HAT-PCR Enables Sensitive Quantification of Minimal Residual Disease in Chronic Lymphocytic Leukemia and Myeloma
Abstract
1. Introduction
2. Results
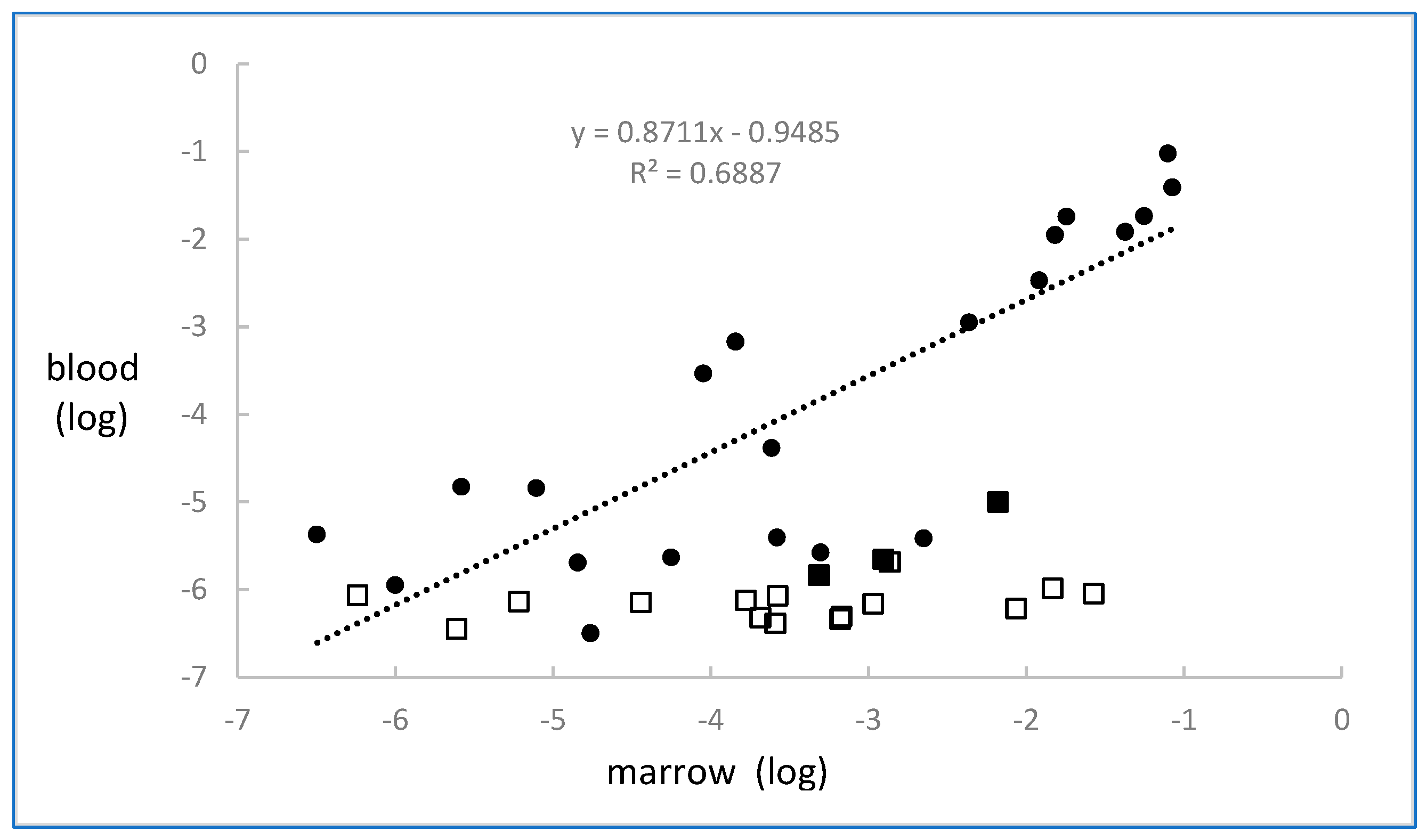
2.1. CLL
2.2. Myeloma
3. Discussion
4. Materials and Methods
4.1. Samples
4.2. Quantification by Flow Cytometry
4.3. Quantification by HAT-PCR
4.3.1. Determining the Sequence
4.3.2. HAT-PCR
- One to four A/T bases at the 3′ end of both the allele-specific oligonucleotide (ASO) and J primers.
- Annealing temperature within 3 °C of critical annealing temperature (Tc, the highest annealing temperature which still gives efficient amplification).
