The Link Between Human Alkyladenine DNA Glycosylase and Cancer Development
Abstract
1. Introduction
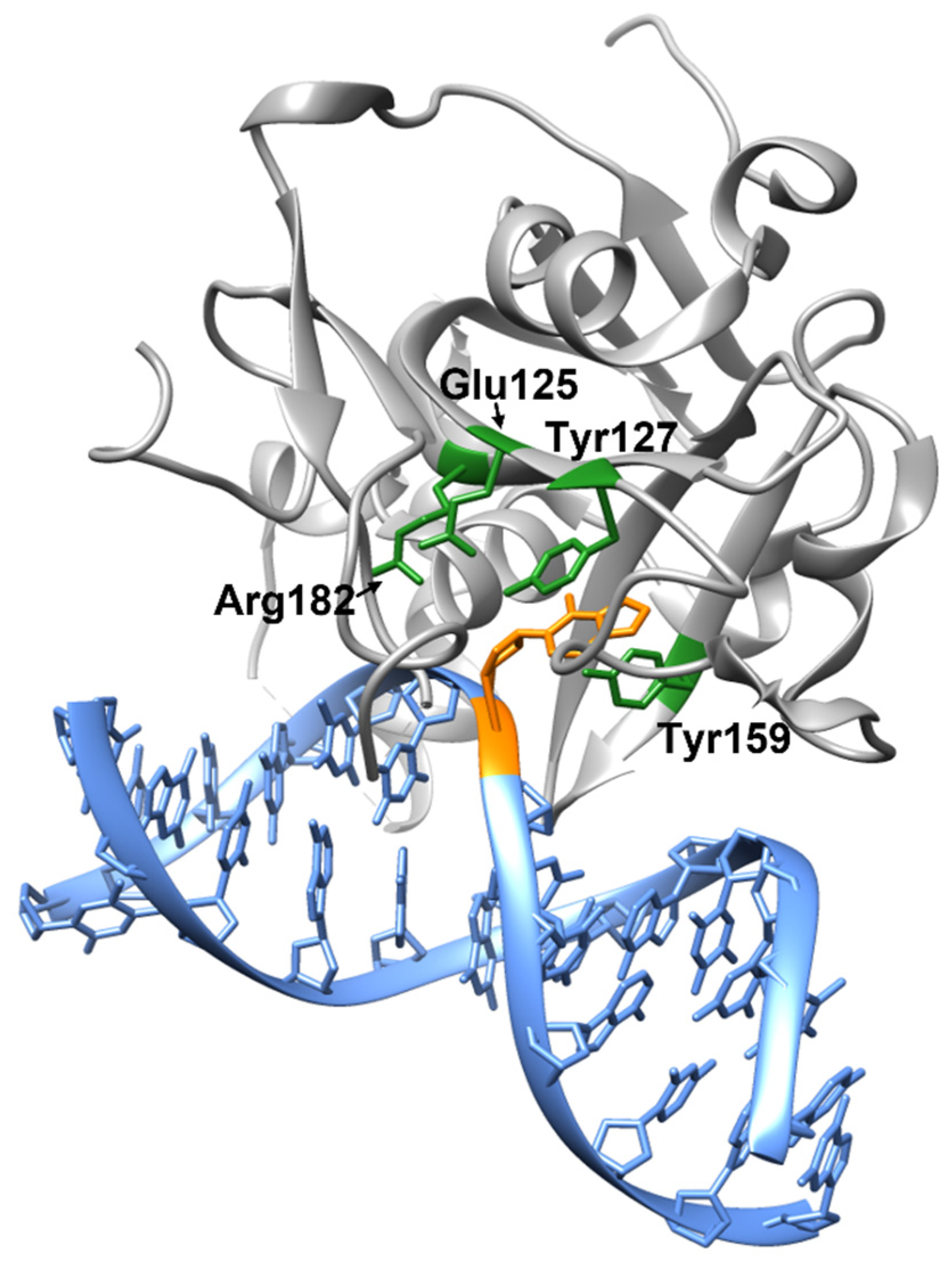
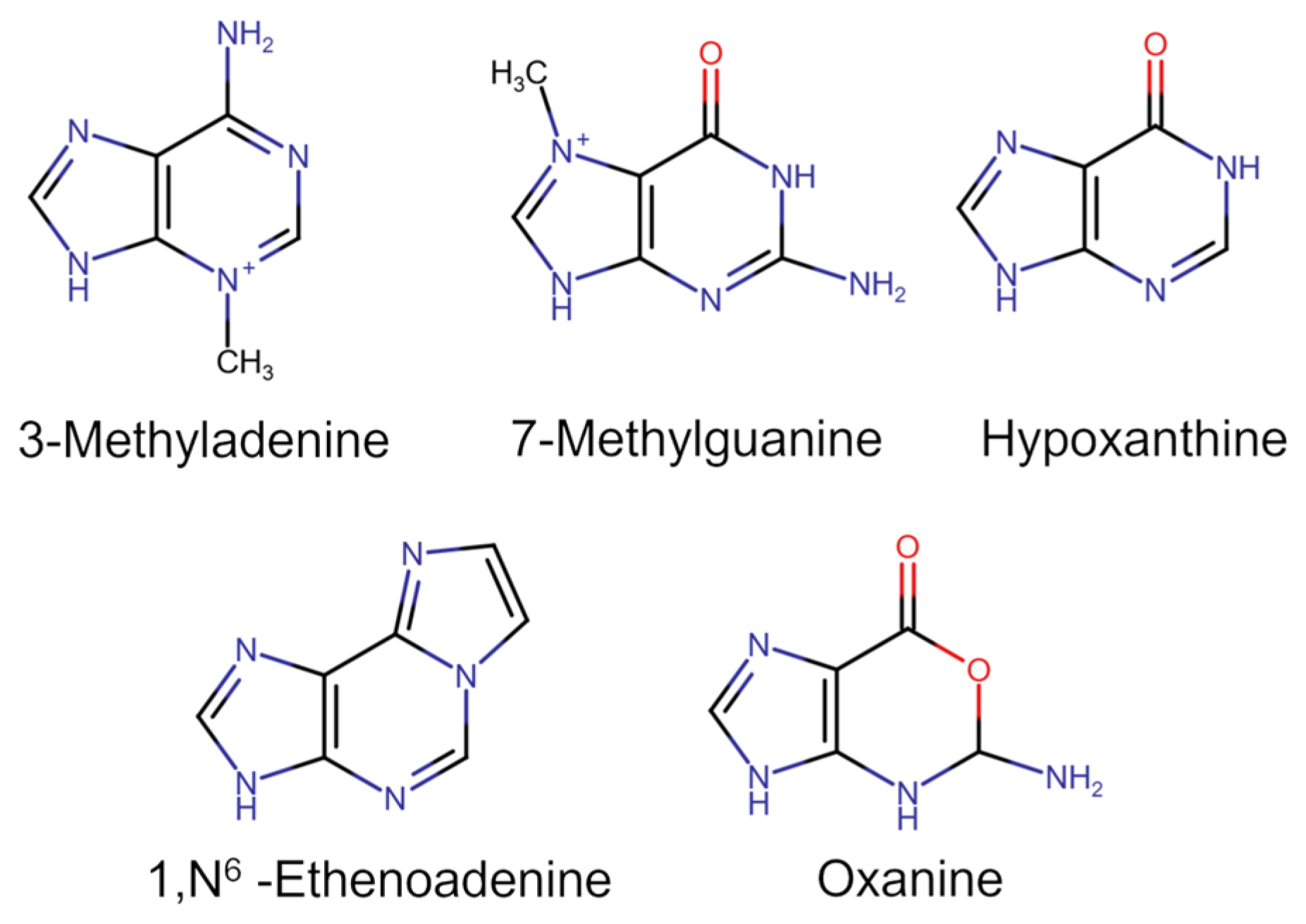
2. Structure and Catalytic Mechanism of AAG
