The DIR Gene Family in Watermelon: Evolution, Stress Expression Profiles, and Functional Exploration of ClDIR8
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification of DIR Gene Family in Watermelon
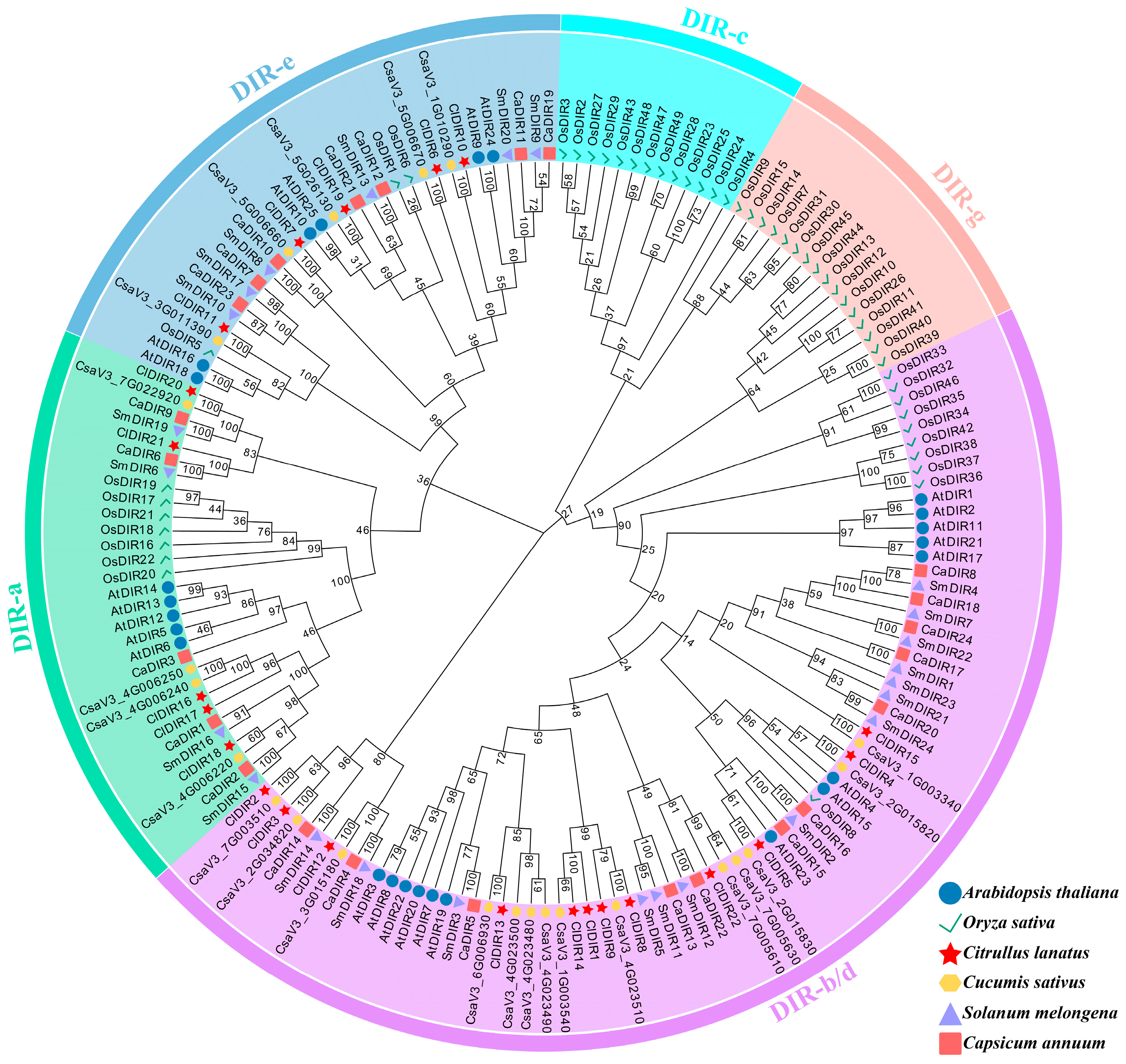
2.2. Phylogenetic Analysis of ClDIR Genes
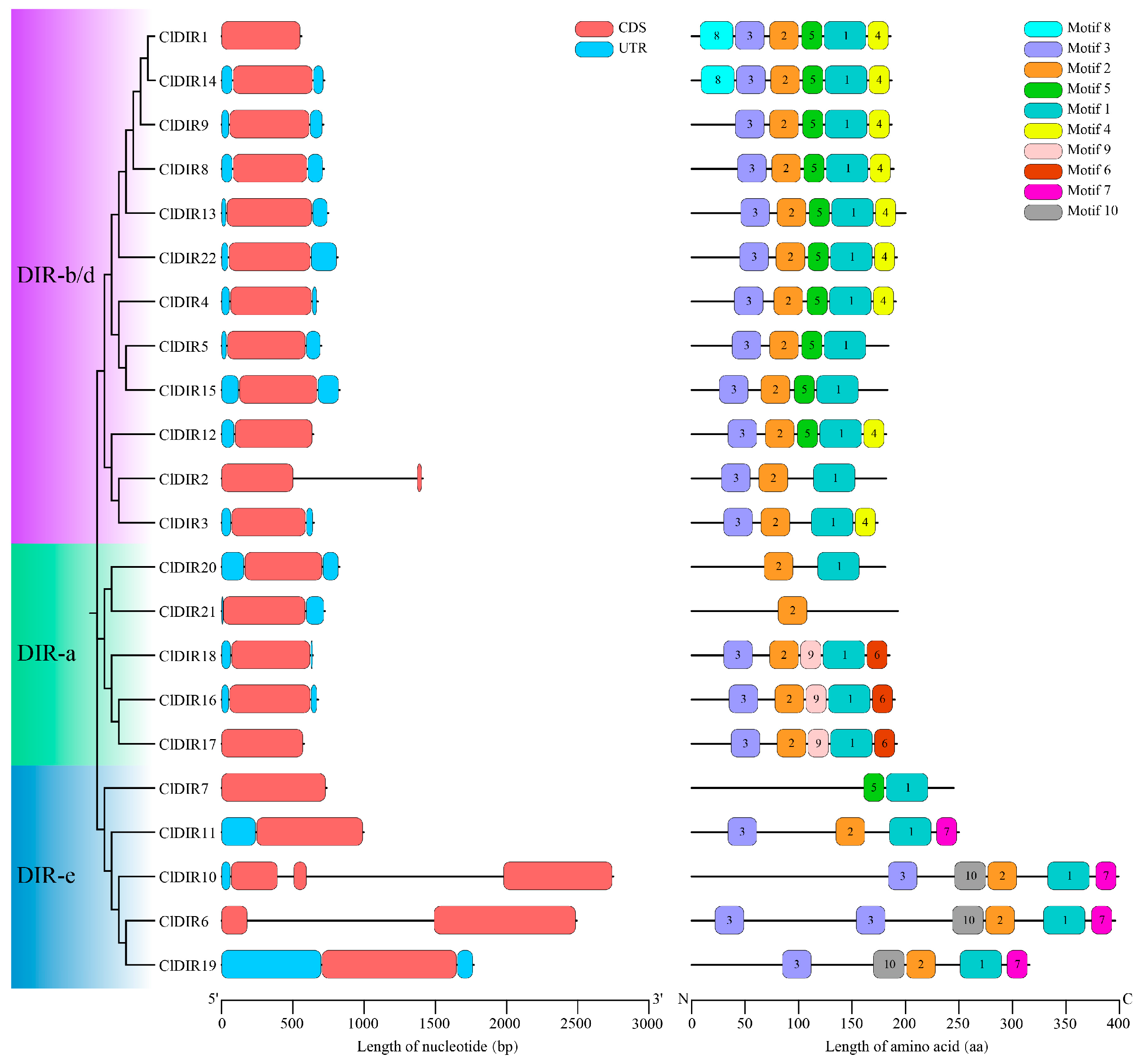
2.3. Gene Structure and Conserved Motif Analyses of ClDIR Genes
2.4. Synteny Analysis of ClDIR Genes
2.5. Analysis of Cis-Acting Elements in ClDIR Promoters
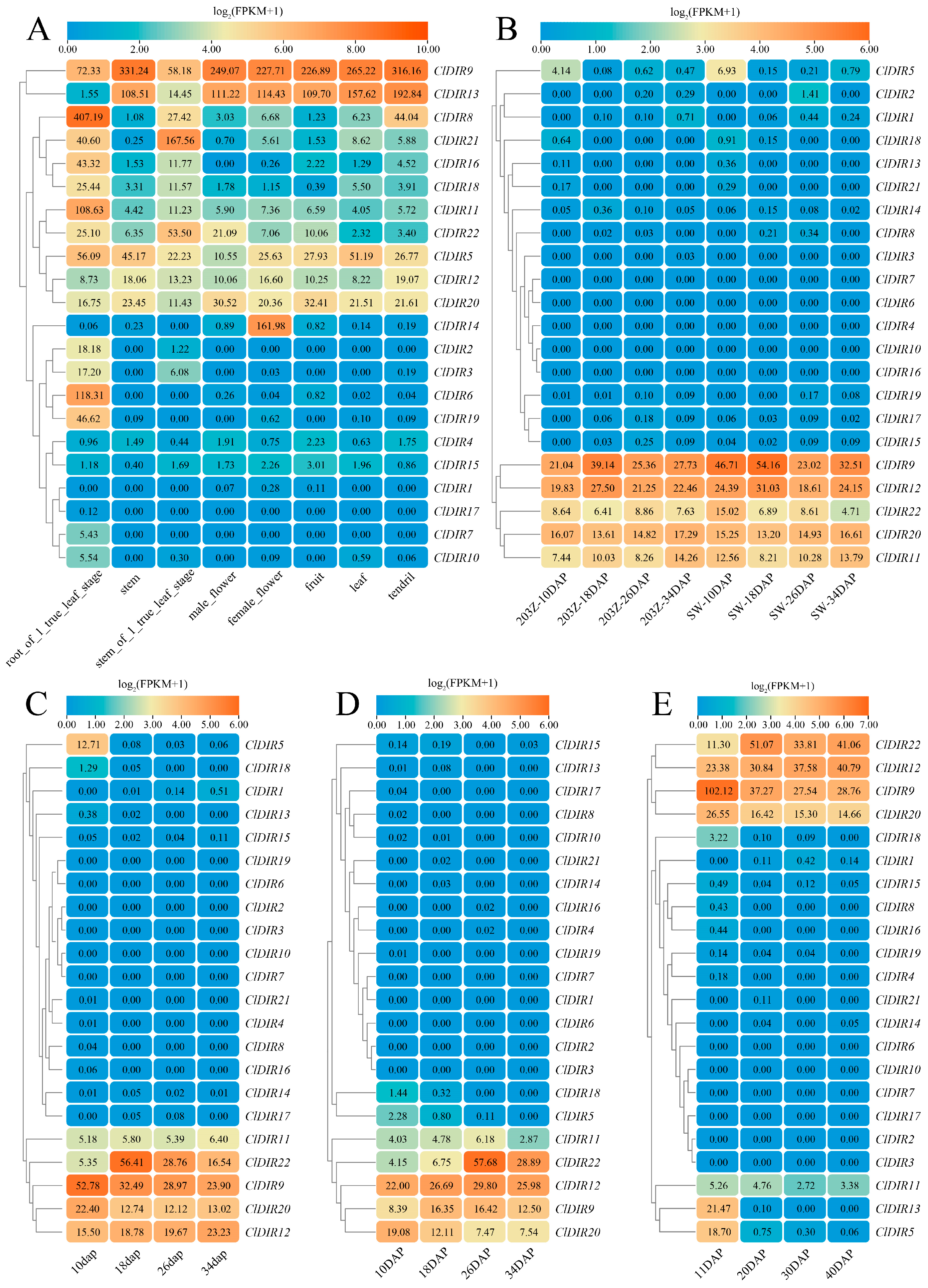
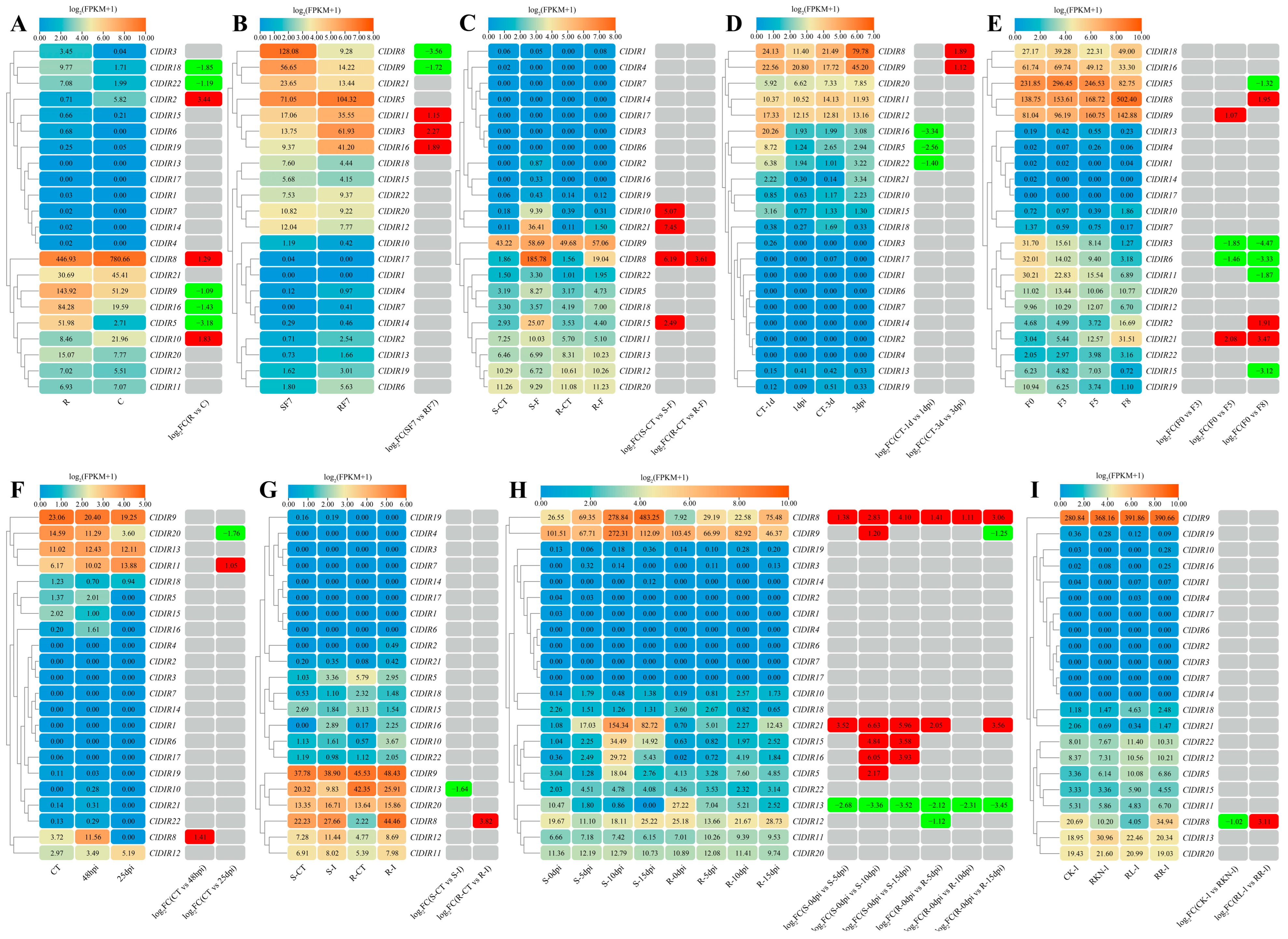
2.6. Tissue-Specific Expression Analysis of ClDIR Genes
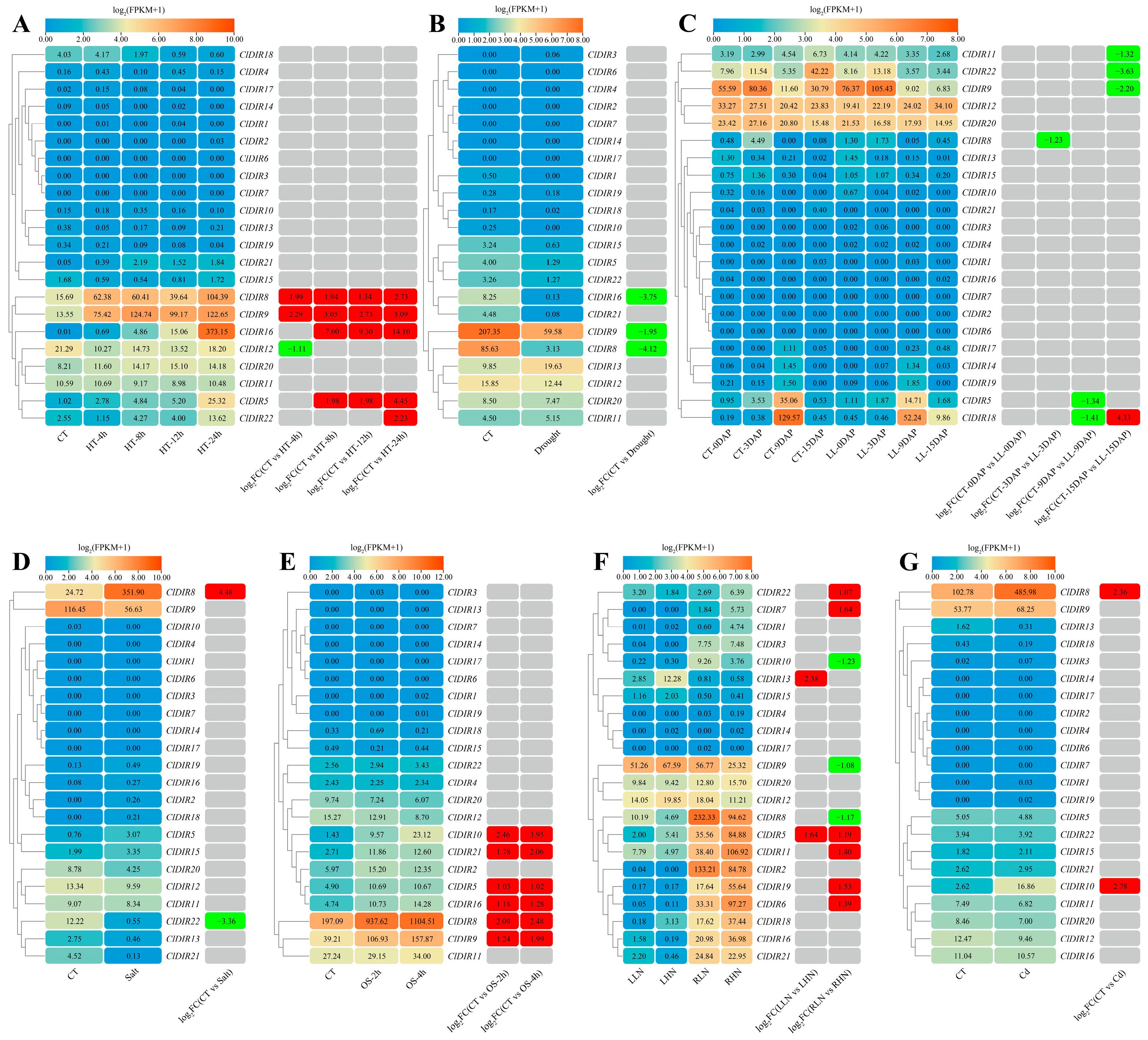
2.7. Expression Analysis of ClDIR Genes Under Abiotic Stresses
2.8. Expression Analysis of ClDIR Genes Under Biotic Stresses
2.9. Expression Patterns of ClDIR Genes Under Abiotic and Biotic Stresses
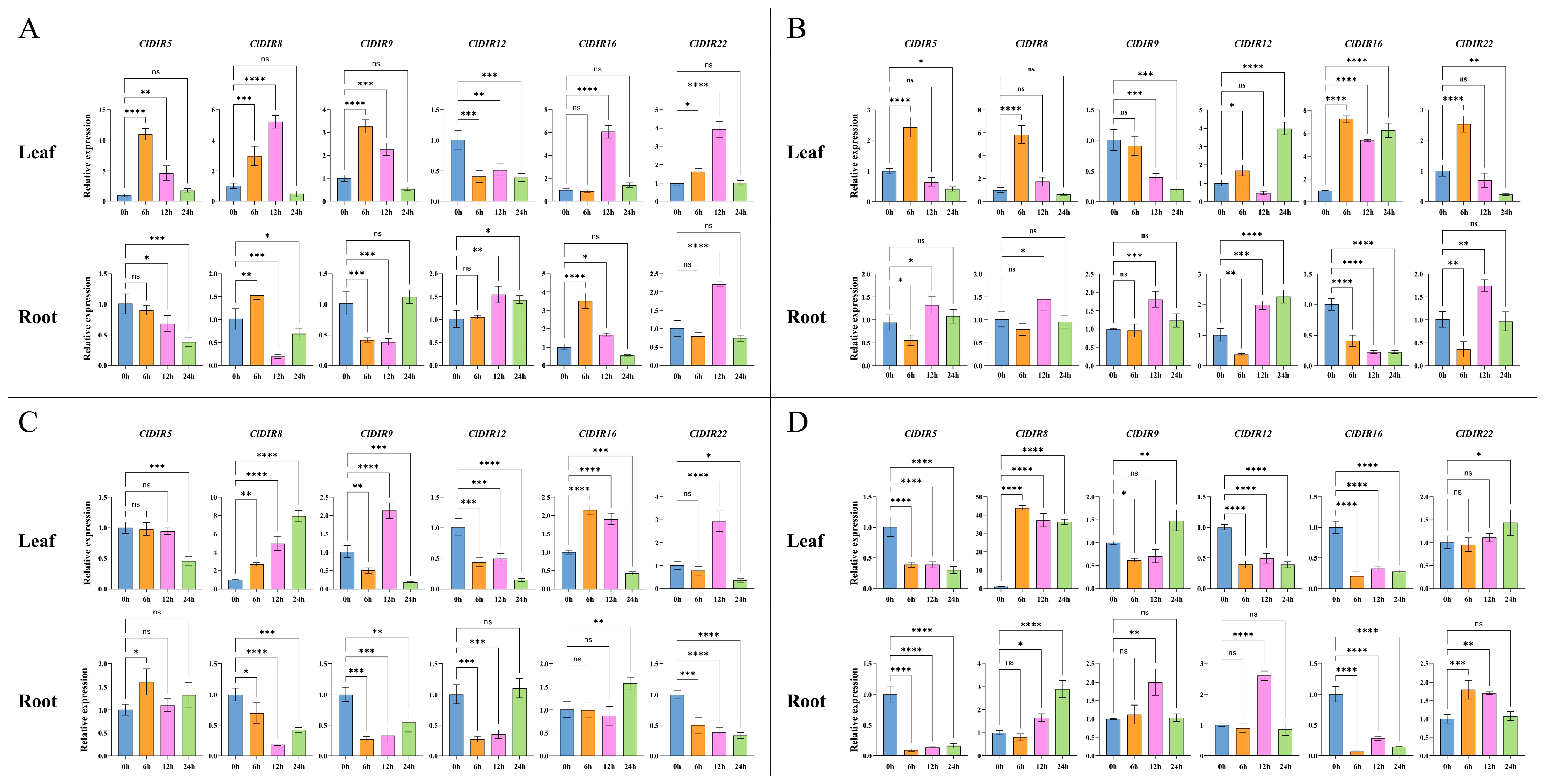
2.10. Validation of the Expression Profiles of ClDIR Genes Under High-Temperature, Drought, Salt and Low-Temperature Stresses
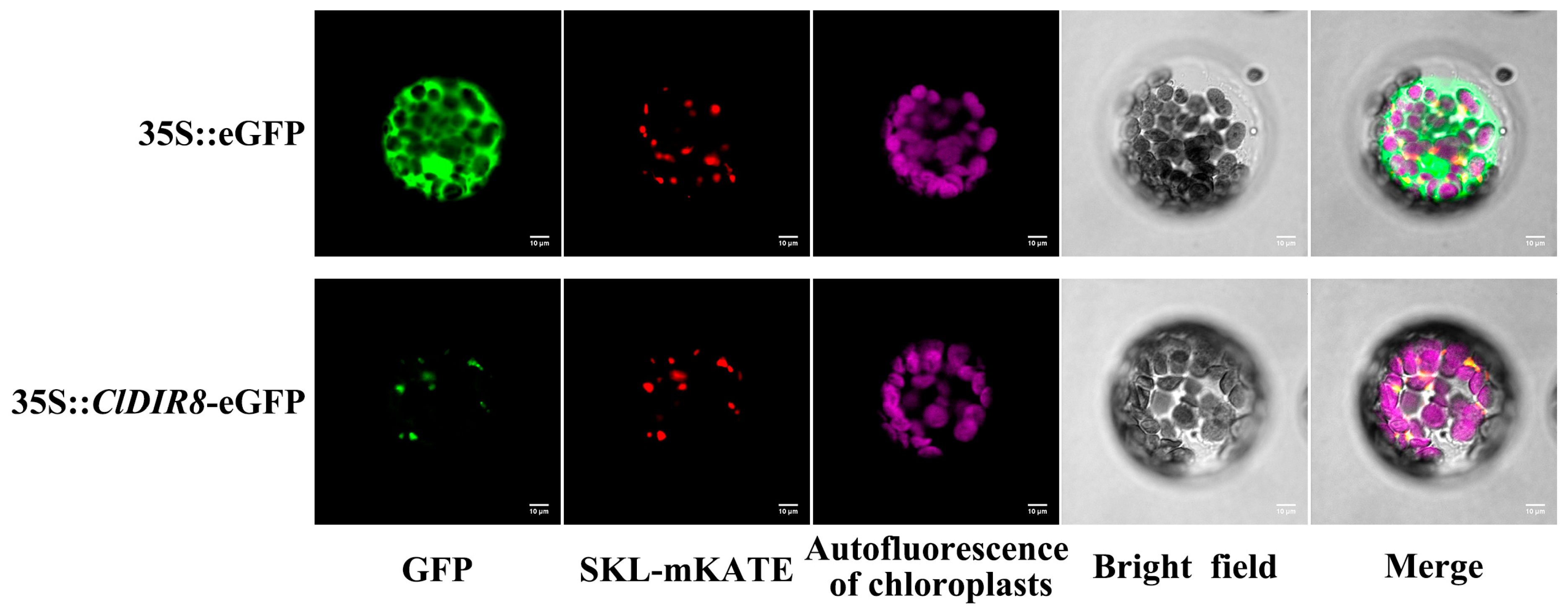
2.11. Subcellular Localization of ClDIR8 Protein
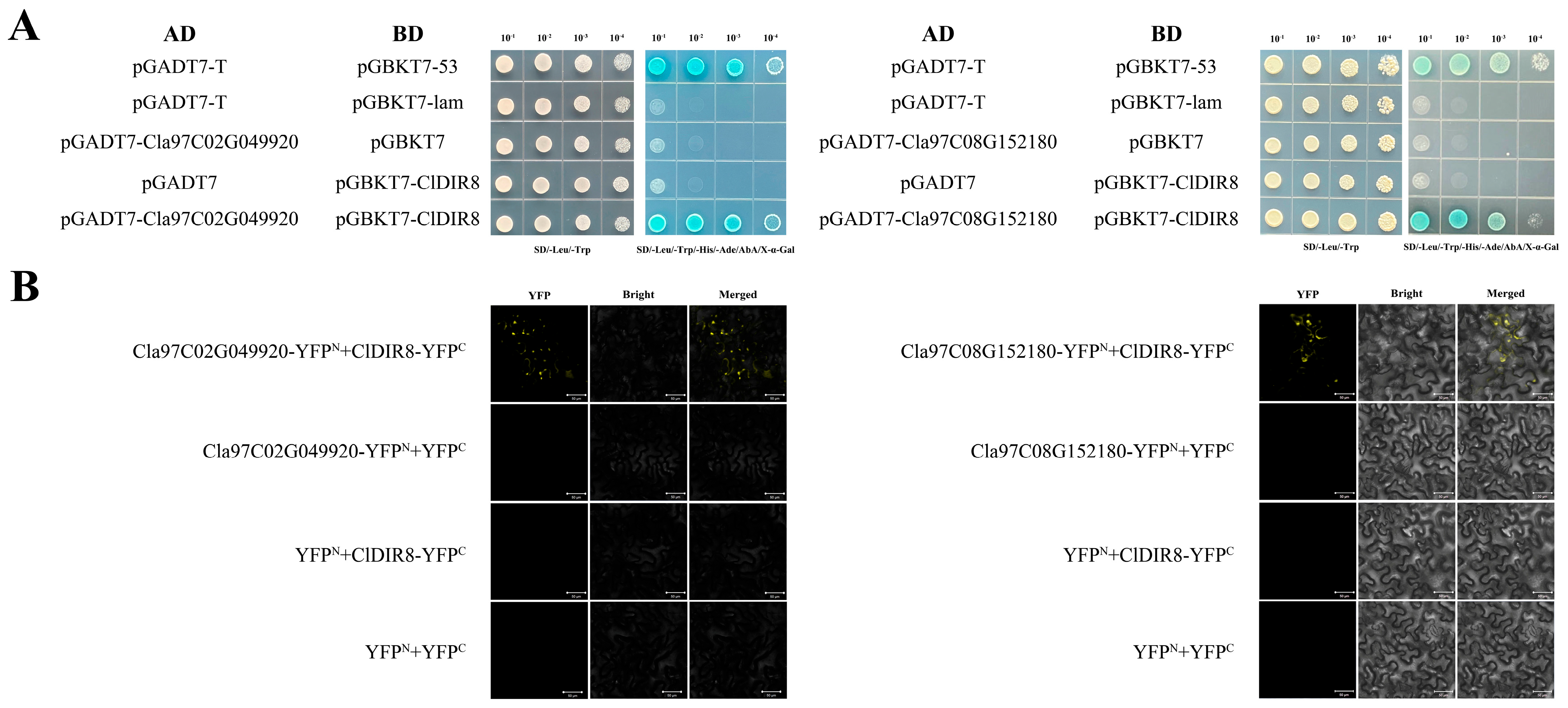
2.12. Screening of Proteins Interacting with ClDIR8 Protein
3. Discussion
3.1. Identification of DIR Gene Family in Watermelon and Its Evolutionary Context
3.2. Mechanisms of DIR Gene Family Expansion in Watermelon
3.3. Phylogenetic Classification and Subfamily-Specific Features of ClDIR Genes
3.4. Gene Structure and Conserved Motifs of ClDIR Genes
3.5. Cis-Acting Elements in ClDIR Promoters
3.6. Tissue-Specific and Fruit Development Expression Patterns of ClDIR Genes
3.7. Expression Profiles of ClDIR Genes Under Abiotic Stresses
3.8. Role of ClDIR Genes in Biotic Stress Responses, with a Focus on ClDIR8
3.9. Subcellular Localization of ClDIR8 and Identification of Interacting Proteins
3.10. Functional Implications of ClDIR8 Interactions with Peroxidase and Catalase, and Future Perspectives
4. Materials and Methods
4.1. Identification of ClDIR Genes and Analysis of Physicochemical Properties of Their Encoded Proteins
4.2. Phylogenetic Tree, Gene Structure, and Conserved Motif Analysis
4.3. Synteny, Gene Duplication, and Selective Pressure Analysis of ClDIR Genes
4.4. Cis-Acting Element Analysis of ClDIR Gene Promoters
4.5. Reanalysis of Watermelon Transcriptome Sequencing Data
4.6. Analysis of ClDIR Gene Expression Patterns in Different Tissues and Under Abiotic and Biotic Stresses
4.7. Plant Materials and Stress Treatments
4.8. Total RNA Extraction and qRT-PCR Analysis
4.9. Subcellular Localization Analysis of ClDIR8 Protein
4.10. Y2H Library Screening
4.11. Y2H Validation and BiFC Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DIR | Dirigent proteins |
| BiFC | Bimolecular fluorescence complementation |
| Y2H | Yeast two-hybrid |
| GRAVY | Grand average of hydropathicity |
| pI | Theoretical isoelectric points |
| Ka | Non-synonymous substitution rates |
| Ks | Synonymous substitution rates |
| ABA | Abscisic acid |
| SA | Salicylic acid |
| MeJA | Methyl jasmonate |
| GA | Gibberellin |
| DAP | Days after pollination |
| dpi | Day post inoculation |
| YFP | Yellow fluorescent protein |
| CDS | Coding sequence |
| NJ | Neighbor-joining method |
| DEGs | Differentially expressed genes |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| eGFP | Enhanced green fluorescent protein |
