Natural Microbiota of Dogs and Cats as a Source and Vector of Resistance Genes—Clinical Significance
Abstract
1. Introduction
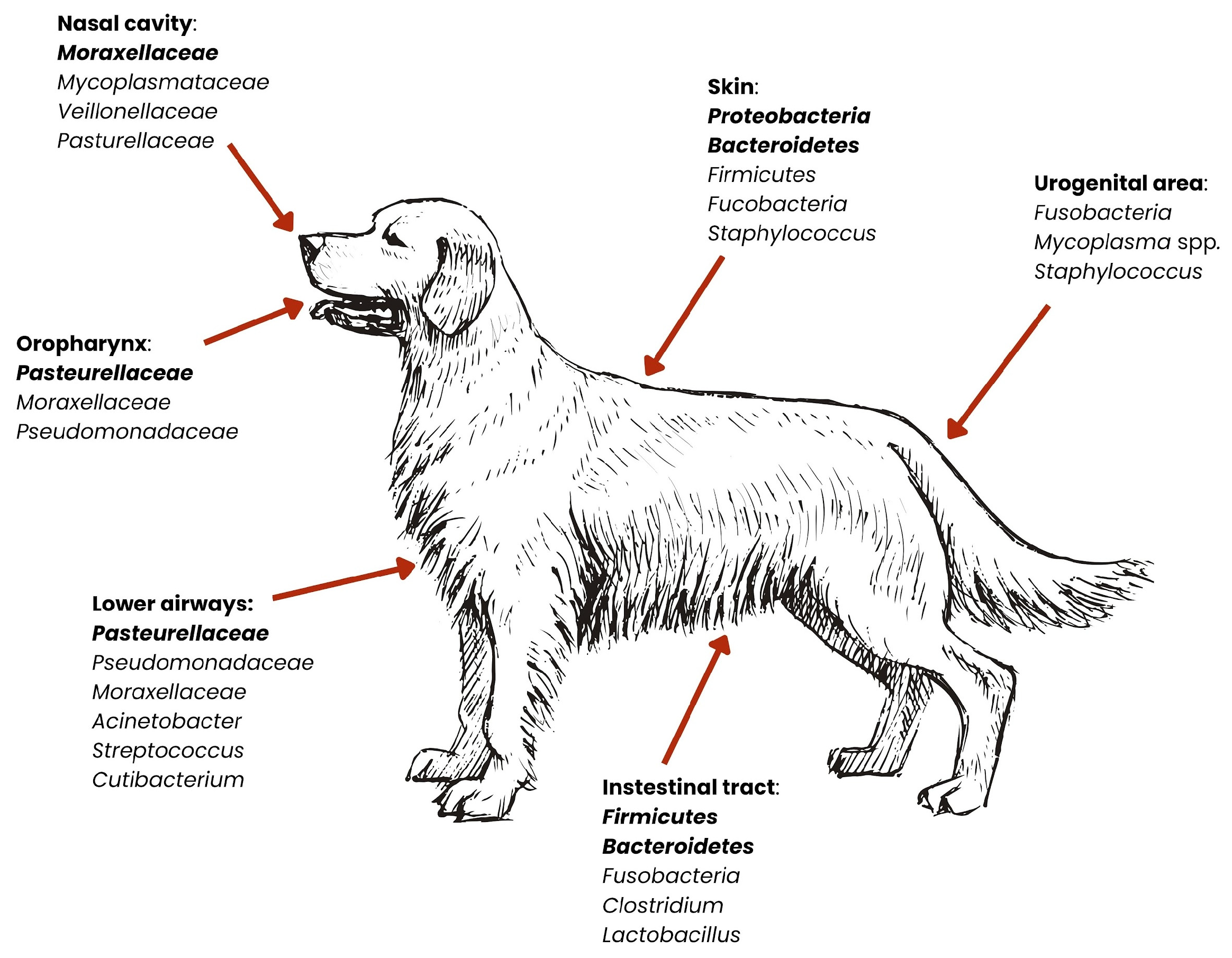
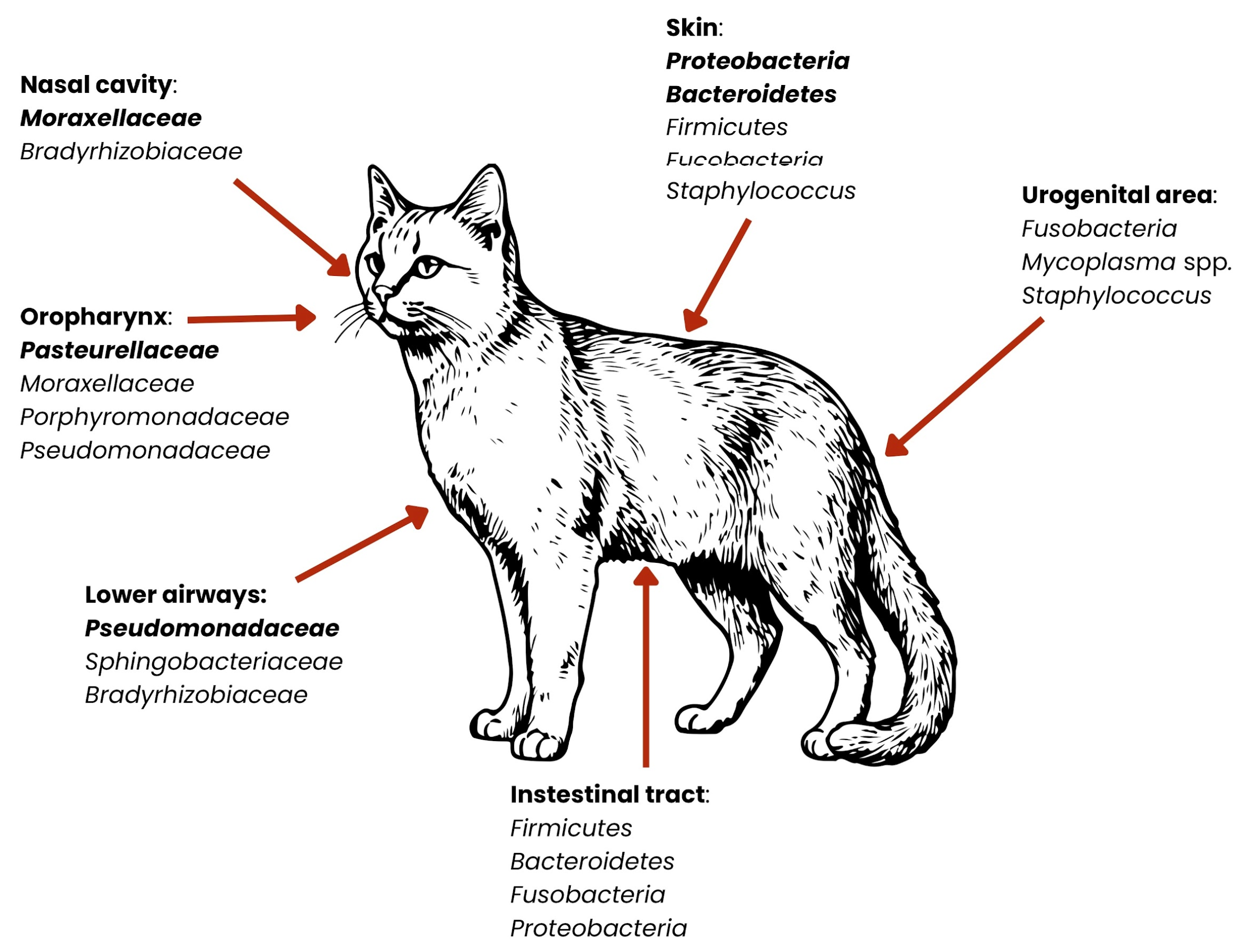
2. Microbiota

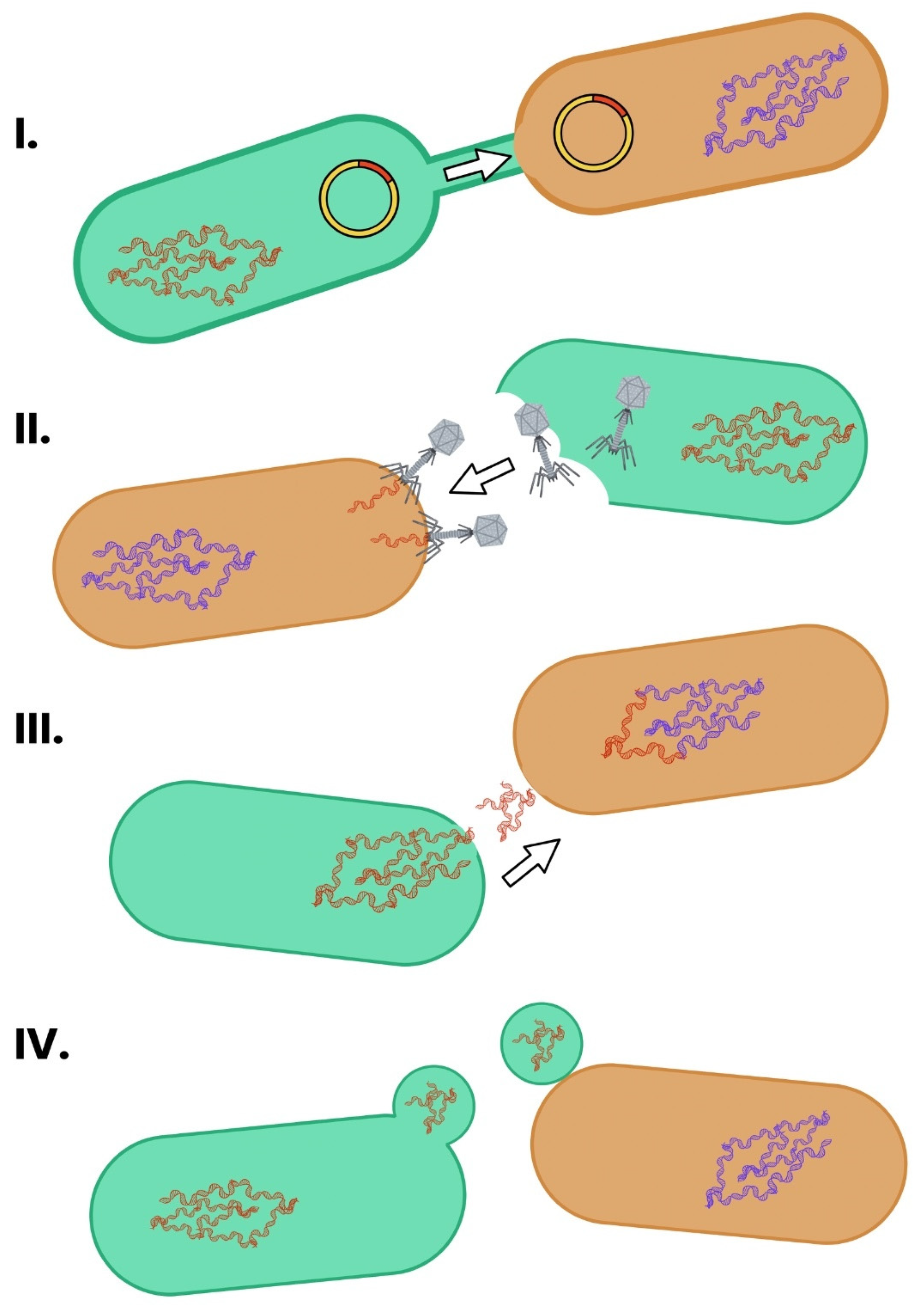
3. Mechanism of Bacterial Resistance Transfer
3.1. Conjugation
3.2. Transduction
3.3. Transformation
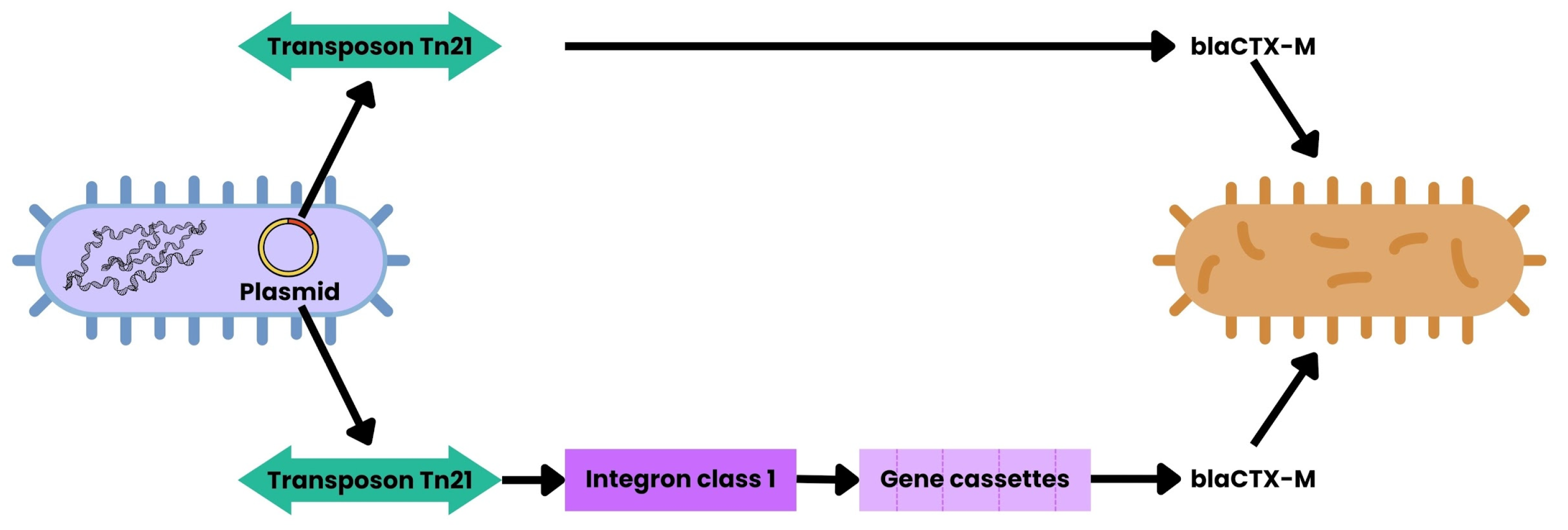
3.4. Transposons
3.5. Integrons
3.6. Outer Membrane Vesicles
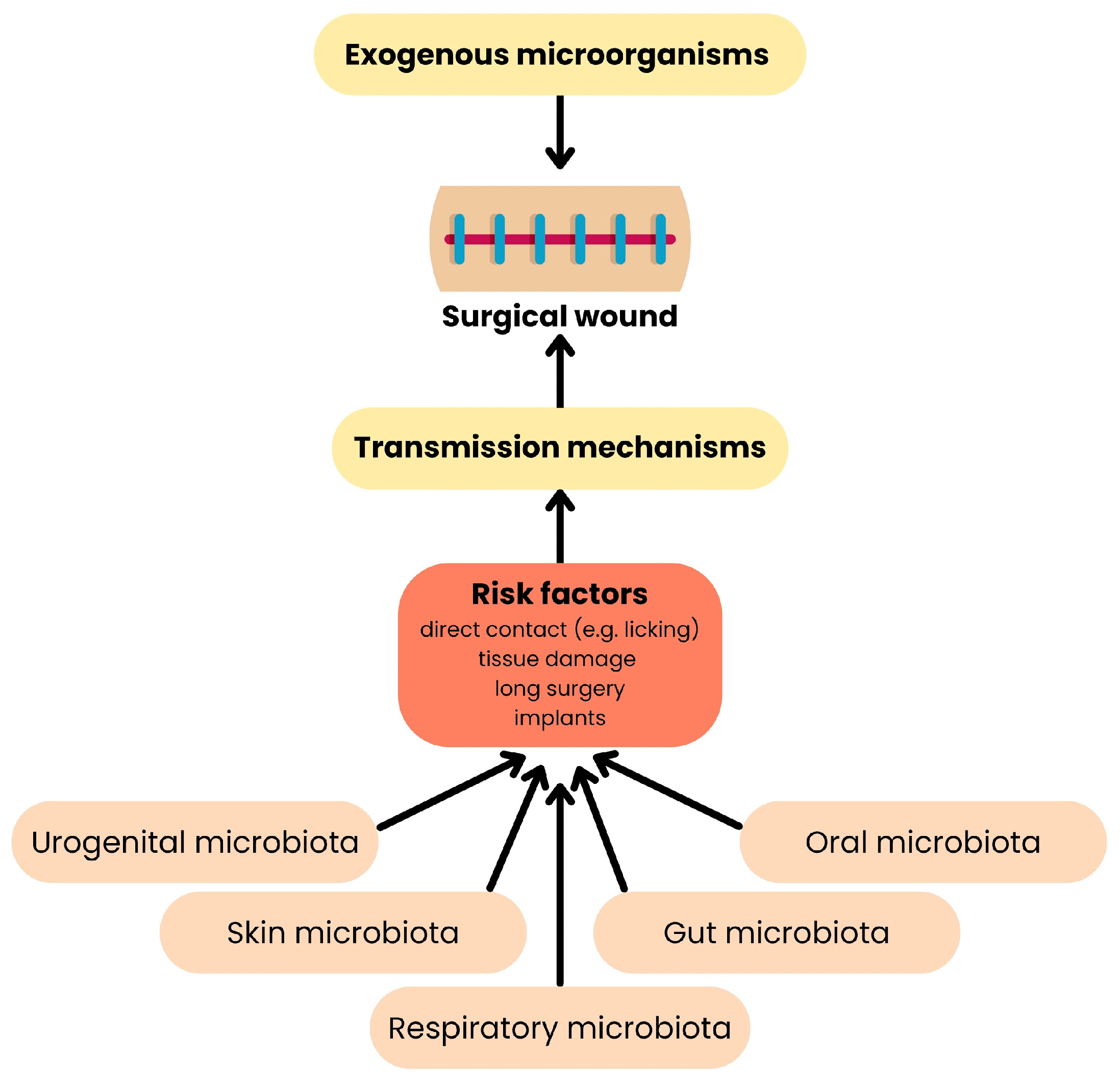
4. Transmission of ARGs Across Hosts
5. Clinical Significance of the Spread of Resistance Among the Natural Microbiota
6. The Importance of Animal Microbiota for Human Health
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AAB | Animal-associated bacteria |
| ARB | Antibiotic-resistant bacteria |
| ARGs | Antibiotic resistance genes |
| AMR | Antimicrobial Resistance |
| BRII | Biofilm-related implant infections |
| BARF | Biologically Appropriate Raw Food |
| DBPs | Disinfection byproducts |
| DI | Dysbiosis index |
| ESBL-producing E. coli | Extended-spectrum β-lactamase-producing E.coli |
| HAB | Human-associated bacteria |
| HGT | Horizontal gene transfer |
| MGEs | Mobile genetic elements |
| MRSA | Methicillin-resistant S. aureus |
| OMVs | Outer membrane vehicles |