3. Polymorphic Variants of AAG and Other DNA Repair Enzymes
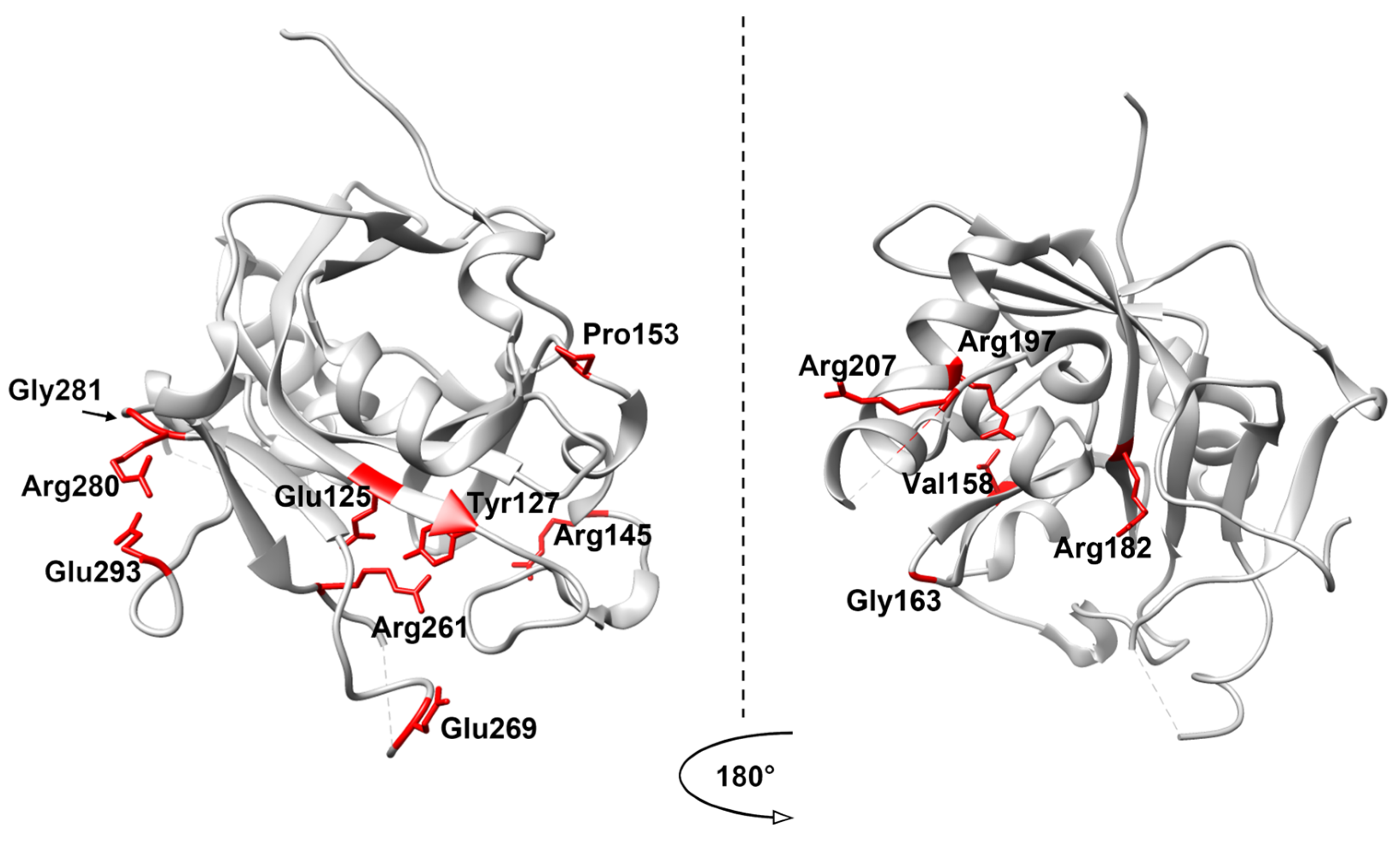
4. Analysis of SNP Variants of AAG
- High negative impact: negative prediction in ≥5 of 6 programs.