References
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, S.; Zhang, Y.; Wang, X.; Li, D.; Li, Q.; Yue, M.; Li, Q.; Zhang, Y.-e.; Xu, Y. Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 2011, 23, 396–411. [Google Scholar] [CrossRef]
- Zhou, J.; Ma, J.; Yang, C.; Zhu, X.; Li, J.; Zheng, X.; Li, X.; Chen, S.; Feng, L.; Wang, P. A non-canonical role of ATG8 in Golgi recovery from heat stress in plants. Nat. Plants 2023, 9, 749–765. [Google Scholar] [CrossRef]
- Imran, Q.M.; Falak, N.; Hussain, A.; Mun, B.-G.; Yun, B.-W. Abiotic stress in plants; stress perception to molecular response and role of biotechnological tools in stress resistance. Agronomy 2021, 11, 1579. [Google Scholar] [CrossRef]
- Yadav, S.; Chattopadhyay, D. Lignin: The building block of defense responses to stress in plants. J. Plant Growth Regul. 2023, 42, 6652–6666. [Google Scholar] [CrossRef]
- Dalisay, D.S.; Kim, K.W.; Lee, C.; Yang, H.; Rübel, O.; Bowen, B.P.; Davin, L.B.; Lewis, N.G. Dirigent protein-mediated lignan and cyanogenic glucoside formation in flax seed: Integrated omics and MALDI mass spectrometry imaging. J. Nat. Prod. 2015, 78, 1231–1242. [Google Scholar] [CrossRef] [PubMed]
- Bardin, M.; Rousselot-Pailley, P.; Tron, T.; Robert, V. Investigation of dirigent like domains from bacterial genomes. BMC Bioinform. 2022, 23, 313. [Google Scholar] [CrossRef]
- Gao, Y.-Q.; Huang, J.-Q.; Reyt, G.; Song, T.; Love, A.; Tiemessen, D.; Xue, P.-Y.; Wu, W.-K.; George, M.W.; Chen, X.-Y. A dirigent protein complex directs lignin polymerization and assembly of the root diffusion barrier. Science 2023, 382, 464–471. [Google Scholar] [CrossRef]
- Li, Q.; Fu, C.; Liang, C.; Ni, X.; Zhao, X.; Chen, M.; Ou, L. Crop lodging and the roles of lignin, cellulose, and hemicellulose in lodging resistance. Agronomy 2022, 12, 1795. [Google Scholar] [CrossRef]
- Guo, Z.; Xu, W.; Wei, D.; Zheng, S.; Liu, L.; Cai, Y. Functional analysis of a dirigent protein AtsDIR23 in Acorus tatarinowii. J. Plant Physiol. 2023, 290, 154098. [Google Scholar] [CrossRef]
- Pei, Y.; Cao, W.; Yu, W.; Peng, C.; Xu, W.; Zuo, Y.; Wu, W.; Hu, Z. Identification and functional characterization of the dirigent gene family in Phryma leptostachya and the contribution of PlDIR1 in lignan biosynthesis. BMC Plant Biol. 2023, 23, 291. [Google Scholar] [CrossRef]
- Shi, Y.; Shen, Y.; Ahmad, B.; Yao, L.; He, T.; Fan, J.; Liu, Y.; Chen, Q.; Wen, Z. Genome-wide identification and expression analysis of dirigent gene family in strawberry (Fragaria vesca) and functional characterization of FvDIR13. Sci. Hortic. 2022, 297, 110913. [Google Scholar] [CrossRef]
- Ma, X.; Xu, W.; Liu, T.; Chen, R.; Zhu, H.; Zhang, H.; Cai, C.; Li, S. Functional characterization of soybean (Glycine max) DIRIGENT genes reveals an important role of GmDIR27 in the regulation of pod dehiscence. Genomics 2021, 113, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Z.; Zhao, H.; Deng, X.; Su, Y.; Li, R.; Chen, B. Overexpression of sugarcane ScDIR genes enhances drought tolerance in Nicotiana benthamiana. Int. J. Mol. Sci. 2022, 23, 5340. [Google Scholar] [CrossRef]
- Zhao, Z.; Guan, Y.; Qin, T.; Zheng, H.; Wang, H.; Xu, W.; Gu, W.; Yu, D.; Wei, J.; Hu, Y. A Dirigent Gene, ZmDIR11, Positively Regulates Drought Tolerance in Maize. Agronomy 2025, 15, 604. [Google Scholar] [CrossRef]