- A relatively high concentration of Taq polymerase.
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brisco, M.J.; Condon, J.; Hughes, E.; Neoh, S.H.; Sykes, P.J.; Seshadri, R.; Morley, A.A.; Toogood, I.; Waters, K.; Tauro, G.; et al. Outcome prediction in childhood acute lymphoblastic leukemia by molecular quantification of residual disease at the end of induction. Lancet 1994, 343, 196–200. [Google Scholar] [CrossRef]
- van der Velden, V.H.; Cazzaniga, G.; Schrauder, A.; Hancock, J.; Bader, P.; Panzer-Grumayer, E.R.; Flohr, T.; Sutton, R.; Cave, H.; Madsen, H.O.; et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: Guidelines for interpretation of real-time quantitative PCR data. Leukemia 2007, 21, 604–611. [Google Scholar] [CrossRef]
- van der Velden, V.H.; van Dongen, J.J. MRD detection in acute lymphoblastic leukemia patients using Ig/TCR gene rearrangements as targets for real-time quantitative PCR. Methods Mol. Biol. 2009, 538, 115–150. [Google Scholar]
- van der Velden, V.H.J.; Dombrink, I.; Alten, J.; Cazzaniga, G.; Clappier, E.; Drandi, D.; Eckert, C.; Fronkova, E.; Hancock, J.; Kotrova, M.; et al. Analysis of measurable residual disease by IG/TR gene rearrangements: Quality assurance and updated EuroMRD guidelines. Leukemia 2024, 38, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Brisco, M.J.; Condon, J.; Sykes, P.J.; Neoh, S.H.; Morley, A.A. Detection and quantitation of neoplastic cells in acute lymphoblastic leukemia, by use of the polymerase chain reaction. Br. J. Haematol. 1991, 79, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Sykes, P.J.; Neoh, S.H.; Brisco, M.J.; Hughes, E.; Condon, J.; Morley, A.A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 1992, 13, 444–449. [Google Scholar] [PubMed]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Della Starza, I.; Nunes, V.; Cavalli, M.; De Novi, L.A.; Ilari, C.; Apicella, V.; Vitale, A.; Testi, A.M.; Del Giudice, I.; Chiaretti, S.; et al. Comparative analysis between RQ-PCR and digital-droplet-PCR of immunoglobulin/T-cell receptor gene rearrangements to monitor minimal residual disease in acute lymphoblastic leukemia. Br. J. Haematol. 2016, 174, 541–549. [Google Scholar] [CrossRef]
- Morley, A.A.; Latham, S.; Brisco, M.J.; Sykes, P.J.; Sutton, R.; Hughes, E.; Wilczek, V.; Budgen, B.; van Zanten, K.; Kuss, B.J.; et al. Sensitive and specific measurement of minimal residual disease in acute lymphoblastic leukemia. J. Mol. Diagn. 2009, 11, 201–210. [Google Scholar] [CrossRef]
- Kotrova, M.; van der Velden, V.H.J.; van Dongen, J.J.M.; Formankova, R.; Sedlacek, P.; Bruggemann, M.; Zuna, J.; Stary, J.; Trka, J.; Fronkova, E. Next-generation sequencing indicates false-positive MRD results and better predicts prognosis after SCT in patients with childhood ALL. Bone Marrow Transplant. 2017, 52, 962–968. [Google Scholar] [CrossRef]
- Rawstron, A.C.; Fazi, C.; Agathangelidis, A.; Villamor, N.; Letestu, R.; Nomdedeu, J.; Palacio, C.; Stehlikova, O.; Kreuzer, K.-A.; Liptrot, S.; et al. A complementary role of multiparameter flow cytometry and high-throughput sequencing for minimal residual disease detection in chronic lymphocytic leukemia: An European Research Initiative on CLL study. Leukemia 2016, 30, 929–936. [Google Scholar] [CrossRef]
- Verbeek, M.W.C.; van der Velden, V.H.J. The Evolving Landscape of Flowcytometric Minimal Residual Disease Monitoring in B-Cell Precursor Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2024, 25, 4881. [Google Scholar] [CrossRef]
- Sherwood, A.M.; Desmarais, C.; Livingston, R.J.; Andriesen, J.; Haussler, M.; Carlson, C.S.; Robins, H. Deep sequencing of the human TCRγ and TCRβ repertoires suggests that TCRβ rearranges after αβ and γδ T cell commitment. Sci. Transl. Med. 2011, 3, 90ra61. [Google Scholar] [CrossRef]
