Genomic Insights into ARR Genes: Key Role in Cotton Leaf Abscission Formation
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of ARR Family Members
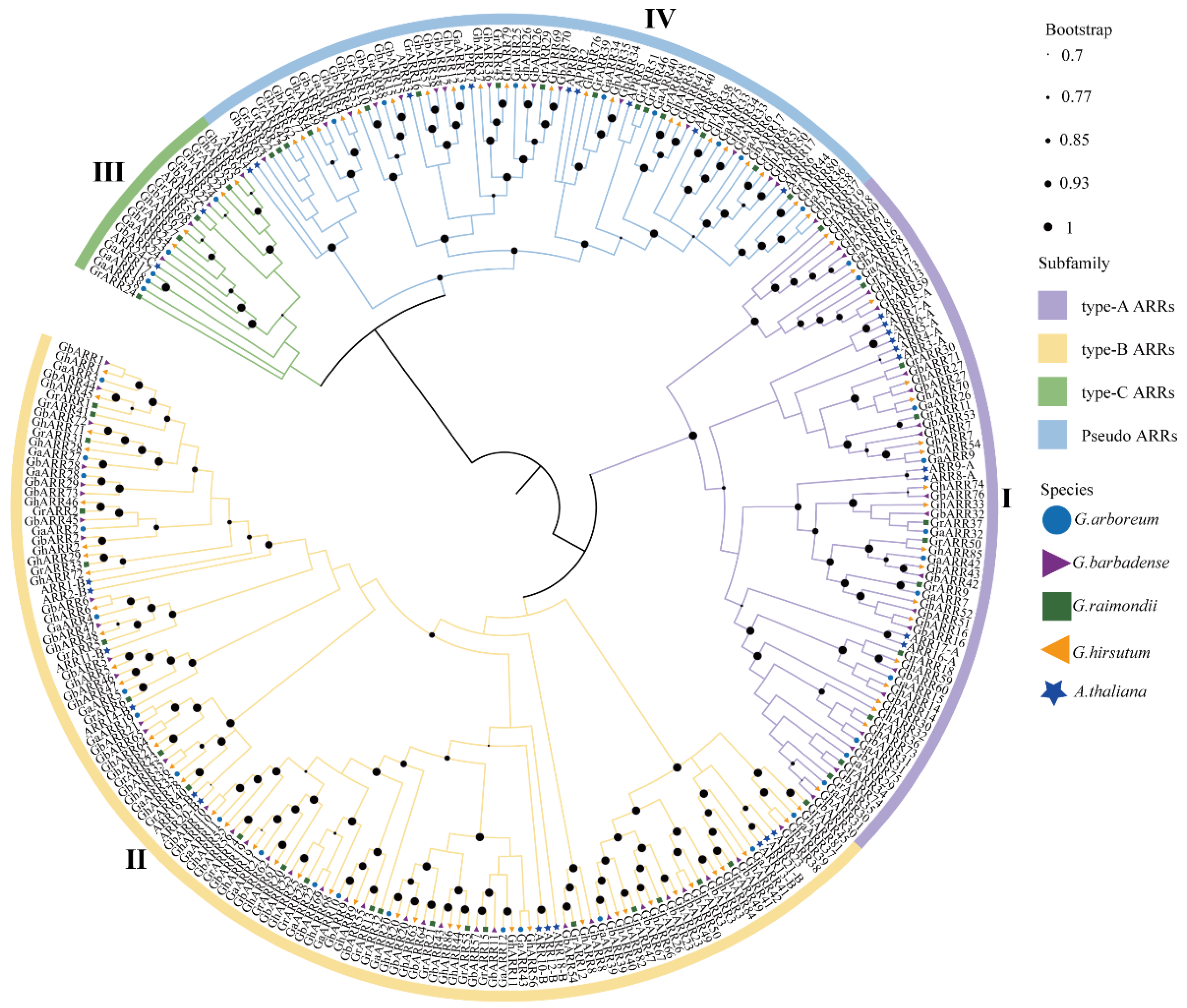
2.2. Phylogenetic Analysis of the ARR Gene Family
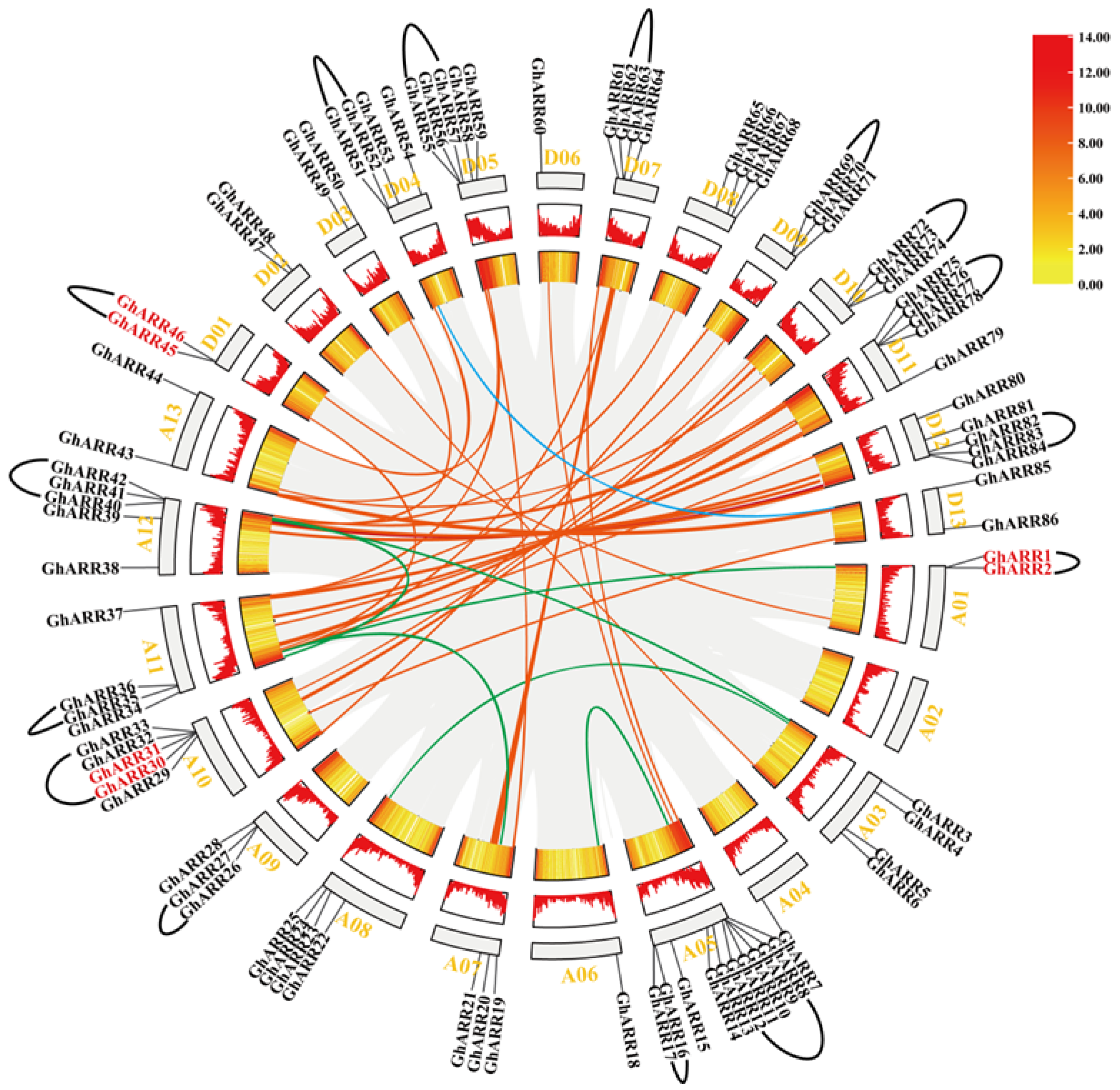
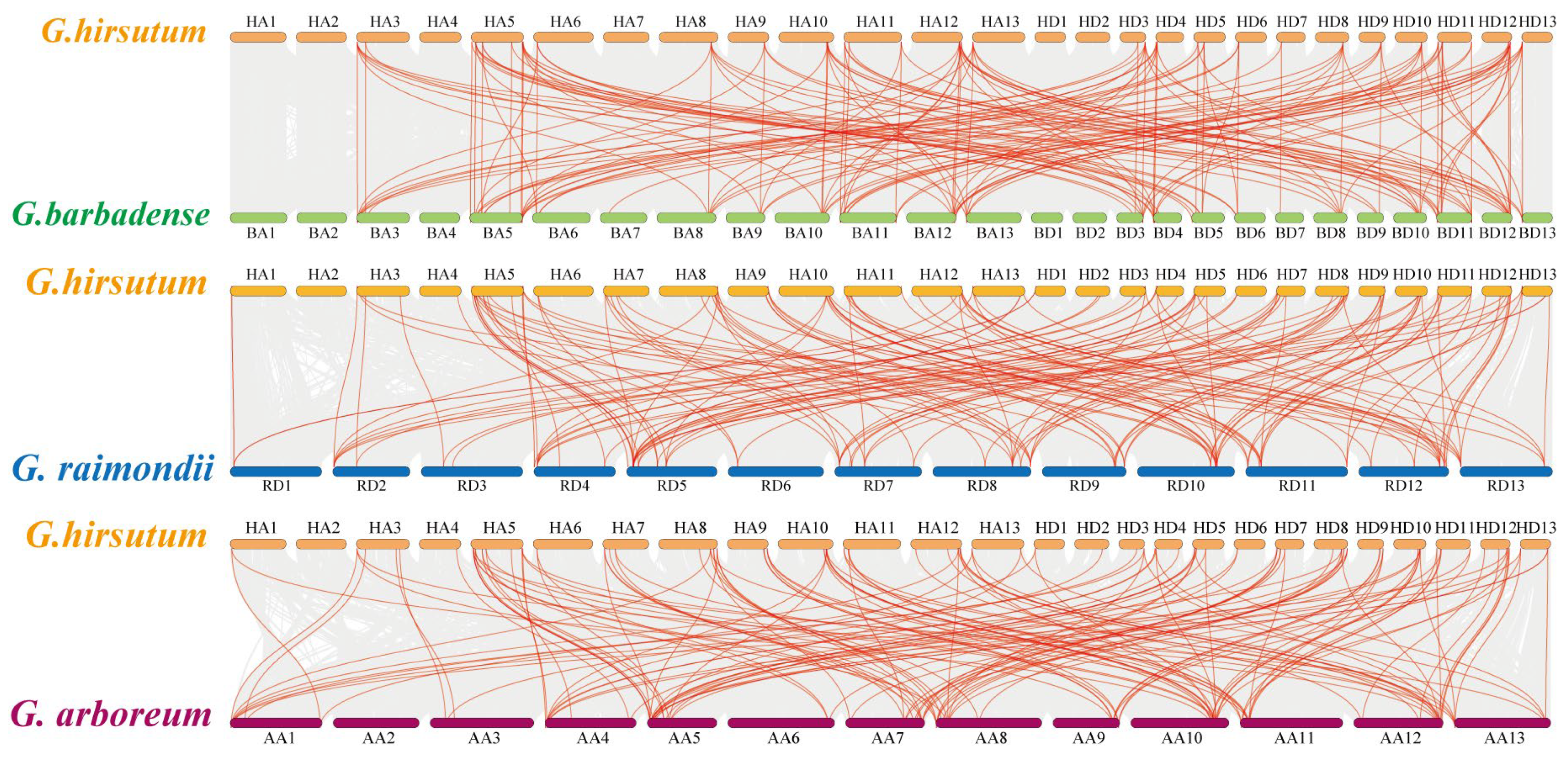
2.3. Chromosomal Distribution and Synteny Analysis of ARR Genes
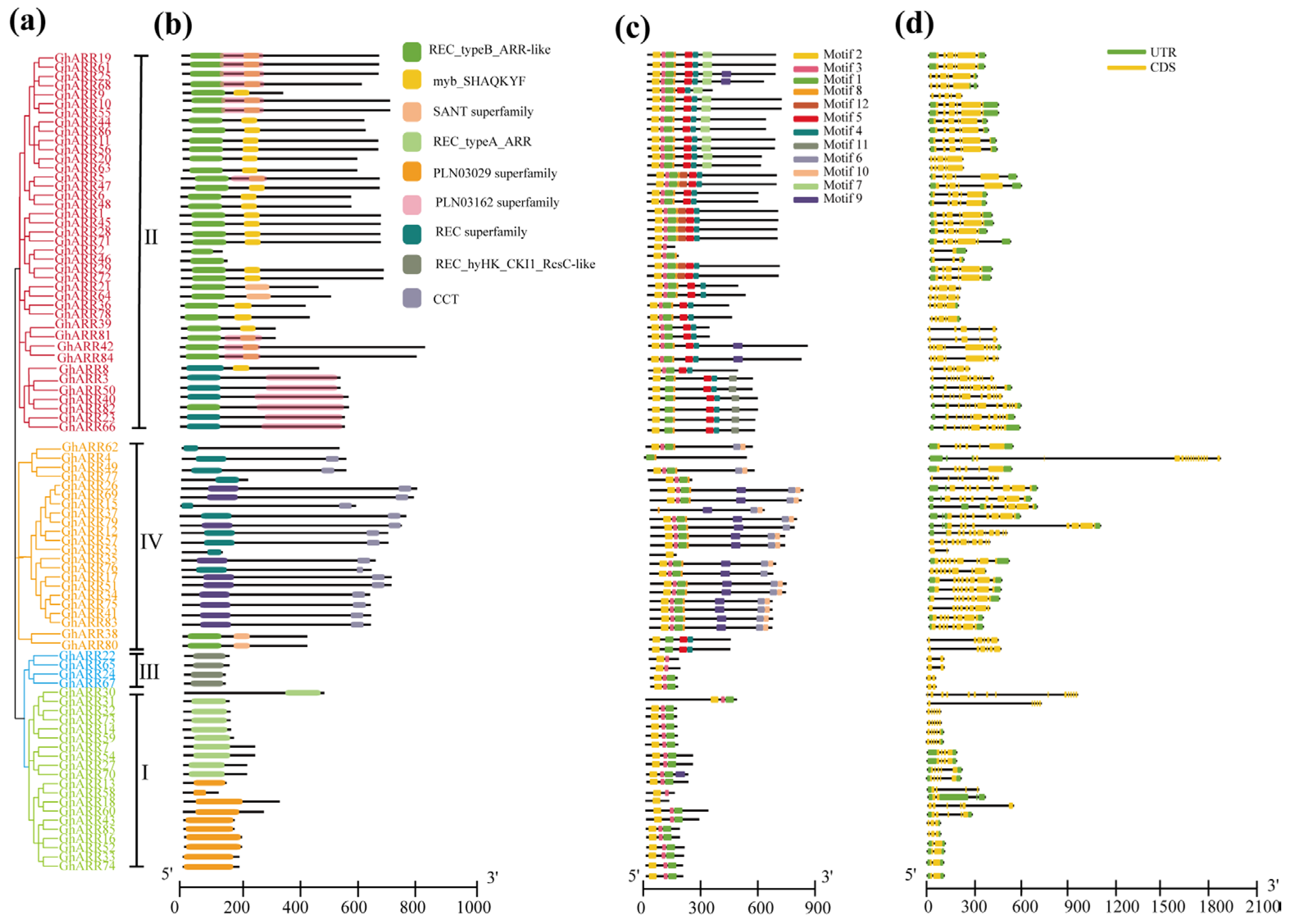
2.4. Gene Structure and Conserved Motif Analysis
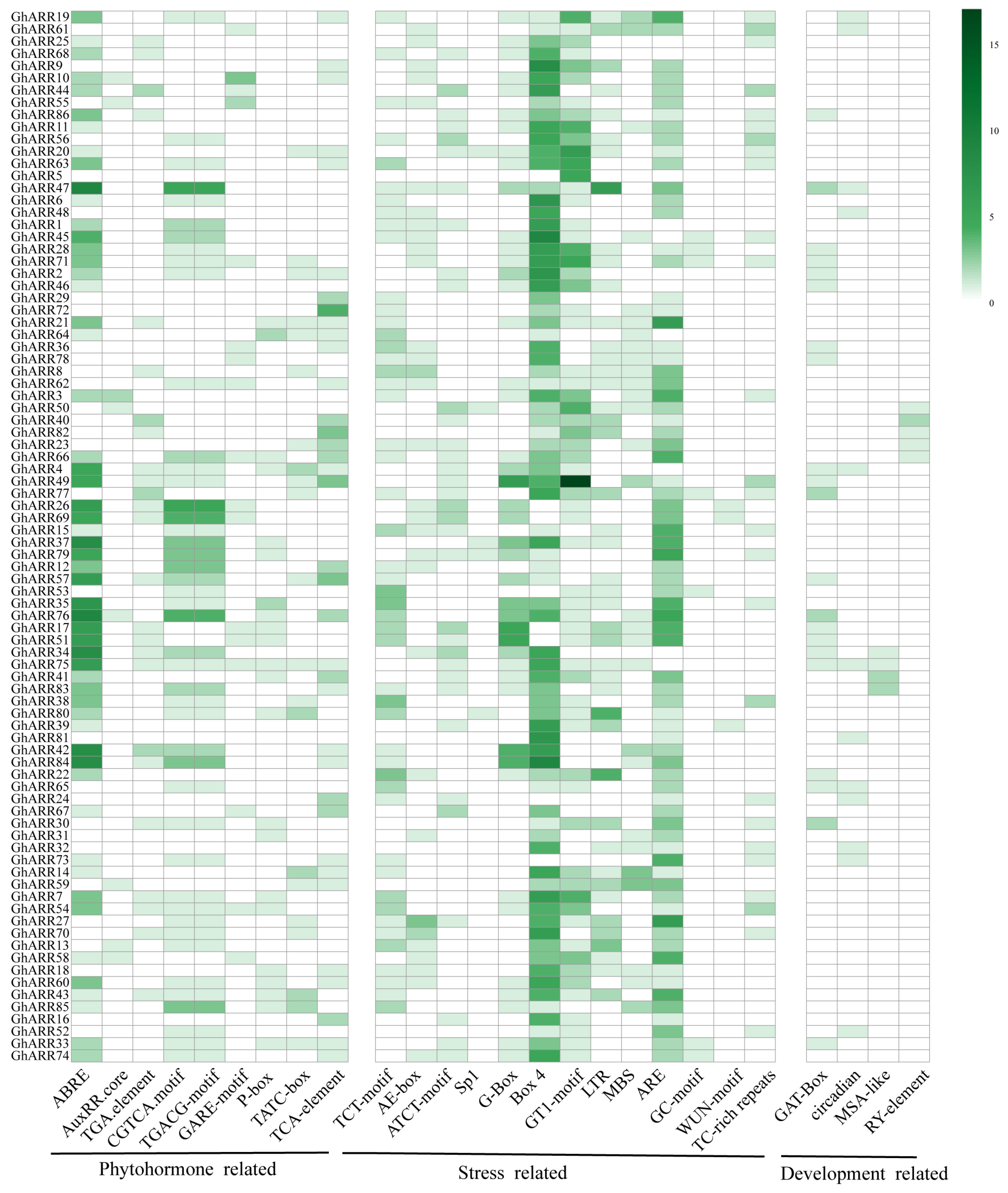
2.5. Analysis of Promoter Cis-Acting Elements
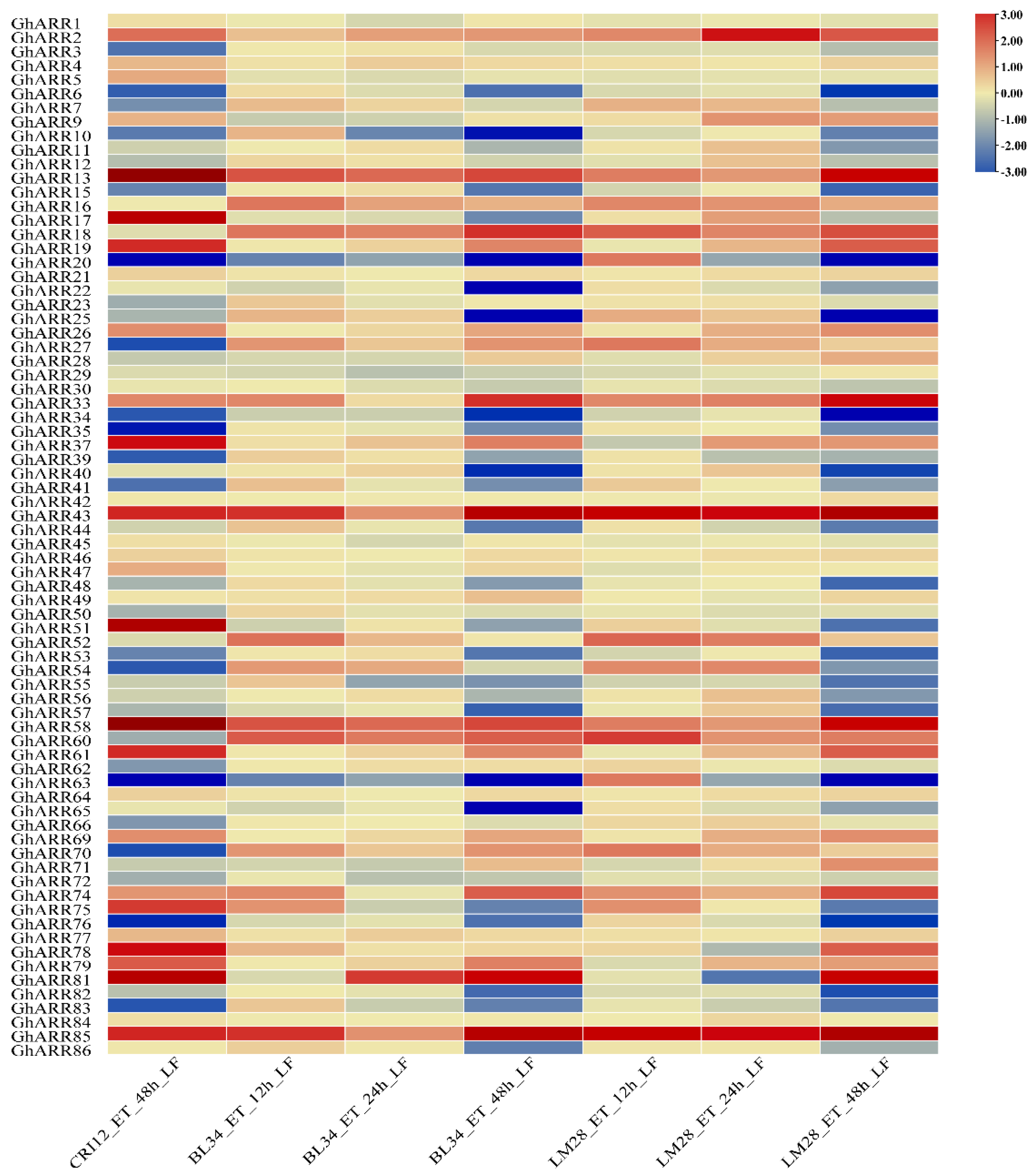
2.6. Expression Profiles of GhARR Genes Under TDZ Stress
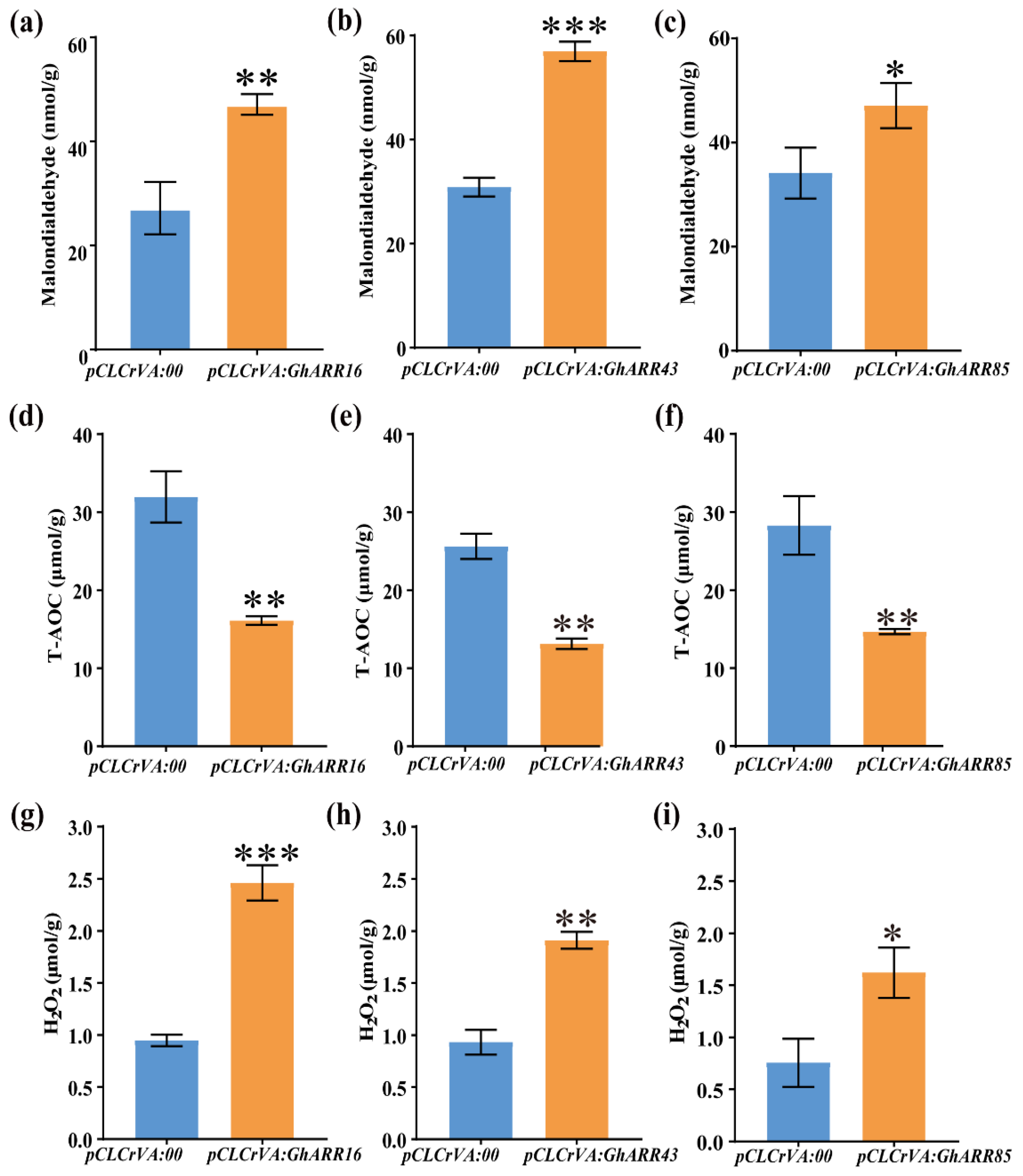
2.7. Functional Analysis of GhARR16, GhARR43 and GhARR85 Gene Silencing in Cotton Under TDZ Stress
3. Discussion
3.1. Diverse Characterization of ARR in G. hirsutum
3.2. Evolution and Expansion of the ARR Gene Family in Cotton
3.3. The Potential Roles of ARR Genes in Response to TDZ Stress
4. Materials and Methods
4.1. Identification and Sequence Analysis of the ARR Gene Family
4.2. Phylogenetic Analysis
4.3. Analysis of Chromosomal Location and Collinearity of ARR Family Genes in G. hirsutum
4.4. Conserved Domain, Gene Structure, and Motif Analysis of the ARRs
4.5. Expression Analysis of GhARRs
4.6. Plant Materials and Growing Conditions
4.7. RNA Extraction and qRT-PCR Analysis
4.8. Virus-Induced Gene Silencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patharkar, O.R.; Walker, J.C. Advances in Abscission Signaling. J. Exp. Bot. 2018, 69, 733–740. [Google Scholar] [CrossRef]
- Yu, Y.; Leyva, P.; Tavares, R.L.; Kellogg, E.A. The Anatomy of Abscission Zones Is Diverse among Grass Species. Am. J Bot. 2020, 107, 549–561. [Google Scholar] [CrossRef]
- Geitmann, A. Bracing for Abscission. Cell 2018, 173, 1320–1322. [Google Scholar] [CrossRef]