- Zhang, Q.; Luo, N.; Dai, X.; Lin, J.; Ahmad, B.; Chen, Q.; Lei, Y.; Wen, Z. Ectopic and transient expression of VvDIR4 gene in Arabidopsis and grapes enhances resistance to anthracnose via affecting hormone signaling pathways and lignin production. BMC Genom. 2024, 25, 895. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.-M.; Liang, X.; Li, Z.-B.; Jie, S.; Yan, H.-B.; Li, S.-X.; Liao, W.-B.; Ming, P. A novel long non-coding RNA, DIR, increases drought tolerance in cassava by modifying stress-related gene expression. J. Integr. Agric. 2022, 21, 2588–2602. [Google Scholar] [CrossRef]
- Dokka, N.; Tyagi, S.; Ramkumar, M.; Rathinam, M.; Senthil, K.; Sreevathsa, R. Genome-wide identification and characterization of DIRIGENT gene family (CcDIR) in pigeonpea (Cajanus cajan L.) provide insights on their spatial expression pattern and relevance to stress response. Gene 2024, 914, 148417. [Google Scholar] [CrossRef]
- Liao, Y.; Liu, S.; Jiang, Y.; Hu, C.; Zhang, X.; Cao, X.; Xu, Z.; Gao, X.; Li, L.; Zhu, J.; et al. Genome-wide analysis and environmental response profiling of dirigent family genes in rice (Oryza sativa). Genes Genom. 2017, 39, 47–62. [Google Scholar] [CrossRef]
- Xu, W.; Liu, T.; Zhang, H.; Zhu, H. Mungbean DIRIGENT gene subfamilies and their expression profiles under salt and drought stresses. Front. Genet. 2021, 12, 658148. [Google Scholar] [CrossRef]
- Deng, J.; Guan, R.; Liang, T.; Su, L.; Ge, F.; Cui, X.; Liu, D. Dirigent gene family is involved in the molecular interaction between Panax notoginseng and root rot pathogen Fusarium solani. Ind. Crops Prod. 2022, 178, 114544. [Google Scholar] [CrossRef]
- Yadav, V.; Wang, Z.; Yang, X.; Wei, C.; Changqing, X.; Zhang, X. Comparative analysis, characterization and evolutionary study of dirigent gene family in cucurbitaceae and expression of novel dirigent peptide against powdery mildew stress. Genes 2021, 12, 326. [Google Scholar] [CrossRef]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E. Dirigent proteins in plants: Modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef]
- Cheng, X.; Su, X.; Muhammad, A.; Li, M.; Zhang, J.; Sun, Y.; Li, G.; Jin, Q.; Cai, Y.; Lin, Y. Molecular characterization, evolution, and expression profiling of the dirigent (DIR) family genes in Chinese white pear (Pyrus bretschneideri). Front. Genet. 2018, 9, 136. [Google Scholar] [CrossRef]
- Gong, L.; Li, B.; Zhu, T.; Xue, B. Genome-wide identification and expression profiling analysis of DIR gene family in Setaria italica. Front. Plant Sci. 2023, 14, 1243806. [Google Scholar] [CrossRef]
- Xuan, X.; Su, S.; Chen, J.; Tan, J.; Yu, Z.; Jiao, Y.; Cai, S.; Zhang, Z.; Ramakrishnan, M. Evolutionary and functional analysis of the DIR gene family in Moso bamboo: Insights into rapid shoot growth and stress responses. Front. Plant Sci. 2025, 16, 1535733. [Google Scholar] [CrossRef]
- Ralph, S.; Park, J.-Y.; Bohlmann, J.; Mansfield, S.D. Dirigent proteins in conifer defense: Gene discovery, phylogeny, and differential wound-and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.). Plant Mol. Biol. 2006, 60, 21–40. [Google Scholar] [CrossRef]
- Khan, A.; Li, R.-J.; Sun, J.-T.; Ma, F.; Zhang, H.-X.; Jin, J.-H.; Ali, M.; Haq, S.u.; Wang, J.-E.; Gong, Z.-H. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 2018, 8, 5500. [Google Scholar] [CrossRef]