- Medium impact: negative prediction in 3–4 programs.
- Low impact: negative prediction in ≤2 programs.
5. The Role of AAG in the Development of Oncological Diseases
6. Discussion
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Friedberg, E.C.; Aguilera, A.; Gellert, M.; Hanawalt, P.C.; Hays, J.B.; Lehmann, A.R.; Lindahl, T.; Lowndes, N.; Sarasin, A.; Wood, R.D. DNA Repair: From Molecular Mechanism to Human Disease. DNA Repair. 2006, 5, 986–996. [Google Scholar] [CrossRef]
- Friedberg, E.C. Inroads into Base Excision Repair II: The Discovery of DNA Glycosylases. DNA Repair. 2004, 3, 1531–1536. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C. How Nucleotide Excision Repair Protects against Cancer. Nat. Rev. Cancer 2001, 1, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA Damage, Repair and Mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235. [Google Scholar] [CrossRef] [PubMed]
- Gohil, D.; Sarker, A.H.; Roy, R. Base Excision Repair: Mechanisms and Impact in Biology, Disease, and Medicine. Int. J. Mol. Sci. 2023, 24, 14186. [Google Scholar] [CrossRef]
- Pascucci, B.; Stucki, M.; Jónsson, Z.O.; Dogliotti, E.; Hübscher, U. Long Patch Base Excision Repair with Purified Human Proteins: DNA Ligase I as Patch Size Mediator for DNA Polymerases δ and ε. J. Biol. Chem. 1999, 274, 33696–33702. [Google Scholar] [CrossRef]
- Krokan, H.E.; Bjørås, M. Base Excision Repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef]
- Fromme, J.C.; Verdine, G.L. Base Excision Repair. Adv. Protein Chem. 2004, 69, 1–41. [Google Scholar] [CrossRef]
- Seeberg, E.; Eide, L.; Bjørås, M. The Base Excision Repair Pathway. Trends Biochem. Sci. 1995, 20, 391–397. [Google Scholar] [CrossRef]
- Jagannathan, I.; Cole, H.A.; Hayes, J.J. Base Excision Repair in Nucleosome Substrates. Chromosome Res. 2006, 14, 27–37. [Google Scholar] [CrossRef]
- Jacobs, A.L.; Schär, P. DNA Glycosylases: In DNA Repair and Beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Visnes, T.; Doseth, B.; Pettersen, H.S.; Hagen, L.; Sousa, M.M.L.; Akbari, M.; Otterlei, M.; Kavli, B.; Slupphaug, G.; Krokan, H.E. Uracil in DNA and Its Processing by Different DNA Glycosylases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Faucher, F.; Doublié, S.; Jia, Z. 8-Oxoguanine DNA Glycosylases: One Lesion, Three Subfamilies. Int. J. Mol. Sci. 2012, 13, 6711–6729. [Google Scholar] [CrossRef] [PubMed]
- Gallinari, P.; Jiricny, J. A New Class of Uracil-DNA Glycosylases Related to Human Thymine-DNA Glycosylase. Nature 1996, 383, 735–738. [Google Scholar] [CrossRef]
- Rosenquist, T.A.; Zaika, E.; Fernandes, A.S.; Zharkov, D.O.; Miller, H.; Grollman, A.P. The Novel DNA Glycosylase, NEIL1, Protects Mammalian Cells from Radiation-Mediated Cell Death. DNA Repair. 2003, 2, 581–591. [Google Scholar] [CrossRef]
- Liu, P.; Burdzy, A.; Sowers, L.C. Substrate Recognition by a Family of Uracil-DNA Glycosylases: UNG, MUG, and TDG. Chem. Res. Toxicol. 2002, 15, 1001–1009. [Google Scholar] [CrossRef]
- Prakash, A.; Doublié, S.; Wallace, S.S. The Fpg/Nei Family of DNA Glycosylases. Prog. Mol. Biol. Transl. Sci. 2012, 110, 71–91. [Google Scholar]
- Demple, B.; Linn, S. DNA N-Glycosylases and UV Repair. Nature 1980, 287, 203–208. [Google Scholar] [CrossRef]
- Krokan, H.E.; Standal, R.; Slupphaug, G. DNA Glycosylases in the Base Excision Repair of DNA. Biochem. J. 1997, 325, 1–16. [Google Scholar] [CrossRef]
- Wallace, S.S. DNA Glycosylases Search for and Remove Oxidized DNA Bases. Environ. Mol. Mutagen. 2013, 54, 691–704. [Google Scholar] [CrossRef]
- Demple, B.; Sung, J.S. Molecular and Biological Roles of Ape1 Protein in Mammalian Base Excision Repair. DNA Repair. 2005, 4, 1442–1449. [Google Scholar] [CrossRef] [PubMed]
- Sobol, R.W.; Wilson, S.H. Mammalian DNA β-Polymerase in Base Excision Repair of Alkylation Damage. Prog. Nucleic Acid. Res. Mol. Biol. 2001, 68, 57–74. [Google Scholar]