| oriT | Origin of transfer |
| PIVC | Peripheral intravenous catheters |
| RAUS | Rational Antibiotic Use System |
| SBP | Spontaneous bacterial peritonitis |
| SCFAs | Short-chain fatty acids |
| SSIs | Surgical site infections |
| VGT | Vertical gene transfer |
| VRE | Vancomycin-resistant Enterococci |
References
- Davies, J. Inactivation of Antibiotics and the Dissemination of Resistance Genes. Science 1994, 264, 375–382. [Google Scholar] [CrossRef]
- Animal Welfare and Antibiotic Resistance in Food Animals. Available online: https://www.reactgroup.org/news-and-views/news-and-opinions/year-2020/animal-welfare-and-antibiotic-resistance-in-food-animals/ (accessed on 29 April 2025).
- Caneschi, A.; Bardhi, A.; Barbarossa, A.; Zaghini, A. The Use of Antibiotics and Antimicrobial Resistance in Veterinary Medicine, a Complex Phenomenon: A Narrative Review. Antibiotics 2023, 12, 487. [Google Scholar] [CrossRef]
- Zhuang, M.; Achmon, Y.; Cao, Y.; Liang, X.; Chen, L.; Wang, H.; Siame, B.A.; Leung, K.Y. Distribution of antibiotic resistance genes in the environment. Environ. Pollut. 2021, 285, 117402. [Google Scholar] [CrossRef]
- Antimicrobial Use and Resistance (AUR) Module. Available online: https://www.cdc.gov/nhsn/pdfs/pscmanual/11pscaurcurrent.pdf (accessed on 29 April 2025).