- Zhang, K.; Xing, W.; Sheng, S.; Yang, D.; Zhen, F.; Jiang, H.; Yan, C.; Jia, L. Genome-wide identification and expression analysis of eggplant DIR gene family in response to biotic and abiotic stresses. Horticulturae 2022, 8, 732. [Google Scholar] [CrossRef]
- Liu, C.; Qin, Z.; Zhou, X.; Xin, M.; Wang, C.; Liu, D.; Li, S. Expression and functional analysis of the Propamocarb-related gene CsDIR16 in cucumbers. BMC Plant Biol. 2018, 18, 16. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Peng, X. Genome-wide identification and characterization of DIR genes in Medicago truncatula. Biochem. Genet. 2019, 57, 487–506. [Google Scholar] [CrossRef]
- Davin, L.B.; Wang, H.-B.; Crowell, A.L.; Bedgar, D.L.; Martin, D.M.; Sarkanen, S.; Lewis, N.G. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 1997, 275, 362–367. [Google Scholar] [CrossRef]
- Kim, K.-W.; Moinuddin, S.G.; Atwell, K.M.; Costa, M.A.; Davin, L.B.; Lewis, N.G. Opposite stereoselectivities of dirigent proteins in Arabidopsis and Schizandra species. J. Biol. Chem. 2012, 287, 33957–33972. [Google Scholar] [CrossRef]
- Wu, R.; Wang, L.; Wang, Z.; Shang, H.; Liu, X.; Zhu, Y.; Qi, D.; Deng, X. Cloning and expression analysis of a dirigent protein gene from the resurrection plant Boea hygrometrica. Prog. Nat. Sci. 2009, 19, 347–352. [Google Scholar] [CrossRef]
- Uchida, K.; Akashi, T.; Aoki, T. The missing link in leguminous pterocarpan biosynthesis is a dirigent domain-containing protein with isoflavanol dehydratase activity. Plant Cell Physiol. 2017, 58, 398–408. [Google Scholar] [CrossRef]
- Corbin, C.; Drouet, S.; Markulin, L.; Auguin, D.; Lainé, É.; Davin, L.B.; Cort, J.R.; Lewis, N.G.; Hano, C. A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: From gene identification and evolution to differential regulation. Plant Mol. Biol. 2018, 97, 73–101. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Peng, J.; Li, Q.; Geng, S.; Zhang, H.; Shu, Y.; Wang, R.; Zhang, B.; Li, C.; Xiang, X. Genome-wide identification and analysis of monocot-specific chimeric jacalins (MCJ) genes in Maize (Zea mays L.). BMC Plant Biol. 2024, 24, 636. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, D.J.; Cicek, M.; Chen, J.; Esen, A. Identification of β-glucosidase aggregating factor (BGAF) and mapping of BGAF binding regions on maize β-glucosidase. J. Biol. Chem. 2001, 276, 11895–11901. [Google Scholar] [CrossRef]
- Kittur, F.S.; Yu, H.Y.; Bevan, D.R.; Esen, A. Deletion of the N-terminal dirigent domain in maize β-glucosidase aggregating factor and its homolog sorghum lectin dramatically alters the sugar-specificities of their lectin domains. Plant Physiol. Bioch. 2010, 48, 731–734. [Google Scholar] [CrossRef]
- Ma, Q.-H. Monocot chimeric jacalins: A novel subfamily of plant lectins. Crit. Rev. Biotechnol. 2014, 34, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zeng, L.; Chen, R.; Wang, Y.; Song, J. Genome-wide identification and characterization of stress-associated protein (SAP) gene family encoding A20/AN1 zinc-finger proteins in Medicago truncatula. Arch. Biol. Sci. 2018, 70, 087–098. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, T.; Zhang, T.; Ma, L. Trends in the evolution of intronless genes in Poaceae. Front. Plant Sci. 2023, 14, 1065631. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wan, C.; Wu, W.; Zhang, Y.; Pan, Y.; Chen, X.; Li, C.; Pi, J.; Wang, Z.; Ye, Y. Methyl jasmonate (MeJA) enhances salt tolerance of okra (Abelmoschus esculentus L.) plants by regulating ABA signaling, osmotic adjustment substances, photosynthesis and ROS metabolism. Sci. Hortic. 2023, 319, 112145. [Google Scholar] [CrossRef]