- Kumar, A.; Widen, S.G.; Williams, K.R.; Kedar, P.; Karpel, R.L.; Wilson, S.H. Studies of the Domain Structure of Mammalian DNA Polymerase Beta. Identification of a Discrete Template Binding Domain. J. Biol. Chem. 1990, 265, 2124–2131. [Google Scholar] [CrossRef]
- Beard, W.A.; Wilson, S.H. DNA Polymerase Beta and Other Gap-Filling Enzymes in Mammalian Base Excision Repair. In Enzymes; Academic Press: Cambridge, MA, USA, 2019; Volume 45, pp. 1–26. ISBN 9780128173961. [Google Scholar]
- Campalans, A.; Marsin, S.; Nakabeppu, Y.; O’Connor, T.R.; Boiteux, S.; Radicella, J.P. XRCC1 Interactions with Multiple DNA Glycosylases: A Model for Its Recruitment to Base Excision Repair. DNA Repair. 2005, 4, 826–835. [Google Scholar] [CrossRef]
- Cannan, W.J.; Rashid, I.; Tomkinson, A.E.; Wallace, S.S.; Pederson, D.S. The Human Ligase IIIα-XRCC1 Protein Complex Performs DNA Nick Repair after Transient Unwrapping of Nucleosomal DNA. J. Biol. Chem. 2017, 292, 5227–5238. [Google Scholar] [CrossRef]
- Dianova, I.I.; Sleeth, K.M.; Allinson, S.L.; Parsons, J.L.; Breslin, C.; Caldecott, K.W.; Dianov, G.L. XRCC1-DNA Polymerase β Interaction Is Required for Efficient Base Excision Repair. Nucleic Acids Res. 2004, 32, 2550–2555. [Google Scholar] [CrossRef]
- Marintchev, A.; Robertson, A.; Dimitriadis, E.K.; Prasad, R.; Wilson, S.H.; Mullen, G.P. Domain Specific Interaction in the XRCC1-DNA Polymerase Beta Complex. Nucleic Acids Res. 2000, 28, 2049–2059. [Google Scholar] [CrossRef]
- Nash, R.A.; Caldecott, K.W.; Barnes, D.E.; Lindahl, T. XRCC1 Protein Interacts with One of Two Distinct Forms of DNA Ligase III. Biochemistry 1997, 36, 5207–5211. [Google Scholar] [CrossRef]
- Odell, I.D.; Barbour, J.-E.; Murphy, D.L.; Della-Maria, J.A.; Sweasy, J.B.; Tomkinson, A.E.; Wallace, S.S.; Pederson, D.S. Nucleosome Disruption by DNA Ligase III-XRCC1 Promotes Efficient Base Excision Repair. Mol. Cell Biol. 2011, 31, 4623–4632. [Google Scholar] [CrossRef]
- Li, Y. DNA Adducts in Cancer Chemotherapy. J. Med. Chem. 2024, 67, 5113–5143. [Google Scholar] [CrossRef]
- Kondo, N.; Takahashi, A.; Ono, K.; Ohnishi, T. DNA Damage Induced by Alkylating Agents and Repair Pathways. J. Nucleic Acids 2010, 2010, 543531. [Google Scholar] [CrossRef] [PubMed]
- Plosky, B.; Samson, L.; Engelward, B.P.; Gold, B.; Schlaen, B.; Millas, T.; Magnotti, M.; Schor, J.; Scicchitano, D.A. Base Excision Repair and Nucleotide Excision Repair Contribute to the Removal of N-Methylpurines from Active Genes. DNA Repair. 2002, 1, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, B.; Bates, P.A.; Paik, J.; Jacobs, S.C.; Lindahl, T. Repair of Alkylated DNA: Recent Advances. DNA Repair. 2007, 6, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Gentil, A.; Cabral-Neto, J.B.; Mariage-Samson, R.; Margot, A.; Imbach, J.L.; Rayner, B.; Sarasin, A. Mutagenicity of a Unique Apurinic/Apyrimidinic Site in Mammalian Cells. J. Mol. Biol. 1992, 227, 981–984. [Google Scholar] [CrossRef]
- Wilson, D.M.; Barsky, D. The Major Human Abasic Endonuclease: Formation, Consequences and Repair of Abasic Lesions in DNA. Mutat. Res.-DNA Repair. 2001, 485, 283–307. [Google Scholar] [CrossRef]
- Carles, J.; Monzo, M.; Amat, M.; Jansa, S.; Artells, R.; Navarro, A.; Foro, P.; Alameda, F.; Gayete, A.; Gel, B.; et al. Single-Nucleotide Polymorphisms in Base Excision Repair, Nucleotide Excision Repair, and Double Strand Break Genes as Markers for Response to Radiotherapy in Patients with Stage I to II Head-and-Neck Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2006, 66, 1022–1030. [Google Scholar] [CrossRef]
- Kiyohara, C.; Takayama, K.; Nakanishi, Y. Association of Genetic Polymorphisms in the Base Excision Repair Pathway with Lung Cancer Risk: A Meta-Analysis. Lung Cancer 2006, 54, 267–283. [Google Scholar] [CrossRef]