- Li, J.; Su, S. Abscission in Plants: From Mechanism to Applications. Adv. Biotechnol. 2024, 2, 27. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Li, Q.; Tan, Y.; Wang, S.; Liu, Y.; Liu, Y. Advances in Understanding the Mechanisms of Organ Abscission in Vivo and in Vitro Plants. Plant Growth Regul. 2024, 103, 293–306. [Google Scholar] [CrossRef]
- Gan, S.; Amasino, R.M. Inhibition of Leaf Senescence by Autoregulated Production of Cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef] [PubMed]
- Hallmark, H.T.; Rashotte, A.M. Cytokinin Isopentenyladenine and Its Glucoside isopentenyladenine-9G Delay Leaf Senescence through Activation of Cytokinin-associated Genes. Plant Direct 2020, 4, e00292. [Google Scholar] [CrossRef]
- Zwack, P.J.; Robinson, B.R.; Risley, M.G.; Rashotte, A.M. Cytokinin Response Factor 6 Negatively Regulates Leaf Senescence and Is Induced in Response to Cytokinin and Numerous Abiotic Stresses. Plant Cell Physiol. 2013, 54, 971–981. [Google Scholar] [CrossRef]
- Kiba, T.; Aoki, K.; Sakakibara, H.; Mizuno, T. Arabidopsis Response Regulator, ARR22, Ectopic Expression of Which Results in Phenotypes Similar to the Wol Cytokinin-Receptor Mutant. Plant Cell Physiol. 2004, 45, 1063–1077. [Google Scholar] [CrossRef]
- Kiba, T.; Yamada, H.; Mizuno, T. Characterization of the ARR15 and ARR16 Response Regulators with Special Reference to the Cytokinin Signaling Pathway Mediated by the AHK4 Histidine Kinase in Roots of Arabidopsis Thaliana. Plant Cell Physiol. 2002, 43, 1059–1066. [Google Scholar] [CrossRef]
- Wurgler-Murphy, S. Two-Component Signal Transducers and MAPK Cascades. Trends Biochem. Sci. 1997, 22, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Chen, H.-C.; Sheen, J. Two-Component Signal Transduction Pathways in Arabidopsis. Plant Physiol. 2002, 129, 500. [Google Scholar] [CrossRef] [PubMed]