- Tunger, O.; Karakaya, Y.; Cetin, C.B.; Dinc, G.; Borand, H. Rational antibiotic use. J. Infect. Dev. Ctries. 2009, 3, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Du, W.; Chen, S.; Guo, X.; Ju, X. Exploring the Impact of the Rational Antibiotic Use System on Hospital Performance: The Direct Effect and the Spillover Effect. Int. J. Environ. Res. Public Health 2019, 16, 3463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Cheng, W. The Mechanism of Bacterial Resistance and Potential Bacteriostatic Strategies. Antibiotics 2022, 11, 1215. [Google Scholar] [CrossRef] [PubMed]
- Baran, A.; Kwiatkowska, A.; Potocki, L. Antibiotics and Bacterial Resistance—A Short Story of an Endless Arms Race. Int. J. Mol. Sci. 2023, 24, 5777. [Google Scholar] [CrossRef]
- Shi, X.; Xia, Y.; Wei, W.; Ni, B. Accelerated spread of antibiotic resistance genes (ARGs) induced by non-antibiotic conditions: Roles and mechanisms. Water Res. 2022, 224, 119060. [Google Scholar] [CrossRef]
- Warinner, C.; Rodrigues, J.; Vyas, R.; Trachsel, C.; Shved, N.; Grossmann, J.; Radini, A.; Hancock, Y.; Tito, R.Y.; Fiddyment, S.; et al. Pathogens and host immunity in the ancient human oral cavity. Nat. Genet. 2014, 46, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Dionisio, F.; Domingues, C.P.F.; Rebelo, J.S.; Monteiro, F.; Nogueira, T. The Impact of Non-Pathogenic Bacteria on the Spread of Virulence and Resistance Genes. Int. J. Mol. Sci. 2023, 24, 1967. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, G.; Wang, F.; Zhang, J.; Xu, L.; Yu, C. The impact of antibiotic exposure on antibiotic resistance gene dynamics in the gut microbiota of inflammatory bowel disease patients. Front. Microbiol. 2024, 15, 1382332. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Wu, Y.; Wang, Y.; Yang, H.; Li, Q.; Gong, X.; Zhang, G.; Zhu, K. Resident bacteria contribute to opportunistic infections of the respiratory tract. PLoS Pathog. 2021, 17, e10094361. [Google Scholar] [CrossRef]
- Lawton, L.E. All in the Family: Pets and Family Structure. Populations 2025, 1, 8. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [PubMed]
- Ottman, N.; Smidt, H.; de Vos, W.M.; Belzer, C. The function of our microbiota: Who is out there and what do they do. Front. Cell. Infect. Microbiol. 2012, 2, 104. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front. Microbiol. 2017, 8, 1162. [Google Scholar] [CrossRef]
- Lucchetti, B.; Lane, S.L.; Koenig, A.; Good, J.; Suchodolski, J.S.; Brainard, B.M. Effects of a perioperative antibiotic and veterinary probiotic on fecal dysbiosis index in dogs. Can. Vet. J. 2021, 62, 240–246. [Google Scholar] [PubMed]
- Campbell, A.G.; Schwientek, P.; Vishnivetskaya, T.; Woyke, T.; Levy, S.; Beall, C.J.; Griffen, A.; Leys, E.; Podar, M. Diversity and genomic insights into the uncultured Chloroflexi from the human microbiota. Environ. Microbiol. 2014, 16, 2635–2643. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.S.; Dowd, S.E.; Suchodolski, J.S.; Middelbos, I.S.; Vester, B.M.; Barry, K.A.; Nelson, K.E.; Torralba, M.; Henrissat, B.; Coutinho, P.M.; et al. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2011, 5, 639–649. [Google Scholar] [CrossRef]
- Bor, B.; Bedree, J.K.; Shi, W.; McLean, J.S.; He, X. Saccharibacteria (TM7) in the Human Oral Microbiome. J. Dent. Res. 2019, 98, 500–509. [Google Scholar] [CrossRef]
- Sturgeon, A.; Pinder, S.L.; Costa, M.C.; Weese, J.S. Characterization of the oral microbiota of healthy cats using next-generation sequencing. Vet. J. 2014, 201, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Miszczak, M.; Korzeniowska-Kowal, A.; Wzorek, A.; Prorok, P.; Szenborn, L.; Rypuła, K.; Bierowiec, K. Staphylococcus aureus and Staphylococcus pseudintermedius isolated from humans and pets—A comparison of drug resistance and risk factors associated with colonisation. J. Vet. Res. 2025, 69, 199–211. [Google Scholar] [CrossRef]
- Belizário, J.E.; Napolitano, M. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol. 2015, 6, 1050. [Google Scholar] [CrossRef]
- You, I.; Kim, M.J. Comparison of Gut Microbiota of 96 Healthy Dogs by Individual Traits: Breed, Age, and Body Condition Score. Animals 2021, 11, 2432. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Goh, T.W.; Kang, M.G.; Choi, H.J.; Yeo, S.Y.; Yang, J.; Huh, C.S.; Kim, Y.Y.; Kim, Y. Perspectives and advances in probiotics and the gut microbiome in companion animals. J. Anim. Sci. Technol. 2022, 64, 197–217. [Google Scholar] [CrossRef]
- Schmitz, S.; Suchodolski, J. Understanding the canine intestinal microbiota and its modification by pro-, pre- and synbiotics—What is the evidence. Vet. Med. Sci. 2016, 2, 71–94. [Google Scholar] [CrossRef]
- Misic, A.M.; Davis, M.F.; Tyldsley, A.S.; Hodkinson, B.P.; Tolomeo, P.; Hu, B.; Nachamkin, I.; Lautenbach, E.; Morris, D.O.; Grice, E.A. The shared microbiota of humans and companion animals as evaluated from Staphylococcus carriage sites. Microbiome 2015, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Alessandri, G.; Milani, C.; Mancabelli, L.; Longhi, G.; Anzalone, R.; Lugli, G.A.; Duranti, S.; Turroni, F.; Ossiprandi, M.C.; van Sinderen, D.; et al. Deciphering the Bifidobacterial Populations within the Canine and Feline Gut Microbiota. Appl. Environ. Microbiol. 2020, 86, e02875-19. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Sun, Q.; Li, Y.; Guan, Z.; Wei, J.; Li, B.; Liu, K.; Shao, D.; Mi, R.; Liu, H.; et al. Analysis and Comparison of Gut Microbiome in Young Detection Dogs. Front. Microbiol. 2022, 13, 872230. [Google Scholar] [CrossRef]
- Ballash, G.A.; Parker, E.M.; Mollenkopf, D.F.; Wittum, T.E. The One Health dissemination of antimicrobial resistance occurs in both natural and clinical environments. J. Am. Vet. Med. Assoc. 2024, 262, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Ruparell, A.; Inui, T.; Staunton, R.; Wallis, C.; Deusch, O.; Holcombe, L.J. The canine oral microbiome: Variation in bacterial populations across different niches. BMC Microbiol. 2020, 20, 42. [Google Scholar] [CrossRef]
- Mei, S.; Cai, M.; Lei, F.; Wang, X.; Yuan, X.; Lin, Y.; Zhu, B. Revealing microbial community characteristics in healthy human, cat and canine salivas and looking for species-specific microbes. Int. J. Leg. Med. 2024, 138, 2259–2269. [Google Scholar] [CrossRef] [PubMed]
- Whittle, M.J.; Castillo-Fernandez, J.; Amos, G.C.A.; Watson, P. Metagenomic characterisation of canine skin reveals a core healthy skin microbiome. Sci. Rep. 2024, 14, 20104. [Google Scholar] [CrossRef]
- Older, C.E.; Diesel, A.; Patterson, A.P.; Meason-Smith, C.; Johnson, T.J.; Mansell, J.; Suchodolski, J.S.; Rodrigues Hoffmann, A. The feline skin microbiota: The bacteria inhabiting the skin of healthy and allergic cats. PLoS ONE 2017, 12, e0178555. [Google Scholar] [CrossRef]
- Alessandri, G.; Argentini, C.; Milani, C.; Turroni, F.; Cristina Ossiprandi, M.; van Sinderen, D.; Ventura, M. Catching a glimpse of the bacterial gut community of companion animals: A canine and feline perspective. Microb. Biotechnol. 2020, 13, 1708–1732. [Google Scholar] [CrossRef]
- Vientós-Plotts, A.I.; Ericsson, A.C.; Reinero, C.R. The respiratory microbiota and its impact on health and disease in dogs and cats: A One Health perspective. J. Vet. Intern. Med. 2023, 37, 1641–1655. [Google Scholar] [CrossRef]
- Cuscó, A.; Sánchez, A.; Altet, L.; Ferrer, L.; Francino, O. Individual Signatures Define Canine Skin Microbiota Composition and Variability. Front. Vet. Sci. 2017, 4, 6. [Google Scholar]
- Banchi, P.; Spanoghe, L.; Maes, D.; Morrell, J.; Van Soom, A. The reproductive microbiome in dogs: Friend or foe. Vet. J. 2024, 304, 106100. [Google Scholar] [CrossRef]
- Banchi, P.; Bertero, A.; Gionechetti, F.; Corrò, M.; Spagnolo, E.; Donato, G.G.; Pallavicini, A.; Rota, A. The vaginal microbiota of healthy female cats. Theriogenology 2024, 224, 134–142. [Google Scholar] [CrossRef]
- Tress, B.; Dorn, E.S.; Suchodolski, J.S.; Nisar, T.; Ravindran, P.; Weber, K.; Hartmann, K.; Schulz, B.S. Bacterial microbiome of the nose of healthy dogs and dogs with nasal disease. PLoS ONE 2017, 12, e0176736. [Google Scholar] [CrossRef]