- Qu, X.; Zou, J.; Wang, J.; Yang, K.; Wang, X.; Le, J. A rice R2R3-type MYB transcription factor OsFLP positively regulates drought stress response via OsNAC. Int. J. Mol. Sci. 2022, 23, 5873. [Google Scholar] [CrossRef]
- Chen, D.; Sun, M.; Yang, Y.; Tan, B.; Ren, D.; Tao, Y.; Li, R.; Zhao, Q. Biochemistry; Comprehensive genome-wide analysis and functional characterization of the DIR gene family in Herpetospermum pedunculosum: Insights from HpDIR16 and HpDIR17. Plant Physiol. Bioch. 2025, 226, 110074. [Google Scholar] [CrossRef]
- Burlat, V.; Kwon, M.; Davin, L.B.; Lewis, N.G. Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 2001, 57, 883–897. [Google Scholar] [CrossRef]
- Gallego-Giraldo, L.; Jikumaru, Y.; Kamiya, Y.; Tang, Y.; Dixon, R.A. Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol. 2011, 190, 627–639. [Google Scholar] [CrossRef]
- Li, X.; Liu, Z.; Wu, H.; Yu, Z.; Meng, J.; Zhao, H.; Deng, X.; Su, Y.; Chen, B.; Li, R. Four sugarcane ScDIR genes contribute to lignin biosynthesis and disease resistance to Sporisorium scitamineum. Phytopathol. Res. 2024, 6, 17. [Google Scholar] [CrossRef]
- Harmatha, J.; Dinan, L. Biological activities of lignans and stilbenoids associated with plant-insect chemical interactions. Phytochem. Rev. 2003, 2, 321–330. [Google Scholar] [CrossRef]
- Shi, H.; Liu, Z.; Zhu, L.; Zhang, C.; Chen, Y.; Zhou, Y.; Li, F.; Li, X. Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Bioch. Bioph. Sin. 2012, 44, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhao, M.; Liu, T.; Dong, L.; Cheng, Q.; Wu, J.; Wang, L.; Chen, X.; Zhang, C.; Lu, W. A novel soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to Phytophthora sojae. Front. Plant Sci. 2017, 8, 1185. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.-H.; Liu, Y.-C. TaDIR13, a dirigent protein from wheat, promotes lignan biosynthesis and enhances pathogen resistance. Plant Mol. Biol. Rep. 2015, 33, 143–152. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, M.; Zhang, H. Comprehensive genome wide analysis of the dirigent (DIR) gene family in Arabidopsis thaliana and Brassica species: Evolutionary dynamics and abiotic stress response mechanisms. Genet. Resour. Crop Evol. 2025, 1–21. [Google Scholar] [CrossRef]
- Zoclanclounon, Y.A.B.; Rostás, M.; Chung, N.-J.; Mo, Y.; Karlovsky, P.; Dossa, K. Characterization of peroxidase and laccase gene families and in silico identification of potential genes involved in upstream steps of lignan formation in sesame. Life 2022, 12, 1200. [Google Scholar] [CrossRef]
- De Oliveira, F.K.; Santos, L.O.; Buffon, J.G. Mechanism of action, sources, and application of peroxidases. Food Res. Int. 2021, 143, 110266. [Google Scholar] [CrossRef]
- Alfonso-Prieto, M.; Biarnés, X.; Vidossich, P.; Rovira, C. The molecular mechanism of the catalase reaction. J. Am. Chem. Soc. 2009, 131, 11751–11761. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, S.; Bai, Y.; Sun, H.; Jiao, C.; Guo, S.; Zhao, K.; Blanca, J.; Zhang, Z.; Huang, S. Cucurbit Genomics Database (CuGenDB): A central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 2019, 47, D1128–D1136. [Google Scholar] [CrossRef]