- Grefen, C.; Harter, K. Plant Two-Component Systems: Principles, Functions, Complexity and Cross Talk. Planta 2004, 219, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Hwang, I. Attenuation of Cytokinin Signaling via Proteolysis of a Type-B Response Regulator. Plant Signal. Behav. 2012, 7, 756–759. [Google Scholar] [CrossRef]
- Schaller, G.E.; Kieber, J.J.; Shiu, S.-H. Two-Component Signaling Elements and Histidyl-Aspartyl Phosphorelays. Arab. Book 2008, 6, e0112. [Google Scholar] [CrossRef]
- He, L.; Zhang, F.; Wu, X.; Hu, Y.; Dong, L.; Dewitte, W.; Wen, B. Genome-Wide Characterization and Expression of Two-Component System Genes in Cytokinin-Regulated Gall Formation in Zizania Latifolia. Plants 2020, 9, 1409. [Google Scholar] [CrossRef]
- Zhang, W.; To, J.P.C.; Cheng, C.; Eric Schaller, G.; Kieber, J.J. Type-A Response Regulators Are Required for Proper Root Apical Meristem Function through Post-transcriptional Regulation of PIN Auxin Efflux Carriers. Plant J. 2011, 68, 1–10. [Google Scholar] [CrossRef]
- Hill, K.; Mathews, D.E.; Kim, H.J.; Street, I.H.; Wildes, S.L.; Chiang, Y.-H.; Mason, M.G.; Alonso, J.M.; Ecker, J.R.; Kieber, J.J.; et al. Functional Characterization of Type-B Response Regulators in the Arabidopsis Cytokinin Response. Plant Physiol. 2013, 162, 212–224. [Google Scholar] [CrossRef]
- Shanks, C.M.; Hecker, A.; Cheng, C.; Brand, L.; Collani, S.; Schmid, M.; Schaller, G.E.; Wanke, D.; Harter, K.; Kieber, J.J. Role of BASIC PENTACYSTEINE Transcription Factors in a Subset of Cytokinin Signaling Responses. Plant J. 2018, 95, 458–473. [Google Scholar] [CrossRef]
- Veerabagu, M.; Elgass, K.; Kirchler, T.; Huppenberger, P.; Harter, K.; Chaban, C.; Mira-Rodado, V. The Arabidopsis B-type Response Regulator 18 Homomerizes and Positively Regulates Cytokinin Responses. Plant J. 2012, 72, 721–731. [Google Scholar] [CrossRef]
- Cho, L.-H.; Yoon, J.; Pasriga, R.; An, G. Homodimerization of Ehd1 Is Required to Induce Flowering in Rice. Plant Physiol 2016, 170, 2159–2171. [Google Scholar] [CrossRef]