- Faham, M.; Zheng, J.; Moorhead, M.; Carlton, V.E.; Stow, P.; Coustan-Smith, E.; Pui, C.-H.; Campana, D. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood 2012, 120, 5173–5180. [Google Scholar] [CrossRef]
- Ching, T.; Duncan, M.E.; Newman-Eerkes, T.; McWhorter, M.M.E.; Tracy, J.M.; Steen, M.S.; Brown, R.P.; Venkatasubbarao, S.; Akers, N.K.; Vignali, M.; et al. Analytical evaluation of the clonoSEQ Assay for establishing measurable (minimal) residual disease in acute lymphoblastic leukemia, chronic lymphocytic leukemia, and multiple myeloma. BMC Cancer 2020, 20, 612. [Google Scholar] [CrossRef]
- Latham, S.; Hughes, E.; Budgen, B.; Ross, D.; Greenwood, M.; Bradstock, K.; Dalla-Pozza, L.; Huang, L.; Law, T.; Doculara, L.; et al. Sensitive Measurement of Minimal Residual Disease in Blood by HAT-PCR. J. Mol. Diagn. 2022, 24, 632–641. [Google Scholar] [CrossRef]
- Latham, S.; Hughes, E.; Budgen, B.; Morley, A. Inhibition of the PCR by genomic DNA. PLoS ONE 2023, 18, e0284538. [Google Scholar] [CrossRef] [PubMed]
- Della Starza, I.; De Novi, L.A.; Elia, L.; Bellomarino, V.; Beldinanzi, M.; Soscia, R.; Cardinali, D.; Chiaretti, S.; Guarini, A.; Foà, R. Optimizing Molecular Minimal Residual Disease Analysis in Adult Acute Lymphoblastic Leukemia. Cancers 2023, 15, 374. [Google Scholar] [CrossRef]
- Saygin, C.; Cannova, J.; Stock, W.; Muffly, L. Measurable residual disease in acute lymphoblastic leukemia: Methods and clinical context in adult patients. Haematologica 2022, 107, 2783–2793. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Henderson, M.J.; Kwan, E.; Beesley, A.H.; Sutton, R.; Bahar, A.Y.; Giles, J.; Venn, N.C.; Pozza, L.D.; Baker, D.L.; et al. Relapse in children with acute lymphoblastic leukemia involving selection of a preexisting drug-resistant subclone. Blood 2007, 110, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Short, N.J.; Kantarjian, H.; Ravandi, F.; Konopleva, M.; Jain, N.; Kanagal-Shamanna, R.; Patel, K.P.; Macaron, W.; Kadia, T.M.; Wang, S.; et al. High-sensitivity next-generation sequencing MRD assessment in ALL identifies patients at very low risk of relapse. Blood Adv. 2022, 6, 4006–4014. [Google Scholar] [CrossRef] [PubMed]
- Ashouri, K.; Nittur, V.; Ginosyan, A.A.; Hwang, J.; Adnani, B.; Chen, D.; Savitala-Damerla, L.; Schiff, K.; Chaudhary, P.; Kovach, A.E.; et al. Concordance of Next-Generation Sequencing and Multiparametric Flow Cytometry Methods for Detecting Measurable Residual Disease in Adult Acute Lymphoblastic Leukemia: Optimizing Prediction of Clinical Outcomes from a Single-Center Study. Clin. Lymphoma Myeloma Leuk. 2024, 24, e59–e66.e2. [Google Scholar] [CrossRef]
- Yan, N.; Wang, Z.L.; Wang, X.J.; Gale, R.P.; Zhou, Y.L.; Zhao, M.Y.; Wu, L.-X.; Liao, M.-Y.; Yang, J.; Wang, C.-Y.; et al. Measurable residual disease testing by next generation sequencing is more accurate compared with multiparameter flow cytometry in adults with B-cell acute lymphoblastic leukemia. Cancer Lett. 2024, 598, 217104. [Google Scholar] [CrossRef] [PubMed]
- Flores-Montero, J.; Sanoja-Flores, L.; Paiva, B.; Puig, N.; García-Sánchez, O.; Böttcher, S.; Van Der Velden, V.H.J.; Pérez-Morán, J.-J.; Vidriales, M.-B.; García-Sanz, R.; et al. Next Generation Flow for highly sensitive and standardized detection of minimal residual disease in multiple myeloma. Leukemia 2017, 31, 2094–2103. [Google Scholar] [CrossRef]