- Vangrinsven, E.; Fastrès, A.; Taminiau, B.; Frédéric, B.; Daube, G.; Clercx, C. Variations in facial conformation are associated with differences in nasal microbiota in healthy dogs. BMC Vet. Res. 2021, 17, 361. [Google Scholar] [CrossRef]
- Dorn, E.S.; Tress, B.; Suchodolski, J.S.; Nisar, T.; Ravindran, P.; Weber, K.; Hartmann, K.; Schulz, B.S. Bacterial microbiome in the nose of healthy cats and in cats with nasal disease. PLoS ONE 2017, 12, e0180299. [Google Scholar] [CrossRef]
- AL-Amrah, H.; Aburokba, R.; Alotiby, A.; AlJuhani, B.; Huri, H.; Algarni, N.; Aljedani, R. The Impact of Dogs Oral Microbiota on Human Health: A review. Biosci. Biotechnol. Res. Asia 2024, 21, 1–9. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Klein, E.A.; Bennett, M.L.; Croft, J.M.; Harris, S.J.; Marshall-Jones, Z.V. The feline oral microbiome: A provisional 16S rRNA gene based taxonomy with full-length reference sequences. Vet. Microbiol. 2015, 175, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Mondo, E.; Marliani, G.; Accorsi, P.A.; Cocchi, M.; Di Leone, A. Role of gut microbiota in dog and cat’s health and diseases. Open Vet. J. 2019, 9, 253–258. [Google Scholar] [CrossRef]
- Suchodolski, J.S. Analysis of the gut microbiome in dogs and cats. Vet. Clin. Pathol. 2022, 50, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.A.; Park, B.; Scarsella, E.; Jospin, G.; Entrolezo, Z.; Jarett, J.K.; Martin, A.; Ganz, H.H. Species-level characterization of the core microbiome in healthy dogs using full-length 16S rRNA gene sequencing. Front. Vet. Sci. 2024, 11, 1405470. [Google Scholar] [CrossRef] [PubMed]
- Langon, X. Validation of method for faecal sampling in cats and dogs for faecal microbiome analysis. BMC Vet. Res. 2023, 19, 274. [Google Scholar] [CrossRef]
- Tokuda, M.; Shintani, M. Microbial evolution through horizontal gene transfer by mobile genetic elements. Microb. Biotechnol. 2024, 17, e14408. [Google Scholar] [CrossRef]
- Bennett, P.M. Plasmid encoded antibiotic resistance: Acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 2008, 153, 347–357. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Selection and Transmission of Antibiotic-Resistant Bacteria. Microbiol. Spectr. 2017, 5, 10.1128. [Google Scholar] [CrossRef]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef]
- Johnston, C.; Martin, B.; Fichant, G.; Polard, P.; Claverys, J.P. Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 2014, 12, 181–196. [Google Scholar] [CrossRef]
- Colavecchio, A.; Cadieux, B.; Lo, A.; Goodridge, L.D. Bacteriophages Contribute to the Spread of Antibiotic Resistance Genes among Foodborne Pathogens of the Enterobacteriaceae Family—A Review. Front. Microbiol. 2017, 8, 1108. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.; Zhang, S.; Li, J.; Mao, L.; Yuan, Z.; Bond, P.L.; Guo, J. Non-antibiotic pharmaceuticals promote the transmission of multidrug resistance plasmids through intra- and intergenera conjugation. ISME J. 2021, 15, 2493–2508. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Lai, Y.; Xiao, W.; Zhong, T.; Liu, F.; Gong, J.; Huang, J. Microbial extracellular vesicles contribute to antimicrobial resistance. PLoS Pathog. 2024, 20, e1012143. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Wang, J.; Huang, M.; Huang, Y.; Zhang, R.; Bu, F.; Yang, B.; Chen, J.; Lin, X.; Hu, X.; et al. Outer membrane vesicles-transmitted virulence genes mediate the emergence of new antimicrobial-resistant hypervirulent Klebsiella pneumoniae. Emerg. Microbes Infect. 2022, 11, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics 2023, 12, 328. [Google Scholar] [CrossRef] [PubMed]
- Domingues, S.; Harms, K.; Fricke, W.F.; Johnsen, P.J.; da Silva, G.J.; Nielsen, K.M. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 2012, 8, e1002837. [Google Scholar] [CrossRef]
- Schjørring, S.; Krogfelt, K.A. Assessment of bacterial antibiotic resistance transfer in the gut. Int. J. Microbiol. 2011, 2011, 312956. [Google Scholar] [CrossRef]
- Ali, N.; Ali, I.; Din, A.U.; Akhtar, K.; He, B.; Wen, R. Integrons in the Age of Antibiotic Resistance: Evolution, Mechanisms, and Environmental Implications: A Review. Microorganisms 2024, 12, 2579. [Google Scholar] [CrossRef]
- García-Cazorla, Y.; Getino, M.; Sanabria-Ríos, D.J.; Carballeira, N.M.; de la Cruz, F.; Arechaga, I.; Cabezón, E. Conjugation inhibitors compete with palmitic acid for binding to the conjugative traffic ATPase TrwD, providing a mechanism to inhibit bacterial conjugation. J. Biol. Chem. 2018, 293, 16923–16930. [Google Scholar] [CrossRef] [PubMed]
- Ott, L.C.; Mellata, M. Short-chain fatty acids inhibit bacterial plasmid transfer through conjugation in vitro and in ex vivo chicken tissue explants. Front. Microbiol. 2024, 15, 1414401. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 10.1128. [Google Scholar] [CrossRef]
- Tang, K.L.; Caffrey, N.P.; Nóbrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D.; et al. Comparison of different approaches to antibiotic restriction in food-producing animals: Stratified results from a systematic review and meta-analysis. BMJ Glob. Health 2019, 4, e001710. [Google Scholar] [CrossRef]
- Lam, T.; Ellison, C.K.; Eddington, D.T.; Brun, Y.V.; Dalia, A.B.; Morrison, D.A. Competence pili in Streptococcus pneumoniae are highly dynamic structures that retract to promote DNA uptake. Mol. Microbiol. 2021, 116, 381–396. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Tetu, S.G.; Gillings, M.R. Predicting the taxonomic and environmental sources of integron gene cassettes using structural and sequence homology of attC sites. Commun. Biol. 2021, 4, 946. [Google Scholar] [CrossRef]
- Souque, C.; Escudero, J.A.; MacLean, R.C. Integron activity accelerates the evolution of antibiotic resistance. Elife 2021, 10, e62474. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, X.; Li, J.; Lv, N.; Liu, F.; Wu, J.; Lin, I.Y.; Wu, N.; Weimer, B.C.; Gao, G.F.; et al. The Bacterial Mobile Resistome Transfer Network Connecting the Animal and Human Microbiomes. Appl. Environ. Microbiol. 2016, 82, 6672–6681. [Google Scholar] [CrossRef]
- Zhao, R.; Hao, J.; Yang, J.; Tong, C.; Xie, L.; Xiao, D.; Zeng, Z.; Xiong, W. The co-occurrence of antibiotic resistance genes between dogs and their owners in families. Imeta 2022, 1, e21. [Google Scholar] [CrossRef]
- Zhang, X.F.; Doi, Y.; Huang, X.; Li, H.Y.; Zhong, L.L.; Zeng, K.J.; Zhang, Y.F.; Patil, S.; Tian, G.B. Possible Transmission of mcr-1-Harboring Escherichia coli between Companion Animals and Human. Emerg. Infect. Dis. 2016, 22, 1679–1681. [Google Scholar] [CrossRef]
- Alessandri, G.; Milani, C.; Mancabelli, L.; Mangifesta, M.; Lugli, G.A.; Viappiani, A.; Duranti, S.; Turroni, F.; Ossiprandi, M.C.; van Sinderen, D.; et al. The impact of human-facilitated selection on the gut microbiota of domesticated mammals. FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef] [PubMed]
- Rantala, M.; Lahti, E.; Kuhalampi, J.; Pesonen, S.; Järvinen, A.K.; Saijonmaa-Koulumies, L.; Honkanen-Buzalski, T. Antimicrobial resistance in Staphylococcus spp., Escherichia coli and Enterococcus spp. in dogs given antibiotics for chronic dermatological disorders, compared with non-treated control dogs. Acta Vet. Scand. 2004, 45, 37. [Google Scholar] [CrossRef]
- Pomba, C.; Rantala, M.; Greko, C.; Baptiste, K.E.; Catry, B.; van Duijkeren, E.; Mateus, A.; Moreno, M.A.; Pyörälä, S.; Ružauskas, M.; et al. Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 2017, 72, 957–968. [Google Scholar] [CrossRef]
- Allcock, S.; Young, E.H.; Holmes, M.; Gurdasani, D.; Dougan, G.; Sandhu, M.S.; Solomon, L.; Török, M.E. Antimicrobial resistance in human populations: Challenges and opportunities. Glob. Health Epidemiol. Genom. 2017, 2, e4. [Google Scholar] [CrossRef] [PubMed]
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE pathogens in the environment: Antibiotic resistance status, community-acquired infection and risk to human health. Int. J. Hyg. Environ. Health 2022, 244, 114006. [Google Scholar] [CrossRef]
- Church, N.A.; McKillip, J.L. Antibiotic resistance crisis: Challenges and imperatives. Biologia 2021, 76, 1535–1550. [Google Scholar] [CrossRef]