- Salomé, P.A.; To, J.P.C.; Kieber, J.J.; McClung, C.R. Arabidopsis Response Regulators ARR3 and ARR4 Play Cytokinin-Independent Roles in the Control of Circadian Period. Plant Cell 2006, 18, 55–69. [Google Scholar] [CrossRef]
- Buechel, S.; Leibfried, A.; To, J.P.C.; Zhao, Z.; Andersen, S.U.; Kieber, J.J.; Lohmann, J.U. Role of A-Type ARABIDOPSIS RESPONSE REGULATORS in Meristem Maintenance and Regeneration. Eur. J. Cell Biol. 2010, 89, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, X.; Gong, Z.; Yang, S.; Shi, Y. ABI4 Represses the Expression of Type-a ARRs to Inhibit Seed Germination in Arabidopsis. Plant J. 2017, 89, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Dutta, S.; Chattopadhyay, S. MYC2 Regulates ARR16, a Component of Cytokinin Signaling Pathways, in Arabidopsis Seedling Development. Plant Direct 2019, 3, e00177. [Google Scholar] [CrossRef] [PubMed]
- Vatén, A.; Soyars, C.L.; Tarr, P.T.; Nimchuk, Z.L.; Bergmann, D.C. Modulation of Asymmetric Division Diversity through Cytokinin and SPEECHLESS Regulatory Interactions in the Arabidopsis Stomatal Lineage. Dev. Cell 2018, 47, 53–66.e5. [Google Scholar] [CrossRef]
- Nguyen, K.H.; Ha, C.V.; Nishiyama, R.; Watanabe, Y.; Leyva-González, M.A.; Fujita, Y.; Tran, U.T.; Li, W.; Tanaka, M.; Seki, M.; et al. Arabidopsis Type B Cytokinin Response Regulators ARR1, ARR10, and ARR12 Negatively Regulate Plant Responses to Drought. Proc. Natl. Acad. Sci. USA 2016, 113, 3090–3095. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J. Arabidopsis Response Regulator1 and Arabidopsis Histidine Phosphotransfer Protein2 (AHP2), AHP3, and AHP5 Function in Cold Signaling. Plant Physiol. 2012, 161, 408–424. [Google Scholar] [CrossRef]
- Du, M.; Li, Y.; Tian, X.; Duan, L.; Zhang, M.; Tan, W.; Xu, D.; Li, Z. The Phytotoxin Coronatine Induces Abscission-Related Gene Expression and Boll Ripening during Defoliation of Cotton. PLoS ONE 2014, 9, e97652. [Google Scholar] [CrossRef]