- Figueroa, J.D.; Malats, N.; Real, F.X.; Silverman, D.; Kogevinas, M.; Chanock, S.; Welch, R.; Dosemeci, M.; Tardón, A.; Serra, C.; et al. Genetic Variation in the Base Excision Repair Pathway and Bladder Cancer Risk. Hum. Genet. 2007, 121, 233–242. [Google Scholar] [CrossRef]
- Jacob, K.D.; Hooten, N.N.; Tadokoro, T.; Lohani, A.; Barnes, J.; Evans, M.K. Alzheimer’s Disease-Associated Polymorphisms in Human OGG1 Alter Catalytic Activity and Sensitize Cells to DNA Damage. Free Radic. Biol. Med. 2013, 63, 115–125. [Google Scholar] [CrossRef]
- Hollis, T.; Ichikawa, Y.; Ellenberger, T. DNA Bending and a Flip-out Mechanism for Base Excision by the Helix-Hairpin-Helix DNA Glycosylase, Escherichia Coli AlkA. EMBO J. 2000, 19, 758–766. [Google Scholar] [CrossRef]
- Labahn, J.; Schärer, O.D.; Long, A.; Ezaz-Nikpay, K.; Verdine, G.L.; Ellenberger, T.E. Structural Basis for the Excision Repair of Alkylation-Damaged DNA. Cell 1996, 86, 321–329. [Google Scholar] [CrossRef]
- Rubinson, E.H.; Gowda, A.S.P.; Spratt, T.E.; Gold, B.; Eichman, B.F. An Unprecedented Nucleic Acid Capture Mechanism for Excision of DNA Damage. Nature 2010, 468, 406–413. [Google Scholar] [CrossRef][Green Version]
- Lau, A.Y.; Schä, O.D.; Samson, L.; Verdine, G.L.; Ellenberger, T. Crystal Structure of a Human Alkylbase-DNA Repair Enzyme Complexed to DNA: Mechanisms for Nucleotide Flipping and Base Excision. Cell 1998, 95, 249–258. [Google Scholar] [CrossRef]
- Thayer, M.M.; Ahern, H.; Xing, D.; Cunningham, R.P.; Tainer, J. A Novel DNA Binding Motifs in the DNA Repair Enzyme Endonuclease III Crystal Structure. EMBO J. 1995, 14, 4108–4120. [Google Scholar] [CrossRef] [PubMed]
- Doherty, A.J.; Serpell, L.C.; Ponting, C.P. The Helix-Hairpin-Helix DNA-Binding Motif: A Structural Basis for Non-Sequence-Specific Recognition of DNA. Nucleic Acids Res. 1996, 24, 2488–2497. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, Y.; Kato, M.; Odawara, K.; Tokuno, Y.; Nakashima, Y.; Matsushima, N.; Yasumura, K.; Tomita, K.; Ihara, K.; Fujii, Y.; et al. Three-Dimensional Structure of a DNA Repair Enzyme, 3-Methyladenine DNA Glycosylase II, from Escherichia Coli. Cell 1996, 86, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Rubinson, E.H.; Eichman, B.F. Nucleic Acid Recognition by Tandem Helical Repeats. Curr. Opin. Struct. Biol. 2012, 22, 101–109. [Google Scholar] [CrossRef][Green Version]
- Izumi, T.; Tatsuka, M.; Tano, K.; Asano, M.; Mitra, S. Molecular Cloning and Characterization of the Promoter of the Human N-Methylpurine-DNA Glycosylase (MPG) Gene. Carcinogenesis 1997, 18, 1837–1839. [Google Scholar] [CrossRef][Green Version]
- Samson, L.; Derfler, B.; Boosalis, M.; Call, K. Cloning and Characterization of a 3-Methyladenine DNA Glycosylase CDNA from Human Cells Whose Gene Maps to Chromosome 16. Proc. Natl. Acad. Sci. USA 1991, 88, 9127–9131. [Google Scholar] [CrossRef]
- Vickers, M.A.; Vyas, P.; Harris, P.C.; Simmons, D.L.; Higgs, D.R. Structure of the Human 3-Methyladenine DNA Glycosylase Gene and Localization Close to the 16p Telomere. Proc. Natl. Acad. Sci. USA 1993, 90, 3437–3441. [Google Scholar] [CrossRef]
- Montaldo, N.P.; Bordin, D.L.; Brambilla, A.; Rösinger, M.; Fordyce Martin, S.L.; Bjørås, K.Ø.; Bradamante, S.; Aas, P.A.; Furrer, A.; Olsen, L.C.; et al. Alkyladenine DNA Glycosylase Associates with Transcription Elongation to Coordinate DNA Repair with Gene Expression. Nat. Commun. 2019, 10, 5460. [Google Scholar] [CrossRef]
- Beranek, D.T. Distribution of Methyl and Ethyl Adducts Following Alkylation with Monofunctional Alkylating Agents. Mutat. Res./Fundam. Mol. Mech. Mutagen. 1990, 231, 11–30. [Google Scholar] [CrossRef]