- Hackmann, C.; Gastmeier, P.; Schwarz, S.; Lübke-Becker, A.; Bischoff, P.; Leistner, R. Pet husbandry as a risk factor for colonization or infection with MDR organisms: A systematic meta-analysis. J. Antimicrob. Chemother. 2021, 76, 1392–1405. [Google Scholar] [CrossRef]
- Bogaerts, P.; Huang, T.D.; Bouchahrouf, W.; Bauraing, C.; Berhin, C.; El Garch, F.; Glupczynski, Y. Characterization of ESBL- and AmpC-Producing Enterobacteriaceae from Diseased Companion Animals in Europe. Microb. Drug Resist. 2015, 21, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Grobbel, M.; Bethe, A.; Wieler, L.H.; Guenther, S. Extended-spectrum beta-lactamases-producing gram-negative bacteria in companion animals: Action is clearly warranted. Berl. Munch. Tierarztl. Wochenschr. 2011, 124, 94–101. [Google Scholar]
- Pomba, C.; Endimiani, A.; Rossano, A.; Saial, D.; Couto, N.; Perreten, V. First report of OXA-23-mediated carbapenem resistance in sequence type 2 multidrug-resistant Acinetobacter baumannii associated with urinary tract infection in a cat. Antimicrob. Agents Chemother. 2014, 58, 1267–1268. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, L.; Gutierrez, B.; Ovejero, C.M.; Carrilero, L.; Matrat, S.; Saba, C.K.; Santos-Lopez, A.; Thomas-Lopez, D.; Hoefer, A.; Suarez, M.; et al. Klebsiella pneumoniae sequence type 11 from companion animals bearing ArmA methyltransferase, DHA-1 β-lactamase, and QnrB4. Antimicrob. Agents Chemother. 2013, 57, 4532–4534. [Google Scholar] [CrossRef]
- Armstrong, J.L.; Shigeno, D.S.; Calomiris, J.J.; Seidler, R.J. Antibiotic-resistant bacteria in drinking water. Appl. Environ. Microbiol. 1981, 42, 277–283. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, J.; Wu, J.; Wang, J.; Luo, Y. Potential risks of microplastics combined with superbugs: Enrichment of antibiotic resistant bacteria on the surface of microplastics in mariculture system. Ecotoxicol. Environ. Saf. 2020, 187, 109852. [Google Scholar] [CrossRef]
- Wright, R.J.; Erni-Cassola, G.; Zadjelovic, V.; Latva, M.; Christie-Oleza, J.A. Marine Plastic Debris: A New Surface for Microbial Colonization. Environ. Sci. Technol. 2020, 54, 11657–11672. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Lu, J.; Fu, S.; Wang, S.; Senehi, N.; Yuan, Q. Enhanced propagation of intracellular and extracellular antibiotic resistance genes in municipal wastewater by microplastics. Environ. Pollut. 2022, 292, 118284. [Google Scholar] [CrossRef]
- Sirelkhatim, A.; Mahmud, S.; Seeni, A.; Kaus, N.H.M.; Ann, L.C.; Bakhori, S.K.M.; Hasan, H.; Mohamad, D. Review on Zinc Oxide Nanoparticles: Antibacterial Activity and Toxicity Mechanism. Nano-Micro Lett. 2015, 7, 219–242. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Mao, D.; Luo, Y. Ionic Liquid Facilitates the Conjugative Transfer of Antibiotic Resistance Genes Mediated by Plasmid RP4. Environ. Sci. Technol. 2015, 49, 8731–8740. [Google Scholar] [CrossRef]
- Liao, J.; Huang, H.; Chen, Y. CO2 promotes the conjugative transfer of multiresistance genes by facilitating cellular contact and plasmid transfer. Environ. Int. 2019, 129, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.H.; Yuan, L.; Geng, Y.K.; Li, N.; Sheng, G.P. Evaluating the effect of gradient applied voltages on antibiotic resistance genes proliferation and biogas production in anaerobic electrochemical membrane bioreactor. J. Hazard. Mater. 2021, 416, 125865. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Song, H.; Ji, Y.; He, Z.; Pu, Y.; Zhou, J.; Xu, J. Ultrasound-mediated DNA transformation in thermophilic gram-positive anaerobes. PLoS ONE 2010, 5, e12582. [Google Scholar] [CrossRef]
- Frieri, M.; Kumar, K.; Boutin, A. Antibiotic resistance. J. Infect. Public Health 2017, 10, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Merchán, E.C.; Davidson, D.J.; Liddle, A.D. Recent Strategies to Combat Infections from Biofilm-Forming Bacteria on Orthopaedic Implants. Int. J. Mol. Sci. 2021, 22, 10243. [Google Scholar] [CrossRef] [PubMed]
- Crisi, P.E.; De Santis, F.; Aste, G.; Tiscar, P.G.; Mosca, F.; Gasparini, A.; Felici, A.; Ferroni, L.; Miglio, A.; Di Tommaso, M.; et al. Inflammatory, Mechanical and Infectious Complications Associated with Peripheral Intravenous Catheters in Dogs and Cats: A Risk Factor Analysis. Vet. Sci. 2022, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Windahl, U.; Bengtsson, B.; Nyman, A.K.; Holst, B.S. The distribution of pathogens and their antimicrobial susceptibility patterns among canine surgical wound infections in Sweden in relation to different risk factors. Acta Vet. Scand. 2015, 57, 11. [Google Scholar] [CrossRef][Green Version]
- Marco-Fuertes, A.; Marin, C.; Lorenzo-Rebenaque, L.; Vega, S.; Montoro-Dasi, L. Antimicrobial Resistance in Companion Animals: A New Challenge for the One Health Approach in the European Union. Vet. Sci. 2022, 9, 208. [Google Scholar] [CrossRef]
- Williams, R.W.; Cole, S.; Holt, D.E. Microorganisms associated with incisional infections after gastrointestinal surgery in dogs and cats. Vet. Surg. 2020, 49, 1301–1306. [Google Scholar] [CrossRef]
- Espinel-Rupérez, J.; Martín-Ríos, M.D.; Salazar, V.; Baquero-Artigao, M.R.; Ortiz-Díez, G. Incidence of surgical site infection in dogs undergoing soft tissue surgery: Risk factors and economic impact. Vet. Rec. Open 2019, 6, e000233. [Google Scholar] [CrossRef]
- Devriendt, N.; Mortier, F.; Rooster, H. Antimicrobial prophylaxis in canine and feline surgery. Vlaams Diergeneeskd. Tijdschr. 2023, 92, 131–141. [Google Scholar] [CrossRef]
- Kent, M.; Boozer, L.; Glass, E.N.; Sanchez, S.; Platt, S.R.; Freeman, L.M. Post-operative Salmonella surgical site infection in a dog. Can. Vet. J. 2017, 58, 936–940. [Google Scholar]
- Gulaydin, A.; Gulaydin, O.; Akgul, M.B. Isolation of aerobic bacteria from surgical site infections following orthopaedic operations in cats and dogs. Vet. Med. 2024, 69, 243–253. [Google Scholar] [CrossRef]
- Radulescu, S.; Allais, M.; Le Gal, A.; Cook, S. Medically managed spontaneous bacterial peritonitis and bacteraemia associated with jugular catheter infection in a dog with tetanus. Vet. Rec. Case Rep. 2022, 10, e354. [Google Scholar] [CrossRef]
- Ameer, M.A.; Foris, L.A.; Mandiga, P.; Haseeb, M. Spontaneous Bacterial Peritonitis (Archived); StatPearls: Treasure Island, FL, USA, 2023. [Google Scholar]
- Martin, T.C.S.; Abdelmalek, J.; Yee, B.; Lavergne, S.; Ritter, M. Pasteurella multocida line infection: A case report and review of literature. BMC Infect. Dis. 2018, 18, 420. [Google Scholar] [CrossRef]
- Pavone, G.; Castellucci, B.; Pavone, S.; Stefanetti, V.; Vitolo, C.; Mangiaterra, S. Unusual Case of Biliary Peritonitis in a Dog Secondary to a Gastric Perforation. Vet. Sci. 2023, 10, 384. [Google Scholar] [CrossRef] [PubMed]
- Aleid, A.; Aldanyowi, S.N.; Aljabr, A.; Alaidarous, H.A.A.; Aleid, Z.; Alharthi, A.; Alsubaie, M.; AlOraini, L.; Almoslem, A.; Al Mutair, A. Effect of preoperative hair removal vs. no removal on surgical site infections: A systematic review and meta-analysis. F1000Research 2024, 13, 1487. [Google Scholar] [CrossRef] [PubMed]
- Thapa, N.; Basukala, S.; Regmi, S.K.; Shrestha, O.; Paudel, S.; Chaudhary, K.; Metha, B.; K. C., M.; Thapa, S.; Bista, S. Postoperative surgical site infection after preoperative use of razor versus clipper for hair removal in inguinal hernia surgery: A quasi-randomized clinical trial. Health Sci. Rep. 2024, 7, e1830. [Google Scholar] [CrossRef] [PubMed]
- Ossai, J.O.; Anele, A.A.; Ekwunife, C.N.; Nwagbara, I.C.; Opara, K.O. Preoperative Shaving with Razor Blades Versus no Preoperative Shaving in Elective Inguinal Hernia Repair: Impact on Surgical Site Infection. Orient J. Med. 2024, 36, 25–34. [Google Scholar]
- Tsai, H.Y.; Liao, W.C.; Wang, M.; Ueng, K.C.; Huang, C.Y.; Tseng, Y.C. Randomized clinical trial of preoperative skin preparation with 2% chlorhexidine versus conventional hair shaving in percutaneous coronary intervention. Medicine 2021, 100, e25304. [Google Scholar] [CrossRef]
- Practical Guide to Use of Antibiotics During Soft Tissue Surgery by Charly McGahan. Available online: https://bvna.org.uk/blog/practical-guide-to-use-of-antibiotics-during-soft-tissue-surgery-by-charly-mcgahan/ (accessed on 27 June 2025).