- Du, M.; Ren, X.; Tian, X.; Duan, L.; Zhang, M.; Tan, W.; Li, Z. Evaluation of Harvest Aid Chemicals for the Cotton-Winter Wheat Double Cropping System. J. Integr. Agric. 2013, 12, 273–282. [Google Scholar] [CrossRef]
- Nisler, J.; Kopečný, D.; Končitíková, R.; Zatloukal, M.; Bazgier, V.; Berka, K.; Zalabák, D.; Briozzo, P.; Strnad, M.; Spíchal, L. Novel Thidiazuron-Derived Inhibitors of Cytokinin Oxidase/Dehydrogenase. Plant Mol Biol 2016, 92, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chen, L.; Sun, H.; Wusiman, N.; Sun, W.; Li, B.; Gao, Y.; Kong, J.; Zhang, D.; Zhang, X.; et al. Crosstalk between Cytokinin and Ethylene Signaling Pathways Regulates Leaf Abscission in Cotton in Response to Chemical Defoliants. J. Exp. Bot. 2019, 70, 1525–1538. [Google Scholar] [CrossRef] [PubMed]
- Jameson, P.E.; Song, J. Cytokinin: A Key Driver of Seed Yield. J. Exp. Bot. 2016, 67, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Heyl, A.; Brault, M.; Frugier, F.; Kuderova, A.; Lindner, A.-C.; Motyka, V.; Rashotte, A.M.; Schwartzenberg, K.V.; Vankova, R.; Schaller, G.E. Nomenclature for Members of the Two-Component Signaling Pathway of Plants. Plant Physiol. 2013, 161, 1063–1065. [Google Scholar] [CrossRef]
- Mizuno, T. Plant Response Regulators Implicated in Signal Transduction and Circadian Rhythm. Curr. Opin. Plant Biol. 2004, 7, 499–505. [Google Scholar] [CrossRef]
- Schaller, G.E.; Doi, K.; Hwang, I.; Kieber, J.J.; Khurana, J.P.; Kurata, N.; Mizuno, T.; Pareek, A.; Shiu, S.-H.; Wu, P.; et al. Nomenclature for Two-Component Signaling Elements of Rice. Plant Physiol. 2007, 143, 555–557. [Google Scholar] [CrossRef]
- Pils, B.; Heyl, A. Unraveling the Evolution of Cytokinin Signaling. Plant Physiol. 2009, 151, 782–791. [Google Scholar] [CrossRef]