- Mao, P.; Brown, A.J.; Malc, E.P.; Mieczkowski, P.A.; Smerdon, M.J.; Roberts, S.A.; Wyrick, J.J. Genome-Wide Maps of Alkylation Damage, Repair, and Mutagenesis in Yeast Reveal Mechanisms of Mutational Heterogeneity. Genome Res. 2017, 27, 1674–1684. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Sun, B.; Guo, W.T.; Zhang, Y.H.; Liu, X.; Xing, Y.; Dong, D.L. 3-Methyladenine Induces Cell Death and Its Interaction with Chemotherapeutic Drugs Is Independent of Autophagy. Biochem. Biophys. Res. Commun. 2013, 432, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.I.; Delaney, J.C.; Kartalou, M.; Lingaraju, G.M.; Maor-Shoshani, A.; Essigmann, J.M.; Samson, L.D. Recognition and Processing of a New Repertoire of DNA Substrates by Human 3-Methyladenine DNA Glycosylase (AAG). Biochemistry 2009, 48, 1850–1861. [Google Scholar] [CrossRef] [PubMed]
- Maher, R.L.; Vallur, A.C.; Feller, J.A.; Bloom, L.B. Slow Base Excision by Human Alkyladenine DNA Glycosylase Limits the Rate of Formation of AP Sites and AP Endonuclease 1 Does Not Stimulate Base Excision. DNA Repair. 2007, 6, 71–81. [Google Scholar] [CrossRef]
- Wang, P.; Guliaev, A.B.; Elder, R.H.; Hang, B. Alkylpurine-DNA-N-Glycosylase Excision of 7-(Hydroxymethyl)-1,N 6-Ethenoadenine, a Glycidaldehyde-Derived DNA Adduct. DNA Repair. 2006, 5, 23–31. [Google Scholar] [CrossRef]
- Hang, B.; Singer, B.; Margison, G.P.; Elder, R.H. Targeted Deletion of Alkylpurine-DNA-N-Glycosylase in Mice Eliminates Repair of 1,N6-Ethenoadenine and Hypoxanthine but Not of 3,N4-Ethenocytosine or 8-Oxoguanine. Proc. Natl. Acad. Sci. USA 1997, 94, 12869–12874. [Google Scholar] [CrossRef]
- Hitchcock, T.M.; Dong, L.; Connor, E.E.; Meira, L.B.; Samson, L.D.; Wyatt, M.D.; Cao, W. Oxanine DNA Glycosylase Activity from Mammalian Alkyladenine Glycosylase. J. Biol. Chem. 2004, 279, 38177–38183. [Google Scholar] [CrossRef]
- Fu, D.; Samson, L.D. Direct Repair of 3,N(4)-Ethenocytosine by the Human ALKBH2 Dioxygenase Is Blocked by the AAG/MPG Glycosylase. DNA Repair 2012, 11, 46–52. [Google Scholar] [CrossRef]
- Gros, L.; Maksimenko, A.V.; Privezentzev, C.V.; Laval, J.; Saparbaev, M.K. Hijacking of the Human Alkyl-N-Purine-DNA Glycosylase by 3,N4-Ethenocytosine, a Lipid Peroxidation-Induced DNA Adduct. J. Biol. Chem. 2004, 279, 17723–17730. [Google Scholar] [CrossRef]
- Nemec, A.A.; Wallace, S.S.; Sweasy, J.B. Variant Base Excision Repair Proteins: Contributors to Genomic Instability. Semin. Cancer Biol. 2010, 20, 320. [Google Scholar] [CrossRef]
- Datkhile, K.D.; Durgawale, P.P.; Patil, M.N.; Gudur, R.A.; Gudur, A.K.; Patil, S.R. Impact of Polymorphism in Base Excision Repair and Nucleotide Excision Repair Genes and Risk of Cervical Cancer: A Case-Control Study. Asian Pac. J. Cancer Prev. 2022, 23, 1291. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yue, H.; Li, S.; Guo, J.; Guan, Z.; Zhu, Z.; Niu, B.; Zhang, T.; Wang, J. Genetic Polymorphisms in DNA Repair Gene APE1/Ref-1 and the Risk of Neural Tube Defects in a High-Risk Area of China. Reprod. Sci. 2021, 28, 2592–2601. [Google Scholar] [CrossRef] [PubMed]
- Starcevic, D.; Dalal, S.; Sweasy, J.B. Is There a Link Between DNA Polymerase Beta and Cancer? Cell Cycle 2004, 3, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.Q.; Wang, T.; Zhao, Q.; Yang, H.Y.; Tan, X.H.; Dong, Z.M. Mutation of DNA Polymerase β in Esophageal Carcinoma of Different Regions. World J. Gastroenterol. WJG 2005, 11, 4618. [Google Scholar] [CrossRef]
- Murphy, D.L.; Donigan, K.A.; Jaeger, J.; Sweasy, J.B. The E288K Colon Tumor Variant of DNA Polymerase β Is a Sequence Specific Mutator. Biochemistry 2012, 51, 5269–5275. [Google Scholar] [CrossRef]
- Galick, H.A.; Marsden, C.G.; Kathe, S.; Dragon, J.A.; Volk, L.; Nemec, A.A.; Wallace, S.S.; Prakash, A.; Doublié, S.; Sweasy, J.B. The NEIL1 G83D Germline DNA Glycosylase Variant Induces Genomic Instability and Cellular Transformation. Oncotarget 2017, 8, 85883. [Google Scholar] [CrossRef]
- Almutairi, F.; Ali Khan Pathan, A.; Alanazi, M.; Shalaby, M.; Alabdulkarim, H.A.; Alamri, A.; Al Naeem, A.; Elrobh, M.; Shaik, J.P.; Khan, W.; et al. Association of DNA Repair Gene APE1 Asp148Glu Polymorphism with Breast Cancer Risk. Dis. Markers 2015, 2015, 869512. [Google Scholar] [CrossRef]