- Maemoto, R.; Noda, H.; Ichida, K.; Miyakura, Y.; Kakizawa, N.; Machida, E.; Aizawa, H.; Kato, T.; Iseki, M.; Fukui, T.; et al. Aqueous Povidone-Iodine Versus Normal Saline For Intraoperative Wound Irrigation on The Incidence of Surgical Site Infection in Clean-Contaminated Wounds After Gastroenterological Surgery: A Single-Institute, Prospective, Blinded-Endpoint, Randomized Controlled Trial. Ann. Surg. 2023, 277, 727–733. [Google Scholar] [PubMed]
- Al-Hajri, A.; Ghabisha, S.; Ahmed, F.; Al-Wageeh, S.; Badheeb, M.; Alyhari, Q.; Altam, A.; Alsharif, A. Identification of predictive factors for surgical site infections in gastrointestinal surgeries: A retrospective cross-sectional study in a resource-limited setting. F1000Research 2023, 12, 733. [Google Scholar] [CrossRef] [PubMed]
- Long, D.R.; Alverdy, J.C.; Vavilala, M.S. Emerging Paradigms in the Prevention of Surgical Site Infection: The Patient Microbiome and Antimicrobial Resistance. Anesthesiology 2022, 137, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Trinh, P.; Zaneveld, J.R.; Safranek, S.; Rabinowitz, P.M. One Health Relationships Between Human, Animal, and Environmental Microbiomes: A Mini-Review. Front. Public Health 2018, 6, 235. [Google Scholar] [CrossRef]
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef]
- Habib, I.; Alshehhi, Z. Zoonotic Disease Management and Infection Control Practices Among Veterinarians in the United Arab Emirates. Vet. Sci. 2021, 8, 82. [Google Scholar] [CrossRef]
- Groat, E.F.; Williams, N.J.; Pinchbeck, G.; Warner, B.; Simpson, A.; Schmidt, V.M. UK dogs eating raw meat diets have higher risk of Salmonella and antimicrobial-resistant Escherichia coli faecal carriage. J. Small Anim. Pract. 2022, 63, 435–441. [Google Scholar] [CrossRef]
- Cella, E.; Giovanetti, M.; Benedetti, F.; Scarpa, F.; Johnston, C.; Borsetti, A.; Ceccarelli, G.; Azarian, T.; Zella, D.; Ciccozzi, M. Joining Forces against Antibiotic Resistance: The One Health Solution. Pathogens 2023, 12, 1074. [Google Scholar] [CrossRef]
- Jin, M.; Osman, M.; Green, B.A.; Yang, Y.; Ahuja, A.; Lu, Z.; Cazer, C.L. Evidence for the transmission of antimicrobial resistant bacteria between humans and companion animals: A scoping review. One Health 2023, 17, 100593. [Google Scholar] [CrossRef] [PubMed]
- Razavi, M.; Marathe, N.P.; Gillings, M.R.; Flach, C.; Kristiansson, E.; Larsson, D.G.J. Discovery of the fourth mobile sulfonamide resistance gene. Microbiome 2017, 5, 160. [Google Scholar] [CrossRef]
- El-Far, S.W.; Abukhatwah, M.W. Prevalence of Aminoglycoside Resistance Genes in Clinical Isolates of Pseudomonas aeruginosa from Taif, Saudi Arabia-An Emergence Indicative Study. Microorganisms 2023, 11, 2293. [Google Scholar] [CrossRef]
- Venkatesan, M.; Fruci, M.; Verellen, L.A.; Skarina, T.; Mesa, N.; Flick, R.; Pham, C.; Mahadevan, R.; Stogios, P.J.; Savchenko, A. Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics. Nat. Commun. 2023, 14, 4031. [Google Scholar] [CrossRef]
- Rashvand, P.; Peymani, A.; Mohammadi, M.; Karami, A.A.; Samimi, R.; Hajian, S.; Porasgari, D.; Habibollah-Pourzereshki, N. Molecular survey of aminoglycoside-resistant Acinetobacter baumannii isolated from tertiary hospitals in Qazvin, Iran. New Microbes New Infect. 2021, 42, 100883. [Google Scholar] [CrossRef] [PubMed]
- Pavelquesi, S.L.S.; de Oliveira Ferreira, A.C.A.; Rodrigues, A.R.M.; de Souza Silva, C.M.; Orsi, D.C.; da Silva, I.C.R. Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review. Antibiotics 2021, 10, 1314. [Google Scholar] [CrossRef]
- Del Grosso, M.; Northwood, J.G.; Farrell, D.J.; Pantosti, A. The macrolide resistance genes erm(B) and mef(E) are carried by Tn2010 in dual-gene Streptococcus pneumoniae isolates belonging to clonal complex CC271. Antimicrob. Agents Chemother. 2007, 51, 4184–4186. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Byrne-Bailey, K.G.; Gaze, W.H.; Kay, P.; Boxall, A.B.; Hawkey, P.M.; Wellington, E.M. Prevalence of sulfonamide resistance genes in bacterial isolates from manured agricultural soils and pig slurry in the United Kingdom. Antimicrob. Agents Chemother. 2009, 53, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Marimón, J.M.; Valiente, A.; Ercibengoa, M.; García-Arenzana, J.M.; Pérez-Trallero, E. Erythromycin resistance and genetic elements carrying macrolide efflux genes in Streptococcus agalactiae. Antimicrob. Agents Chemother. 2005, 49, 5069–5074. [Google Scholar] [CrossRef][Green Version]
- Marshall, C.G.; Lessard, I.A.; Park, I.; Wright, G.D. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 1998, 42, 2215–2220. [Google Scholar] [CrossRef] [PubMed]
- Alifano, P.; Palumbo, C.; Pasanisi, D.; Talà, A. Rifampicin-resistance, rpoB polymorphism and RNA polymerase genetic engineering. J. Biotechnol. 2015, 202, 60–77. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Cheng, H.; Liang, Y.; Yu, S.; Yu, T.; Fang, J.; Zhu, C. Diverse Mobile Genetic Elements and Conjugal Transferability of Sulfonamide Resistance Genes (sul1, sul2, and sul3) in Escherichia coli Isolates From Penaeus vannamei and Pork From Large Markets in Zhejiang, China. Front. Microbiol. 2019, 10, 1787. [Google Scholar] [CrossRef]
- Cascone, C.; Santagati, M.; Noviello, S.; Iannelli, F.; Esposito, S.; Pozzi, G.; Stefani, S. Macrolide-resistance genes in clinical isolates of Streptococcus pyogenes. Microb. Drug Resist. 2002, 8, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.E.; Nash, A.K.; Zanghi, B.M.; Otto, C.M.; Perry, E.B. An Assessment of the Stability of the Canine Oral Microbiota After Probiotic Administration in Healthy Dogs Over Time. Front. Vet. Sci. 2020, 7, 616. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Deplano, A.; Meghraoui, A.; Dodémont, M.; Heinrichs, A.; Denis, O.; Nonhoff, C.; Roisin, S. Bacteria from Animals as a Pool of Antimicrobial Resistance Genes. Antibiotics 2017, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hu, X.; Cai, S.; Hu, N.; Yuan, Y.; Wu, Y.; Wang, Y.; Mi, J.; Liao, X. Pet cats may shape the antibiotic resistome of their owner’s gut and living environment. Microbiome 2023, 11, 235. [Google Scholar] [CrossRef]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L.H. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Schwarz, S.; Lloyd, D.H. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 2004, 54, 321–332. [Google Scholar] [CrossRef]
- Blázquez, J.; Couce, A.; Rodríguez-Beltrán, J.; Rodríguez-Rojas, A. Antimicrobials as promoters of genetic variation. Curr. Opin. Microbiol. 2012, 15, 561–569. [Google Scholar] [CrossRef]
- Rzezutka, A.; Cook, N. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 2004, 28, 441–453. [Google Scholar] [CrossRef]
- Poirel, L.; Cattoir, V.; Nordmann, P. Plasmid-Mediated Quinolone Resistance; Interactions between Human, Animal, and Environmental Ecologies. Front. Microbiol. 2012, 3, 24. [Google Scholar] [CrossRef]
- Valiquette, L.; Cossette, B.; Garant, M.P.; Diab, H.; Pépin, J. Impact of a reduction in the use of high-risk antibiotics on the course of an epidemic of Clostridium difficile-associated disease caused by the hypervirulent NAP1/027 strain. Clin. Infect. Dis. 2007, 45 (Suppl. S2), S112–S121. [Google Scholar] [CrossRef]
- Weese, J.S.; van Duijkeren, E. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet. Microbiol. 2010, 140, 418–429. [Google Scholar] [CrossRef]
- Chomel, B.B.; Sun, B. Zoonoses in the bedroom. Emerg. Infect. Dis. 2011, 17, 167–172. [Google Scholar] [CrossRef]
- Gorzelanna, Z.; Miszczak, M. Through the Intestines to the Head? That Is, How the Gastrointestinal Microbiota Affects the Behavior of Companion Animals. Pets 2024, 1, 201–215. [Google Scholar] [CrossRef]
- Kobayaa, H.; Souki, R.R.; Trust, S.; Domachowske, J.B. Pasteurella multocida meningitis in newborns after incidental animal exposure. Pediatr. Infect. Dis. J. 2009, 28, 928–929. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Karasawa, T.; Piao, C.; Itoda, I.; Hidai, H.; Yamaura, H.; Totsuka, K.; Morikawa, T.; Takayama, M. Molecular confirmation of transmission route of Staphylococcus intermedius in mastoid cavity infection from dog saliva. J. Infect. Chemother. 2004, 10, 46–48. [Google Scholar] [CrossRef]
- Kempker, R.; Mangalat, D.; Kongphet-Tran, T.; Eaton, M. Beware of the pet dog: A case of Staphylococcus intermedius infection. Am. J. Med. Sci. 2009, 338, 425–427. [Google Scholar] [CrossRef]
- Rutland, B.E.; Weese, J.S.; Bolin, C.; Au, J.; Malani, A.N. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 2009, 15, 1328–1330. [Google Scholar] [CrossRef] [PubMed]
- Bazzi, R.; Alaboudi, A.; Rácz, G. The role of veterinarians in the One Health approach to antimicrobial resistance perspectives in Jordan. Anim. Dis. 2022, 2, 1. [Google Scholar] [CrossRef]
- Vercelli, C.; Gambino, G.; Amadori, M.; Re, G. Implications of Veterinary Medicine in the comprehension and stewardship of antimicrobial resistance phenomenon. From the origin till nowadays. Vet. Anim. Sci. 2022, 16, 100249. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance. Available online: https://www.woah.org/en/what-we-do/global-initiatives/antimicrobial-resistance/ (accessed on 22 July 2025).
