sulfate sodium induced CRC mouse model | ↓ tumorigenesis; ↓ number of tumors. Anti-angiogenesis effect. ⊝ proliferation, migration, and tube formation of HUVECs. | This combination may represent an important starting point for more investigations. | Liu et al., [81] |
| Salinomycin | Primary tumor-initiating cells derived from CRC patients | Cell death. ⊝ proliferation; ⊝ respiratory chain complex II. ↓ ATP production; ↓ expression of SOD1; ↑ ROS and dysfunctional mitochondria. | These studies underline the need for additional research about analogs of salinomycin or combinations with this challenging toxic drug. | Klose et al., [82] |
| Semi-synthetic salinomycin | SW480 and SW620 human CRC cell lines | ↑ effects at lower concentrations compared to salinomycin. | Klose et al., [83] | |
| Salinomycin and sulforaphane | Human CRC Caco-2 and CX-1 cell lines | Antiproliferative and proapoptotic effects. ↑ p53 expression; ↓ Bcl-2 expression. | Liu et al., [84] | |
| Nigericin | SW620 and KM12 human CRC cell lines | ↑ tumor apoptosis; ↓ tumor cell proliferation; ↓ β-catenin and TCF-1 expressions; ↓ cyclinD1, Survivin, Axin2, MMP-7/-9, and c-Myc. | Nigericin was efficient in the suppression of both tumor growth and metastasis in CRC cells. | Liu et al., [85] |
| HCT-116 cell lines | Apoptosis and autophagy inhibition of GSK-3β and JAK3 kinases. | Garcia-Princival et al., [86] | ||
| Tigecycline | HCT-116 cell lines Colitis-associated CRC murine model | ↓ tumorigenesis; ↓ proliferation (⊝ STAT3 activation and Wnt/β-catenin pathway); ↓ cancer-associated inflammation; ↑ cytotoxic T lymphocytes activity; ↑ apoptosis (↑ CASP7); ↑ protective anti-tumor bacterial genera and species (Akkermansia, Parabacteroides distasonis). | This study demonstrates the potential benefits of using tigecycline and brings up a very promising drug against CRC. | Ruiz-Malagón et al., [87] |
| Minocycline | SW480 and SW620 cells | ↓ metastasis; ⊝ LYN activity; ⊝ STAT3 signaling; ⊝ epithelial–mesenchymal transition. | Via direct binding to LYN, an enzyme correlated with increased activity and metastasis development in CRC, minocycline may serve as an effective treatment. | Yang et al., [88] |
| Antimycin A | HCT-116 cells | ↑ p53 and CASP-9 gene expression; ↓ MAPK, PARP, and NF-kB genes. | The first study to investigate the antimycin A apoptotic pathways in CRC cells. | Kabir et al., [89] |
| Clarithromycin | HCT-116 cells | ↑ 5-Fluorouracil cytotoxic effect; ↑ apoptosis and cytosolic autophagosomes; ⊝ the assembly of a macromolecular structure between hERG1 and p85 component of PI3K. | As hERG1 is increased in aggressive primary CRC, clarithromycin or other drugs aiming hERG1 need further investigations in clinical trials. | Petroni et al., [90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volovat, C.C.; Cosovanu, M.A.; Ostafe, M.-R.; Augustin, I.G.; Volovat, C.; Georgescu, B.; Volovat, S.R. Bacteriophages, Antibiotics and Probiotics: Exploring the Microbial Battlefield of Colorectal Cancer. Int. J. Mol. Sci. 2025, 26, 7837. https://doi.org/10.3390/ijms26167837
Volovat CC, Cosovanu MA, Ostafe M-R, Augustin IG, Volovat C, Georgescu B, Volovat SR. Bacteriophages, Antibiotics and Probiotics: Exploring the Microbial Battlefield of Colorectal Cancer. International Journal of Molecular Sciences. 2025; 26(16):7837. https://doi.org/10.3390/ijms26167837
Chicago/Turabian StyleVolovat, Cristian Constantin, Mihai Andrei Cosovanu, Madalina-Raluca Ostafe, Iolanda Georgiana Augustin, Constantin Volovat, Bogdan Georgescu, and Simona Ruxandra Volovat. 2025. "Bacteriophages, Antibiotics and Probiotics: Exploring the Microbial Battlefield of Colorectal Cancer" International Journal of Molecular Sciences 26, no. 16: 7837. https://doi.org/10.3390/ijms26167837
APA StyleVolovat, C. C., Cosovanu, M. A., Ostafe, M.-R., Augustin, I. G., Volovat, C., Georgescu, B., & Volovat, S. R. (2025). Bacteriophages, Antibiotics and Probiotics: Exploring the Microbial Battlefield of Colorectal Cancer. International Journal of Molecular Sciences, 26(16), 7837. https://doi.org/10.3390/ijms26167837