- Bateman, A.; Haft, D.H. HMM-based databases in InterPro. Brief. Bioinform. 2002, 3, 236–245. [Google Scholar] [CrossRef][Green Version]
- Lowry, J.A.; Atchley, W.R. Molecular evolution of the GATA family of transcription factors: Conservation within the DNA-binding domain. J. Mol. Evol. 2000, 50, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Studholme, D.J.; Rawlings, N.D.; Barrett, A.J.; Bateman, A. A comparison of Pfam and MEROPS: Two databases, one comprehensive, and one specialised. BMC Bioinform. 2003, 4, 17. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhang, K.; He, S.; Jia, L.; Hu, Y.; Yang, D.; Lu, X.; Zhang, Q.; Yan, C. Genome-wide identification and expression analysis of DIR gene family in Cucumber. Sci. Agric. Sin. 2023, 56, 711–728. [Google Scholar]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.-Q.; Wang, J.; Wong, G.K.-S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom Proteom Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Umer, M.J.; Bin Safdar, L.; Gebremeskel, H.; Zhao, S.; Yuan, P.; Zhu, H.; Kaseb, M.; Anees, M.; Lu, X.; He, N. Identification of key gene networks controlling organic acid and sugar metabolism during watermelon fruit development by integrating metabolic phenotypes and gene expression profiles. Hortic. Res. 2020, 7, 193. [Google Scholar] [CrossRef]
- Gong, C.; Diao, W.; Zhu, H.; Umer, M.J.; Zhao, S.; He, N.; Lu, X.; Yuan, P.; Anees, M.; Yang, D. Metabolome and transcriptome integration reveals insights into flavor formation of ‘Crimson’watermelon flesh during fruit development. Front. Plant Sci. 2021, 12, 629361. [Google Scholar] [CrossRef]
- Wu, S.; Wang, X.; Reddy, U.; Sun, H.; Bao, K.; Gao, L.; Mao, L.; Patel, T.; Ortiz, C.; Abburi, V.L. Genome of ‘Charleston Gray’, the principal American watermelon cultivar, and genetic characterization of 1,365 accessions in the US National Plant Germplasm System watermelon collection. Plant Biotechnol. J. 2019, 17, 2246–2258. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Joshi, M.; DiPiazza, J.; Joshi, V. Functional relevance of citrulline in the vegetative tissues of watermelon during abiotic stresses. Front. Plant Sci. 2020, 11, 512. [Google Scholar] [CrossRef]
- Gao, W.; She, F.; Sun, Y.; Han, B.; Wang, X.; Xu, G. Transcriptome analysis reveals the genes related to water-melon fruit expansion under low-light stress. Plants 2023, 12, 935. [Google Scholar] [CrossRef]
- Song, Q.; Joshi, M.; Joshi, V. Transcriptomic analysis of short-term salt stress response in watermelon seedlings. Int. J. Mol. Sci. 2020, 21, 6036. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Li, M.; Yan, M.; Qiao, F.; Jiang, X. Integrated transcriptome analysis and single-base resolution methylomes of watermelon (Citrullus lanatus) reveal epigenome modifications in response to osmotic stress. Front. Plant Sci. 2021, 12, 769712. [Google Scholar] [CrossRef]
- Nawaz, M.A.; Chen, C.; Shireen, F.; Zheng, Z.; Sohail, H.; Afzal, M.; Ali, M.A.; Bie, Z.; Huang, Y. Genome-wide expression profiling of leaves and roots of watermelon in response to low nitrogen. BMC Genom. 2018, 19, 456. [Google Scholar] [CrossRef]