- Ito, Y.; Kurata, N. Identification and Characterization of Cytokinin-Signalling Gene Families in Rice. Gene 2006, 382, 57–65. [Google Scholar] [CrossRef]
- Asakura, Y.; Hagino, T.; Ohta, Y.; Aoki, K.; Yonekura-Sakakibara, K.; Deji, A.; Yamaya, T.; Sugiyama, T. Molecular Characterization of His-Asp Phosphorelay Signaling Factors in Maize Leaves: Implications of the Signal Divergence by Cytokinin-Inducible Response Regulators in the Cytosol and the Nuclei. Plant Mol. Biol. 2003, 52, 331–341. [Google Scholar] [CrossRef]
- Gahlaut, V.; Mathur, S.; Dhariwal, R.; Khurana, J.P.; Tyagi, A.K.; Balyan, H.S.; Gupta, P.K. A Multi-Step Phosphorelay Two-Component System Impacts on Tolerance against Dehydration Stress in Common Wheat. Funct. Integr. Genom. 2014, 14, 707–716. [Google Scholar] [CrossRef]
- Hosoda, K.; Imamura, A.; Katoh, E.; Hatta, T.; Tachiki, M.; Yamada, H.; Mizuno, T.; Yamazaki, T. Molecular Structure of the GARP Family of Plant Myb-Related DNA Binding Motifs of the Arabidopsis Response Regulators. Plant Cell 2002, 14, 2015–2029. [Google Scholar] [CrossRef]
- Chen, C.; Liu, A.; Ren, H.; Yu, Y.; Duanmu, H.; Duan, X.; Sun, X.; Liu, B.; Zhu, Y. Genome-Wide Analysis of Glycine Soja Response Regulator GsRR Genes under Alkali and Salt Stresses. Front. Plant Sci. 2018, 9, 1306. [Google Scholar] [CrossRef]
- Chu, Z.X.; Ma, Q.; Lin, Y.X.; Tang, X.L.; Zhou, Y.Q.; Zhu, S.W.; Fan, J.; Cheng, B.J. Genome-Wide Identification, Classification, and Analysis of Two-Component Signal System Genes in Maize. Genet. Mol. Res. 2011, 10, 3316–3330. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Xu, Y.; Tian, S.; Unver, T.; Liu, Z.; Zhou, Z.; Cai, X.; Wang, K.; Wei, Y.; Liu, Y.; et al. Evolutionary Divergence of Duplicated Genomes in Newly Described Allotetraploid Cottons. Proc. Natl. Acad. Sci. USA 2022, 119, e2208496119. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Li, X.; Gao, Y.; Xie, W.; Zhang, L.; Song, J.; Li, S.; Zhao, Z. Genome-Wide Identification of the RR Gene Family and Its Expression Analysis in Response to TDZ Induction in Rhododendron Delavayi. Plants 2023, 12, 3250. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, G.; Shen, H.; Li, Z.; Li, H.; Xiao, G. Genome-Wide Identification and Expression Profiles Analysis of the Authentic Response Regulator Gene Family in Licorice. Front. Plant Sci. 2024, 14, 1309802. [Google Scholar] [CrossRef]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene Duplication as a Major Force in Evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, J.; Long, M.; Fan, C. gKaKs: The Pipeline for Genome-Level Ka/Ks Calculation. Bioinformatics 2013, 29, 645–646. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and Validation of Promoters and Cis-Acting Regulatory Elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Zhang, B.; Yue, D.; Han, B.; Bao, D.; Zhang, X.; Hao, X.; Lin, X.; Lindsey, K.; Zhu, L.; Jin, S.; et al. RAPID LEAF FALLING 1 Facilitates Chemical Defoliation and Mechanical Harvesting in Cotton. Mol. Plant 2025, 18, 765–782. [Google Scholar] [CrossRef]
- Pokimica, N.; Ćosić, T.; Uzelac, B.; Ninković, S.; Raspor, M. Dissecting the Roles of the Cytokinin Signaling Network: The Case of de Novo Shoot Apical Meristem Formation. Biomolecules 2024, 14, 381. [Google Scholar] [CrossRef]
- Li, F.; Wu, Q.; Liao, B.; Yu, K.; Huo, Y.; Meng, L.; Wang, S.; Wang, B.; Du, M.; Tian, X.; et al. Thidiazuron Promotes Leaf Abscission by Regulating the Crosstalk Complexities between Ethylene, Auxin, and Cytokinin in Cotton. IJMS 2022, 23, 2696. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as Designed by Its Users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An Online Tool for Phylogenetic Tree Display and Annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Wang, X.; Sun, Y.; Joseph, P.V.; Paterson, A.H. Detection of Colinear Blocks and Synteny and Evolutionary Analyses Based on Utilization of MCScanX. Nat. Protoc. 