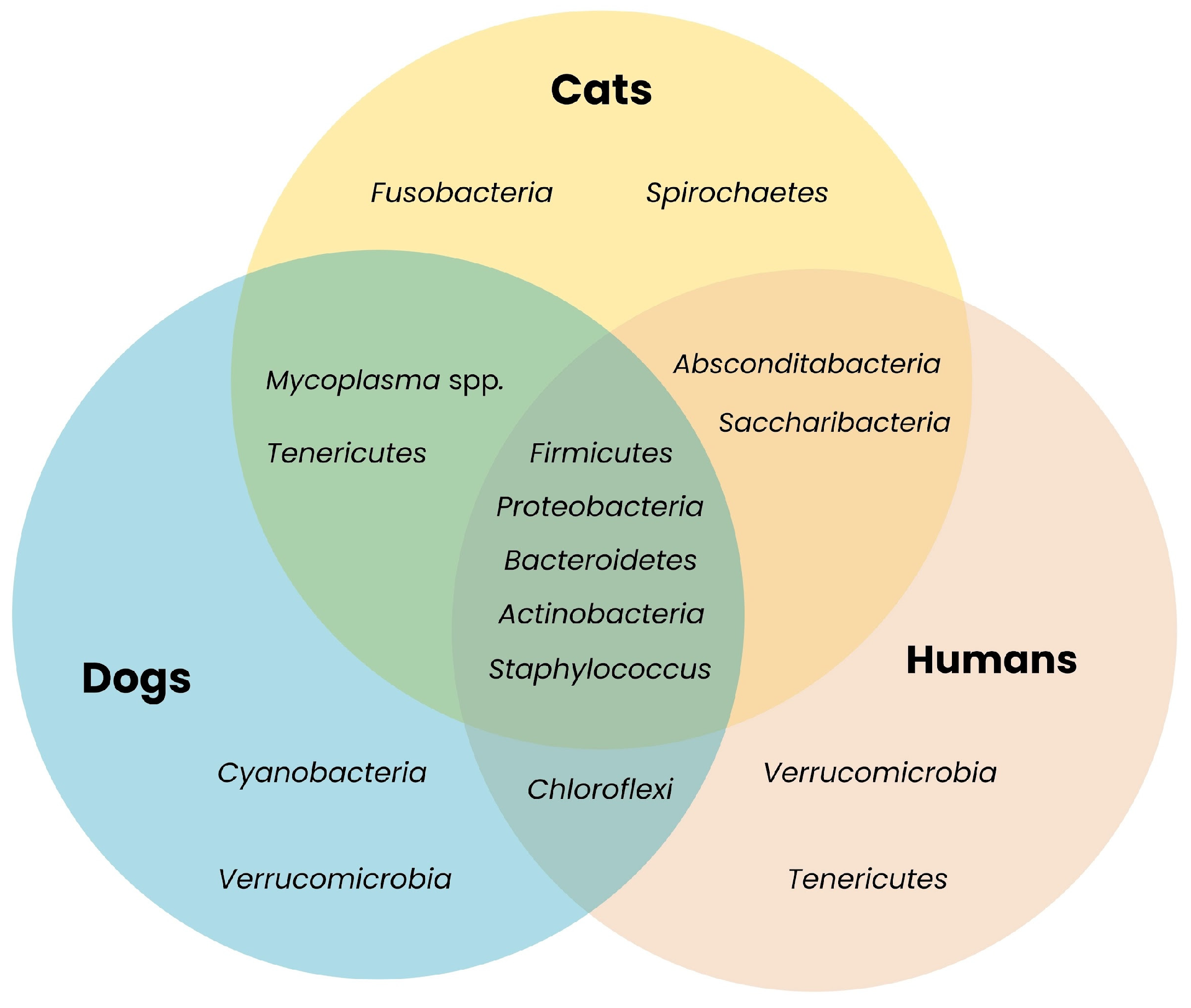
| Phylum | Species Found in Dogs | Species Found in Cats |
|---|---|---|
| Proteobacteria | Capnocytophaga canimorsus, C. canis, Paracoccus marcusii, Sphingomonas aerolata | (less frequently identified at species level) |
| Bacteroidota | Porphyromonas gulae, P. canoris | - |
| Actinobacteria | Cutibacterium spp. | Cutibacterium spp. |
| Firmicutes | Staphylococcus pseudintermedius, Streptococcus canis | Staphylococcus felis and other Staph. spp., Streptococcus spp. |
| System | Species/Type | Breed/Skull Type | Anatomical Niche | Dominant Bacteria (Phyla/Genera) | Clinical/Zoonotic Significance | Citations |
|---|---|---|---|---|---|---|
| Nasal | Dog | Mesocephalic/dolichocephalic | Nasal cavity | ↑ Proteobacteria ↓ Firmicutes, Actinomycetota | Typical, balanced microbiota; may contribute to respiratory immune defense | [43,45,46,47] |
| Brachycephalic | Nasal cavity | ↑ Firmicutes, Actinomycetota ↓ Proteobacteria | Microbiota shifts linked to anatomical traits; higher infection risk | [43,45,46,47] | ||
| Cat | – (no breed-specific data) | Nasal cavity | ↑ Proteobacteria medium: Firmicutes no data: Actinomycetota | Similar to dogs; breed-related studies are lacking | [27,30,36,37,38,39,40,41,46,47] | |
| Oral | Dog | Maltese, Teddy (toy breeds) | Saliva | Porphyromonas, Moraxella | Porphyromonas—linked to periodontal disease; Moraxella—opportunistic respiratory pathogen; zoonotic potential | [27,29,36,37,38,39,40,41] |
| Golden Retriever | Saliva | Neisseria, Streptococcus | Streptococcus—may cause infections in humans; Neisseria—some species are pathogenic | [27,29,36,37,38,39,40,41] | ||
| Cat | British Shorthair, Ragdoll | Saliva | Porphyromonas, Moraxella | Similar to toy dog breeds; risk of periodontal infections and bacterial transmission | [27,30,36,37,38,39,40,41] | |
| Chinese garden cat | Saliva | Porphyromonas, Fusobacterium | Fusobacterium—anaerobes associated with inflammation and oral infections | [27,30,36,37,38,39,40,41] | ||
| Dog and cat | - | Supragingival plaque | Proteobacteria, Bacteroidota, Firmicutes, ↑ Actinobacteria, Saccharibacteria | Highest bacterial diversity; key site for periodontal disease | [27,29,30,36,37,38,39,40,41] | |
| - | Buccal mucosa | Proteobacteria, Firmicutes | Less diverse; relatively stable and less pathogenic flora | [27,29,30,36,37,38,39,40,41] | ||
| - | Dorsum of tongue | Bacteroidota, Firmicutes | Anaerobes may thrive; possible source of halitosis | [27,29,30,36,37,38,39,40,41] | ||
| - | Saliva | Proteobacteria, Bacteroidota, Firmicutes | Breed-dependent differences; influences oral and general health, as well as zoonotic risk | [27,29,30,36,37,38,39,40,41] |
| Procedure | Risk Description | Preventive Recommendations | Citations |
|---|---|---|---|
| Use of Intravenous Catheters (especially in the cervical region) | Prolonged catheter use, particularly in the neck area, can cause local infections, external jugular vein thrombosis, and bacteremia, potentially leading to spontaneous bacterial peritonitis (SBP). | Maintain strict aseptic technique; regularly monitor catheter sites; avoid prolonged use when possible | [107,109] |
| Endoscopic procedures | May cause transient bacteremia due to mucosal damage and bacterial translocation from the oral or gastrointestinal tract into the bloodstream. | Ensure aseptic conditions during procedures; consider antibiotic prophylaxis in high-risk cases. | [107] |
| Paracentesis (abdominal fluid aspiration) | Risk of introducing bacteria into the abdominal cavity, especially if performed non-aseptically; bacteremia may lead to SBP. | Perform under aseptic conditions; monitor patients closely after the procedure. | [107,108] |
| Use of peritoneal dialysis catheters | Can introduce bacterial infections, especially with poor hygiene; contact with pets may be a source of zoonotic transmission. | Ensure strict hygiene and aseptic handling; limit contact with potential infection sources (e.g., other animals or humans with infections). | [107,108] |
| Urological procedures (e.g., bladder catheterization) | Can lead to urinary tract infections and bacteremia, which may result in SBP. | Maintain aseptic technique during catheterization; minimize catheter duration; monitor for signs of infection. | [107] |
| Preoperative shaving | Can cause microtraumas, which become entry points for bacteria, increasing the risk of SSIs. | Avoid shaving with razors; prefer clipping or depilatory creams; perform hair removal immediately before surgery. | [111,112,113] |
| Use of razors for shaving | Causes epidermal damage, increasing the likelihood of SSIs. | Use safer hair removal methods (e.g., clippers); clipping of the surgical site <4 h prior to the procedure; avoid using razors perioperatively. | [112] |
| Skin infections as a source of endogenous infection | Naturally occurring skin bacteria may cause infections if the skin is damaged or contaminated during procedures. | Conduct dermatological evaluations prior to surgery; treat pre-existing skin infections before performing procedures. | [114] |
| Wound Classification | Clinical Description | Antibiotic Therapy Recommendations |
|---|---|---|
| Clean wound | Planned surgical procedure with no entry into the respiratory, gastrointestinal, or genitourinary tract; no infection or trauma. | Antibiotics generally not required, unless the procedure exceeds 90 min or involves implants. |
| Clean-contaminated wound | Entry into the respiratory, gastrointestinal, or genitourinary tract under controlled conditions without major contamination. | Prophylactic antibiotics recommended, e.g., for cystotomy or gastrotomy. |
| Contaminated wound | Fresh traumatic wounds, spillage from the gastrointestinal tract, major break in sterile technique, or acute non-purulent inflammation. | Therapeutic antibiotics indicated, especially if infection is suspected before surgery. |
| Dirty wound | Clinical infection present, necrotic tissue, foreign material, or chronic wound contamination. | Intensive antibiotic therapy required, based on culture and sensitivity results. |
| Drug Class | Drug Example | Resistance Gene(s) | Resistance Mechanism | Bacteria Example | Citations |
|---|---|---|---|---|---|
| β-lactams | Ampicillin | blaTEM, blaSHV | Bacteria produce β-lactamases—enzymes that hydrolyze the β-lactam ring, rendering the antibiotic ineffective. | E. coli, Klebsiella pneumoniae | [125] |
| Cefotaxime | blaCTX-M | Bacteria produce ESBLs that can hydrolyze a wide range of cephalosporins. | E. coli, Enterobacter spp. | [125] | |
| Oxacillin | mecA | mecA encodes a modified penicillin-binding protein (PBP2a) with low affinity for β-lactams, allowing cell wall synthesis to continue | Staphylococcus aureus (MRSA) | [125] | |
| Carbapenems | Imipenem, Meropenem | blaKPC, blaNDM, blaVIM, blaOXA-48 | Bacteria produce carbapenemases—enzymes capable of hydrolyzing carbapenems, among the most potent β-lactams. | K. pneumoniae, P. aeruginosa, A. baumannii | [127] |
| Aminoglycosides | Gentamicin | aac(6′)-Ib, aph(3′), ant(2″) | Bacterial enzymes chemically modify the antibiotic using acetylation or phosphorylation, preventing ribosome binding. | E. coli, P. aeruginosa, Enterococcus spp. | [125,126,128] |
| Tetracyclines | Doxycycline | tet(A), tet(B), tet(M) | Efflux pumps remove the antibiotic from the cell; ribosomal protection proteins prevent drug binding to the ribosome. | E. coli, Streptococcus spp., Enterococcus spp. | [129] |
| Quinolones | Ciprofloxacin | qnrA, qnrB, qnrS, aac(6′)-Ib-cr | Qnr proteins protect DNA gyrase from inhibition; modifying enzymes reduce the antibiotic activity. | E. coli, Salmonella spp., Klebsiella spp. | [126,127] |
| Mutations in gyrA, parC | Mutations alter the structure of DNA gyrase or topoisomerase IV, preventing the antibiotic from binding effectively. | Campylobacter spp., Neisseria gonorrhoeae | [126,127] | ||
| Macrolides | Azithromycin | erm(B), mef(A/E) | erm genes encode rRNA methylation (modifying the binding site), and mef genes encode efflux pumps that expel the drug. | S. pneumoniae, S. pyogenes, Mycoplasma spp. | [127,130,136] |