- Zhu, F.; Wang, Z.; Su, W.; Tong, J.; Fang, Y.; Luo, Z.; Yuan, F.; Xiang, J.; Chen, X.; Wang, R. Study on the role of salicylic acid in watermelon-resistant fusarium wilt under different growth conditions. Plants 2022, 11, 293. [Google Scholar] [CrossRef]
- Zhu, F.; Wang, Z.; Fang, Y.; Tong, J.; Xiang, J.; Yang, K.; Wang, R. Study on the role of phytohormones in resistance to watermelon fusarium wilt. Plants 2022, 11, 156. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, Z.; Xu, W. Bacillus velezensis WB induces systemic resistance in watermelon against Fusarium wilt. Pest Manag. Sci. 2024, 80, 1423–1434. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, J.; Yang, K.; Wang, Y.; Tian, Y.; Liang, Z. Transcriptomic and metabonomic insights into the biocontrol mechanism of Trichoderma asperellum M45a against watermelon Fusarium wilt. PLoS ONE 2022, 17, e0272702. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Wang, Z.; Guo, Y.; Zhang, X. Comparative transcriptome profiling reveals the role of phytohormones and phenylpropanoid pathway in early-stage resistance against powdery mildew in watermelon (Citrullus lanatus L.). Front. Plant Sci. 2022, 13, 1016822. [Google Scholar] [CrossRef]
- Lv, G.; Zheng, X.; Duan, Y.; Wen, Y.; Zeng, B.; Ai, M.; He, B. The GRAS gene family in watermelons: Identification, characterization and expression analysis of different tissues and root-knot nematode infestations. Peer J. 2021, 9, e11526. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Sparkes, I.; Hawes, C.; del Río, L.A.; Sandalio, L.M. Peroxisome dynamics in Arabidopsis plants under oxidative stress induced by cadmium. Free Radic Biol. Med. 2009, 47, 1632–1639. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Gene ID | CDS Size (bp) | Number of Amino Acids (aa) | Molecular Weight (kDa) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|---|---|
| ClDIR1 | Cla97C01G006790 | 561 | 186 | 20.36 | 8.89 | 29.32 | 90.70 | 0.118 |
| ClDIR2 | Cla97C02G028400 | 549 | 182 | 19.88 | 6.71 | 24.00 | 96.98 | 0.332 |
| ClDIR3 | Cla97C02G035380 | 525 | 174 | 18.44 | 9.64 | 9.42 | 99.77 | 0.272 |
| ClDIR4 | Cla97C02G037290 | 576 | 191 | 21.00 | 9.10 | 30.27 | 86.75 | −0.127 |
| ClDIR5 | Cla97C02G037300 | 555 | 184 | 19.80 | 9.19 | 30.59 | 92.66 | 0.147 |
| ClDIR6 | Cla97C02G039150 | 1191 | 396 | 41.46 | 4.36 | 43.18 | 79.34 | −0.140 |
| ClDIR7 | Cla97C02G039160 | 738 | 245 | 27.50 | 9.63 | 33.34 | 68.86 | −0.329 |
| ClDIR8 | Cla97C02G050020 | 570 | 189 | 20.91 | 9.39 | 28.23 | 94.87 | 0.145 |
| ClDIR9 | Cla97C02G050030 | 564 | 187 | 20.45 | 10.04 | 29.08 | 101.18 | 0.167 |
| ClDIR10 | Cla97C03G055500 | 1200 | 399 | 43.45 | 6.14 | 52.00 | 77.74 | −0.337 |
| ClDIR11 | Cla97C05G084350 | 753 | 250 | 25.62 | 5.00 | 36.26 | 88.60 | 0.224 |
| ClDIR12 | Cla97C05G088170 | 549 | 182 | 20.06 | 5.41 | 38.82 | 93.63 | 0.116 |
| ClDIR13 | Cla97C06G110310 | 603 | 200 | 22.53 | 6.51 | 42.14 | 89.80 | −0.007 |
| ClDIR14 | Cla97C06G124580 | 564 | 187 | 20.62 | 8.47 | 44.13 | 86.52 | 0.016 |
| ClDIR15 | Cla97C06G124950 | 552 | 183 | 20.04 | 7.89 | 36.89 | 92.19 | 0.236 |
| ClDIR16 | Cla97C07G134890 | 573 | 190 | 21.15 | 8.38 | 22.75 | 83.74 | 0.127 |
| ClDIR17 | Cla97C07G134900 | 579 | 192 | 21.16 | 7.74 | 44.41 | 86.35 | 0.169 |
| ClDIR18 | Cla97C07G134920 | 558 | 185 | 20.59 | 8.95 | 32.29 | 84.43 | 0.039 |
| ClDIR19 | Cla97C09G181680 | 951 | 316 | 32.50 | 4.95 | 34.91 | 80.22 | 0.002 |