2024, 19, 2206–2229. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Jin, D.; Wang, X.; Xu, Y.; Gui, H.; Zhang, H.; Dong, Q.; Sikder, R.K.; Yang, G.; Song, M. Chemical Defoliant Promotes Leaf Abscission by Altering ROS Metabolism and Photosynthetic Efficiency in Gossypium hirsutum. IJMS 2020, 21, 2738. [Google Scholar] [CrossRef]
- Li, H.; Wang, X.; Qin, N.; Hu, D.; Jia, Y.; Sun, G.; He, L.; Zhang, H.; Dai, P.; Peng, Z.; et al. Genomic Loci Associated with Leaf Abscission Contribute to Machine Picking and Environmental Adaptability in Upland Cotton (Gossypium hirsutum L.). J. Adv. Res. 2024, 58, 31–43. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, H.; Wang, Z.; Zhang, Y.; Cheng, G.; Huang, P.; Yang, L.; Tan, S.; Cao, X.; Pei, X.; Liang, Y.; et al. Genomic Insights into ARR Genes: Key Role in Cotton Leaf Abscission Formation. Int. J. Mol. Sci. 2025, 26, 7161. https://doi.org/10.3390/ijms26157161
Shi H, Wang Z, Zhang Y, Cheng G, Huang P, Yang L, Tan S, Cao X, Pei X, Liang Y, et al. Genomic Insights into ARR Genes: Key Role in Cotton Leaf Abscission Formation. International Journal of Molecular Sciences. 2025; 26(15):7161. https://doi.org/10.3390/ijms26157161
Chicago/Turabian StyleShi, Hongyan, Zhenyu Wang, Yuzhi Zhang, Gongye Cheng, Peijun Huang, Li Yang, Songjuan Tan, Xiaoyu Cao, Xiaoyu Pei, Yu Liang, and et al. 2025. "Genomic Insights into ARR Genes: Key Role in Cotton Leaf Abscission Formation" International Journal of Molecular Sciences 26, no. 15: 7161. https://doi.org/10.3390/ijms26157161
APA StyleShi, H., Wang, Z., Zhang, Y., Cheng, G., Huang, P., Yang, L., Tan, S., Cao, X., Pei, X., Liang, Y., Gao, Y., Ren, X., Chen, Q., & Ma, X. (2025). Genomic Insights into ARR Genes: Key Role in Cotton Leaf Abscission Formation. International Journal of Molecular Sciences, 26(15), 7161. https://doi.org/10.3390/ijms26157161