| Glycopeptides | Vancomycin | vanA, vanB | The bacteria alter their cell wall precursors from D-Ala-D-Ala to D-Ala-D-Lac, which reduces vancomycin binding. | E. faecium, E. faecalis | [133] |
| Sulfonamides | Sulfamethoxazole | sul1, sul2, sul3 | The genes encode an alternative dihydropteroate synthase that is not inhibited by the antibiotic—allowing folic acid synthesis to continue. | E. coli, Salmonella spp., Shigella spp. | [125,127,129,131,135] |
| Chloramphenicol | Chloramphenicol | cat, floR, cmlA | The antibiotic is inactivated by acetylation, and efflux pumps remove it from the cell. | Salmonella spp., E. coli, K. pneumoniae | [132] |
| Rifampin | Rifampin | rpoB mutations | Mutations in rpoB alter the binding site on RNA polymerase, making it resistant to inhibition. | Mycobacterium tuberculosis, S. aureus | [125,134] |
| ARG | Antibiotic Resistance | Colonized Species | Typical Anatomical Locations (Dog/Cat/Human) | Typical Microbiota | Notes | Citations |
|---|---|---|---|---|---|---|
| blaCTX-M | ESBL —cephalosporins | Dog: yes (frequent) | Dog: intestines, urinary tract | E. coli, Enterobacteriaceae | Plasmid-mediated; widespread. ST131 in humans genetically related to canine isolates. | [32,138,139,140] |
| Cat: yes (less common) | Cat: intestines | |||||
| Human: yes (frequent) | Human: intestines, urinary tract | |||||
| blaTEM, blaSHV | Penicillins, early cephalosporins | Dog: yes | Dog: intestines | Enterobacteriaceae | Common in both commensal and pathogenic strains. | [32,138,139,140,146] |
| Cat: yes | Cat: intestines | |||||
| Human: yes | Human: intestines | |||||
| mecA | Methicillin —β-lactams | Dog: yes | Dog: skin, nose | S. pseudintermedius, S. aureus | Zoonotic transfer risk; MRSP/MRSA detected across species. | [32,138,139,141,142,146] |
| Cat: less common | Cat: skin, nose | |||||
| Human: yes (carrier state) | Human: nose, skin | |||||
| blaZ | Penicillins | Dog: very common | Dog: skin, nose | Staphylococcus spp. | Highly prevalent in canine staphylococci. | [32,138,139,146] |
| Cat: yes | Cat: skin, nose | |||||
| Human: yes | Human: skin, nose | |||||
| tet(M), tet(A), tet(B) | Tetracyclines | Dog: yes | Dog: oral cavity, intestines, nose | Porphyromonas, Fusobacterium, E. coli | Frequently associated with mobile genetic elements; found in biofilms. | [32,137,138,139,146] |
| Cat: yes | Cat: oral cavity, intestines | |||||
| Human: yes | Human: oral cavity, intestines | |||||
| erm(B), erm(C) | Macrolides, lincosamides | Dog: yes | Dog: oral cavity, intestines, nose | Enterococcus, Staphylococcus, anaerobes | Broadly present in multiple genera, especially anaerobes and staphylococci. | [32,137,138,139,143,145,146] |
| Cat: yes | Cat: oral cavity, intestines | |||||
| Human: yes | Human: oral cavity, intestines, nose | |||||
| aac(6′)-Ib, aph(3′)-IIIa | Aminoglycosides | Dog: yes | Dog: intestines, nose | E. coli, Enterococcus | Found in multidrug-resistant isolates from animals and humans. | [32,138,139,144,146] |
| Cat: less common | Cat: intestines | |||||
| Human: yes | Human: intestines, nose | |||||
| qnr genes (qnrS, qnrB) | Fluoroquinolones | Dog: yes | Dog: urinary tract, intestines | Enterobacteriaceae | Plasmid-mediated quinolone resistance; detected across species. | [32,138,139,144,145,146] |
| Cat: yes | Cat: urinary tract, intestines | |||||
| Human: yes | Human: intestines, urine | |||||
| sul1, sul2 | Sulfonamides | Dog: yes | Dog: intestines, urine | E. coli, Bacteroides | Commonly associated with class 1 integrons; easily disseminated. | [32,138,139] |
| Cat: yes | Cat: intestines, urine | |||||
| Human: yes | Human: intestines, urine | |||||
| dfrA | Trimethoprim | Dog: yes | Dog: intestines | Enterococcus, E. coli | Often co-selected with sul genes (sul1, sul2). | [32,138,139] |
| Cat: yes | Cat: intestines | |||||
| Human: yes | Human: intestines |
| Bacteria | Direction of Transfer | Host(s) | Details | Citations |
|---|---|---|---|---|
| Pasteurella multocida | Cat → Human | Cat and woman with sinusitis | Woman had daily contact with a cat that licked her; nasal and saliva swabs showed biochemically and genotypically similar strains. | [147] |
| Dog → Infant | Dogs and infants | Infants developed infections, including meningitis, after dogs licked them or shared beds. | [149] | |
| Staphylococcus intermedius | Dog → Human | Dog and 51-year-old woman with ear infection | Dog licked patient’s ears; identical bacterial strains isolated from dog’s saliva and patient’s ear. | [150] |
| Staphylococcus intermedius (MR strain) | Dog → Human | Dog with pyoderma and 28-year-old woman with methicillin-resistant sinusitis | Frequent face contact; bacterial strains from dog’s skin and woman’s nose were identical. | [151] |
| Methicillin-resistant Staphylococcus aureus | Human → Dog, then Dog → Human | 76-year-old man and his dog with severe cellulitis after surgery | Man had recurring MRSA; dog, recovering from surgery, became infected. Genetic testing confirmed identical multidrug-resistant strains in both, suggesting bidirectional transmission. | [152] |
| At the Owner Level | At the Veterinary Level | Citations | ||
|---|---|---|---|---|
| Owner | Veterinarian | |||
| Antibiotic use | Regular preventive veterinary check-ups should ensure routine health monitoring, and vaccination helps to reduce the incidence of infections and the need for antibiotic use. | Antibiotic therapy should be based on microbiological diagnostics; antibiotic treatment should be applied only following culture and susceptibility testing. | [153,154,156] | [154,156] |
| Improper disposal of antibiotics should be avoided; unused antibiotics should be returned to designated disposal points (e.g., pharmacies or clinics). | Critically important antibiotics should be prescribed only when strictly necessary. | [154,156] | [154,156] | |
| Veterinarians should educate owners; communication strategies should be implemented to inform owners about the risks of inappropriate antibiotic use. | [153,154] | |||
| Preventive measures, biosecurity | Close physical contact should be limited; sharing beds, food, or allowing animals to lick the face should be avoided to minimize the risk of transmission of resistant bacteria. | Biosecurity protocols should be implemented in veterinary settings; infected animals should be isolated, and disinfection procedures should be strictly followed. | [155,156] | [156] |
| Hand hygiene should be practiced after animal contact; handwashing is recommended after handling, feeding, or cleaning up after animals to reduce the risk of microbial transmission. | [155,156] | |||
| The domestic environment should be regularly cleaned; food and water bowls, bedding, and litter boxes should be disinfected regularly, particularly during illness. | Participation in training and awareness programs should be encouraged; continuous education on responsible antibiotic use should be promoted among veterinary professionals. | [153,155,156] | [153,156] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horodyska, I.; Kasperska, P.; Michalski, K.; Bubak, J.; Herman, I.; Miszczak, M. Natural Microbiota of Dogs and Cats as a Source and Vector of Resistance Genes—Clinical Significance. Int. J. Mol. Sci. 2025, 26, 7717. https://doi.org/10.3390/ijms26167717
Horodyska I, Kasperska P, Michalski K, Bubak J, Herman I, Miszczak M. Natural Microbiota of Dogs and Cats as a Source and Vector of Resistance Genes—Clinical Significance. International Journal of Molecular Sciences. 2025; 26(16):7717. https://doi.org/10.3390/ijms26167717
Chicago/Turabian StyleHorodyska, Iga, Patrycja Kasperska, Kacper Michalski, Joanna Bubak, Izabela Herman, and Marta Miszczak. 2025. "Natural Microbiota of Dogs and Cats as a Source and Vector of Resistance Genes—Clinical Significance" International Journal of Molecular Sciences 26, no. 16: 7717. https://doi.org/10.3390/ijms26167717
APA StyleHorodyska, I., Kasperska, P., Michalski, K., Bubak, J., Herman, I., & Miszczak, M. (2025). Natural Microbiota of Dogs and Cats as a Source and Vector of Resistance Genes—Clinical Significance. International Journal of Molecular Sciences, 26(16), 7717. https://doi.org/10.3390/ijms26167717