- Medina, A.; Puig, N.; Flores-Montero, J.; Jimenez, C.; Sarasquete, M.E.; Garcia-Alvarez, M.; Prieto-Conde, I.; Chillon, C.; Alcoceba, M.; Gutierrez, N.C.; et al. Comparison of next-generation sequencing (NGS) and next-generation flow (NGF) for minimal residual disease (MRD) assessment in multiple myeloma. Blood Cancer J. 2020, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- Oliva, S.; Genuardi, E.; Paris, L.; D’AGostino, M.; Rogers, J.; Rota-Scalabrini, D.; Jacob, A.P.; Patriarca, F.; Luppi, M.; Bertazzoni, P.; et al. Prospective evaluation of minimal residual disease in the phase II FORTE trial: A head-to-head comparison between multiparameter flow cytometry and next-generation sequencing. eClinicalMedicine 2023, 60, 102016. [Google Scholar] [CrossRef] [PubMed]
- Yoroidaka, T.; Takamatsu, H.; Urushihara, R.; Itagaki, M.; Yoshihara, S.; Sato, K.; Takezako, N.; Ozaki, S.; Suzuki, K.; Kohno, K.; et al. Prognostic value of minimal residual disease detected by EuroFlow next-generation flow cytometry and next-generation sequencing in patients with multiple myeloma achieving complete response and receiving lenalidomide maintenance after autotransplant: A prospective comparison study. Haematologica 2025. ahead of print. [Google Scholar] [CrossRef]
- Lasa, M.; Notarfranchi, L.; Agullo, C.; Gonzalez, C.; Castro, S.; Perez, J.J.; Burgos, L.; Guerrero, C.; Calasanz, M.J.; Flores-Montero, J.; et al. Minimally Invasive Assessment of Peripheral Residual Disease During Maintenance or Observation in Transplant-Eligible Patients With Multiple Myeloma. J. Clin. Oncol. 2024, 43, 125–132. [Google Scholar] [CrossRef]
- Sartor, M.M.; Gottlieb, D.J. A single tube 10-color flow cytometry assay optimizes detection of minimal residual disease in chronic lymphocytic leukemia. Cytom. B Clin. Cytom. 2013, 84, 96–103. [Google Scholar] [CrossRef]
- Siegel, S. Nonparametric Statistics for the Behavioural Sciences; McGraw-Hill: New York, NY, USA, 1956. [Google Scholar]
- Szczepanski, T.; Willemse, M.J.; van Wering, E.R.; van Weerden, J.F.; Kamps, W.A.; van Dongen, J.J. Precursor-B-ALL with D(H)-J(H) gene rearrangements have an immature immunogenotype with a high frequency of oligoclonality and hyperdiploidy of chromosome 14. Leukemia 2001, 15, 1415–1423. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hughes, E.; Latham, S.; Kuss, B.; Grist, S.; Hall, R.; Khong, T.; Gorniak, M.; Spencer, A.; Tam, C.; Mulligan, S.; et al. HAT-PCR Enables Sensitive Quantification of Minimal Residual Disease in Chronic Lymphocytic Leukemia and Myeloma. Int. J. Mol. Sci. 2025, 26, 7720. https://doi.org/10.3390/ijms26167720
Hughes E, Latham S, Kuss B, Grist S, Hall R, Khong T, Gorniak M, Spencer A, Tam C, Mulligan S, et al. HAT-PCR Enables Sensitive Quantification of Minimal Residual Disease in Chronic Lymphocytic Leukemia and Myeloma. International Journal of Molecular Sciences. 2025; 26(16):7720. https://doi.org/10.3390/ijms26167720
Chicago/Turabian StyleHughes, Elizabeth, Sue Latham, Bryone Kuss, Scott Grist, Rachel Hall, Tiffany Khong, Malgorzata Gorniak, Andrew Spencer, Constantine Tam, Stephen Mulligan, and et al. 2025. "HAT-PCR Enables Sensitive Quantification of Minimal Residual Disease in Chronic Lymphocytic Leukemia and Myeloma" International Journal of Molecular Sciences 26, no. 16: 7720. https://doi.org/10.3390/ijms26167720
APA StyleHughes, E., Latham, S., Kuss, B., Grist, S., Hall, R., Khong, T., Gorniak, M., Spencer, A., Tam, C., Mulligan, S., Bailey, S., Sartor, M., Carney, D., Cull, G., Gottlieb, D., & Morley, A. (2025). HAT-PCR Enables Sensitive Quantification of Minimal Residual Disease in Chronic Lymphocytic Leukemia and Myeloma. International Journal of Molecular Sciences, 26(16), 7720. https://doi.org/10.3390/ijms26167720