| ClDIR20 | Cla97C09G184270 | 546 | 181 | 20.24 | 9.69 | 38.17 | 85.75 | −0.223 |
| ClDIR21 | Cla97C10G184750 | 582 | 193 | 21.15 | 7.34 | 27.10 | 94.87 | 0.172 |
| ClDIR22 | Cla97C10G200650 | 579 | 192 | 20.89 | 9.69 | 54.36 | 93.44 | 0.223 |
| Duplicated Gene Pairs | Ka | Ks | Ka/Ks | Duplication | Selection Pattern |
|---|---|---|---|---|---|
| ClDIR4/ClDIR5 | 0.48 | 1.33 | 0.36 | Tandem | Purifying selection |
| ClDIR8/ClDIR9 | 0.29 | 1.09 | 0.26 | Tandem | Purifying selection |
| ClDIR16/ClDIR17 | 0.19 | 0.55 | 0.35 | Tandem | Purifying selection |
| ClDIR2/ClDIR3 | 0.43 | 2.06 | 0.21 | Segmental | Purifying selection |
| ClDIR6/ClDIR7 | 1.06 | 0.83 | 1.28 | Tandem | Positive selection |
| Project | No. | Experiment | Accession Number | Sampled Tissue | Reference |
|---|---|---|---|---|---|
| Tissue-specific expression | 1 | Different tissues | PRJNA1031825 | Root, stem, leaf, male flower, female flower, fruit, tendril | - |
| 2 | Fruit development | PRJNA407607 | Fruit flesh | [77] | |
| 3 | Fruit development | PRJNA718123 | Fruit flesh | - | |
| 4 | Fruit development | PRJNA703434 | Fruit flesh | [78] | |
| 5 | Fruit development | PRJNA520808 | Fruit flesh | [79] | |
| Abiotic stresses | 6 | High temperature | PRJNA504354 | Ovule | - |
| 7 | Drought | PRJNA604984 | Leaf | [80] | |
| 8 | Low light | PRJNA602124 | Fruit flesh | [81] | |
| 9 | Salt | PRJNA609260 | Leaf | [82] | |
| 10 | Osmotic stress | PRJNA770012 | Leaf | [83] | |
| 11 | Nitrogen treatment | PRJNA422970 | Leaf, root | [84] | |
| 12 | Cadmium stress | PRJNA1079538 | Leaf | - | |
| Biotic stresses | 13 | Fusarium wilt | PRJNA641525 | Root | [85] |
| 14 | Fusarium wilt | PRJNA794199 | Root | [86] | |
| 15 | Fusarium wilt | PRJNA973274 | Root | - | |
| 16 | Fusarium wilt | PRJNA929333 | Leaf | [87] | |
| 17 | Fusarium wilt | PRJNA783543 | Root | [88] | |
| 18 | Cucumber green mottle mosaic virus | PRJNA534308 | Leaf | - | |
| 19 | Powdery mildew | PRJNA881394 | Leaf | [89] | |
| 20 | Squash vein yellowing virus | PRJNA1086032 | Leaf | - | |
| 21 | Root-knot nematodes | PRJCA001078 | Leaf | [90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Wang, Z.; Tian, H.; Gao, J.; Cui, R.; Shu, Y.; Ding, Q.; Jia, L.; Yan, C. The DIR Gene Family in Watermelon: Evolution, Stress Expression Profiles, and Functional Exploration of ClDIR8. Int. J. Mol. Sci. 2025, 26, 7730. https://doi.org/10.3390/ijms26167730
Zhang K, Wang Z, Tian H, Gao J, Cui R, Shu Y, Ding Q, Jia L, Yan C. The DIR Gene Family in Watermelon: Evolution, Stress Expression Profiles, and Functional Exploration of ClDIR8. International Journal of Molecular Sciences. 2025; 26(16):7730. https://doi.org/10.3390/ijms26167730
Chicago/Turabian StyleZhang, Kaijing, Zhu Wang, Huiyu Tian, Jiong Gao, Rongjing Cui, Yingjie Shu, Qiangqiang Ding, Li Jia, and Congsheng Yan. 2025. "The DIR Gene Family in Watermelon: Evolution, Stress Expression Profiles, and Functional Exploration of ClDIR8" International Journal of Molecular Sciences 26, no. 16: 7730. https://doi.org/10.3390/ijms26167730
APA StyleZhang, K., Wang, Z., Tian, H., Gao, J., Cui, R., Shu, Y., Ding, Q., Jia, L., & Yan, C. (2025). The DIR Gene Family in Watermelon: Evolution, Stress Expression Profiles, and Functional Exploration of ClDIR8. International Journal of Molecular Sciences, 26(16), 7730. https://doi.org/10.3390/ijms26167730