- Dai, Z.J.; Shao, Y.P.; Kang, H.F.; Tang, W.; Xu, D.; Zhao, Y.; Liu, D.; Wang, M.; Yang, P.T.; Wang, X.J. Relationship between Apurinic Endonuclease 1 Asp148Glu Polymorphism and Gastrointestinal Cancer Risk: An Updated Meta-Analysis. World J. Gastroenterol. 2015, 21, 5081–5089. [Google Scholar] [CrossRef]
- Cai, L.; Fu, Y.; Zhang, Y. APE1 Asp148Glu Polymorphism and Lung Cancer Susceptibility. Tumor Biol. 2014, 35, 5237–5244. [Google Scholar] [CrossRef] [PubMed]
- Illuzzi, J.L.; Harris, N.A.; Manvilla, B.A.; Kim, D.; Li, M.; Drohat, A.C.; Wilson, D.M. Functional Assessment of Population and Tumor-Associated APE1 Protein Variants. PLoS ONE 2013, 8, e65922. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, S.; Chetram, M.A.; Woodrick, J.; Mitra, P.S.; Manthena, P.V.; Khatkar, P.; Dakshanamurthy, S.; Dixon, M.; Karmahapatra, S.K.; Nuthalapati, N.K.; et al. Germ Line Variants of Human N-Methylpurine DNA Glycosylase Show Impaired DNA Repair Activity and Facilitate 1,N6-Ethenoadenine-Induced Mutations. J. Biol. Chem. 2015, 290, 4966. [Google Scholar] [CrossRef]
- Huang, C.M.; Chen, S.Y.; Huang, P.H.; Tsai, F.J. Effect of MPG Gene Rs2858056 Polymorphism, Copy Number Variation, and Level of Serum MPG Protein on the Risk for Rheumatoid Arthritis. PLoS ONE 2015, 10, e0120699. [Google Scholar] [CrossRef]
- Chen, S.Y.; Wan, L.; Huang, C.M.; Huang, Y.C.; Sheu, J.J.C.; Lin, Y.J.; Liu, S.P.; Lan, Y.C.; Lai, C.H.; Lin, C.W.; et al. Genetic Polymorphisms of the DNA Repair Gene MPG May Be Associated with Susceptibility to Rheumatoid Arthritis. J. Appl. Genet. 2010, 51, 519–521. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. Predicting Deleterious Amino Acid Substitutions. Genome Res. 2001, 11, 863. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting Amino Acid Changes That Affect Protein Function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- Ramensky, V.; Bork, P.; Sunyaev, S. Human Non-Synonymous SNPs: Server and Survey. Nucleic Acids Res. 2002, 30, 3894–3900. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. In Current Protocols in Human Genetics; Wiley: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Martin, K.; Daniela, M.W.; Preti, J.; Brain, J.O.; Gregory, M.C.; Jay, S. A General Framework for Estimating the Relative Pathogenicity of Human Genetic Variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Schubach, M.; Maass, T.; Nazaretyan, L.; Röner, S.; Kircher, M. CADD v1.7: Using Protein Language Models, Regulatory CNNs and Other Nucleotide-Level Scores to Improve Genome-Wide Variant Predictions. Nucleic Acids Res. 2024, 52, D1143–D1154. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation Prediction for the Deep-Sequencing Age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Rödelsperger, C.; Schuelke, M.; Seelow, D. MutationTaster Evaluates Disease-Causing Potential of Sequence Alterations. Nat. Methods 2010, 7, 575–576. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhi, D.; Wang, K.; Liu, X. MetaRNN: Differentiating Rare Pathogenic and Rare Benign Missense SNVs and InDels Using Deep Learning. Genome Med. 2022, 14, 115. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
- Choi, Y.; Chan, A.P. PROVEAN Web Server: A Tool to Predict the Functional Effect of Amino Acid Substitutions and Indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
- Wallace, S.S.; Murphy, D.L.; Sweasy, J.B. Base Excision Repair and Cancer. Cancer Lett. 2012, 327, 73–89. [Google Scholar] [CrossRef]
- Ibeanu, G.; Hartenstein, B.; Dunn, W.C.; Chang, L.Y.; Hofmann, E.; Coquerelle, T.; Mitra, S.; Kaina, B. Overexpression of Human DNA Repair Protein N-Methylpurine-DNA Glycosylase Results in the Increased Removal of N-Methylpurines in DNA without a Concomitant Increase in Resistance to Alkylating Agents in Chinese Hamster Ovary Cells. Carcinogenesis 1992, 13, 1989–1995. [Google Scholar] [CrossRef]
- Habraken, Y.; Laval, F. Increased Resistance of the Chinese Hamster Mutant Irsl Cells to Monofunctional Alkylating Agents by Transfection of the E. Coli or Mammalian N3-Methyladenine-DNA-Glycosylase Genes. Mutat. Res.-DNA Repair. 1993, 293, 187–195. [Google Scholar] [CrossRef]
- Klapacz, J.; Lingaraju, G.M.; Guo, H.H.; Shah, D.; Moar-Shoshani, A.; Loeb, L.A.; Samson, L.D. Frameshift Mutagenesis and Microsatellite Instability Induced by Human Alkyladenine DNA Glycosylase. Mol. Cell 2010, 37, 843. [Google Scholar] [CrossRef][Green Version]
- Sobol, R.W.; Prasad, R.; Evenski, A.; Baker, A.; Yang, X.P.; Horton, J.K.; Wilson, S.H. The Lyase Activity of the DNA Repair Protein β-Polymerase Protects from DNA-Damage-Induced Cytotoxicity. Nature 2000, 405, 807–810. [Google Scholar] [CrossRef]
- Kothandapani, A.; Dangeti, V.S.M.N.; Brown, A.R.; Banze, L.A.; Wang, X.H.; Sobol, R.W.; Patrick, S.M. Novel Role of Base Excision Repair in Mediating Cisplatin Cytotoxicity. J. Biol. Chem. 2011, 286, 14564–14574. [Google Scholar] [CrossRef]
- Cerda, S.R.; Turk, P.W.; Thor, A.D.; Weitzman, S.A. Altered Expression of the DNA Repair Protein, N-Methylpurine-DNA Glycosylase (MPG), in Breast Cancer. FEBS Lett. 1998, 431, 12–18. [Google Scholar] [CrossRef]
- Leitner-Dagan, Y.; Sevilya, Z.; Pinchev, M.; Kramer, R.; Elinger, D.; Roisman, L.C.; Rennert, H.S.; Schechtman, E.; Freedman, L.; Rennert, G.; et al. N-Methylpurine DNA Glycosylase and OGG1 DNA Repair Activities: Opposite Associations with Lung Cancer Risk. J. Natl. Cancer Inst. 2012, 104, 1765–1769. [Google Scholar] [CrossRef]
- Liu, C.; Tu, Y.; Yuan, J.; Mao, X.; He, S.; Wang, L.; Fu, G.; Zong, J.; Zhang, Y. Aberrant Expression of N-Methylpurine-DNA Glycosylase Influences Patient Survival in Malignant Gliomas. J. Biomed. Biotechnol. 2012, 2012, 760679. [Google Scholar] [CrossRef]
- Tang, J.B.; Svilar, D.; Trivedi, R.N.; Wang, X.H.; Goellner, E.M.; Moore, B.; Hamilton, R.L.; Banze, L.A.; Brown, A.R.; Sobol, R.W. N-Methylpurine DNA Glycosylase and DNA Polymerase Beta Modulate BER Inhibitor Potentiation of Glioma Cells to Temozolomide. Neuro Oncol. 2011, 13, 471–486. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Heras, G.; Castro-Robles, B.; Romero-Sánchez, C.M.; Carrión, B.; Barbella-Aponte, R.; Sandoval, H.; Segura, T. Involvement of N-Methylpurine DNA Glycosylase in Resistance to Temozolomide in Patient-Derived Glioma Cells. Sci. Rep. 2020, 10, 22185. [Google Scholar] [CrossRef] [PubMed]
- Hofseth, L.J.; Khan, M.A.; Ambrose, M.; Nikolayeva, O.; Xu-Welliver, M.; Kartalou, M.; Hussain, S.P.; Roth, R.B.; Zhou, X.; Mechanic, L.E.; et al. The Adaptive Imbalance in Base Excision–Repair Enzymes Generates Microsatellite Instability in Chronic Inflammation. J. Clin. Investig. 2003, 112, 1887. [Google Scholar] [CrossRef] [PubMed]
- Meira, L.B.; Bugni, J.M.; Green, S.L.; Lee, C.-W.; Pang, B.; Borenshtein, D.; Rickman, B.H.; Rogers, A.B.; Moroski-Erkul, C.A.; McFaline, J.L.; et al. DNA Damage Induced by Chronic Inflammation Contributes to Colon Carcinogenesis in Mice. J. Clin. Investig. 2008, 118, 2516–2525. [Google Scholar] [CrossRef]
- Speina, E.; Zielińska, M.; Barbin, A.; Gackowski, D.; Kowalewski, J.; Graziewicz, M.A.; Siedlecki, J.A.; Oliński, R.; Tudek, B. Decreased Repair Activities of 1,N6-Ethenoadenine and 3,N4-Ethenocytosine in Lung Adenocarcinoma Patients1. Cancer Res. 2003, 63, 4351–4357. Available online: https://aacrjournals.org/cancerres/article/63/15/4351/510303/Decreased-Repair-Activities-of-1-N6-Ethenoadenine (accessed on 7 February 2025).
| Cancers by Body Location/System | AAG Mutations |
|---|---|
| Breast | S47T, R100W, R120C, M151I |
| Digestive/Gastrointestinal | A5T, Q20H, R24Q, P25A, A28T, D36N, A37T, S47T, R60C, P64L, R72S, I74M, E89D, R100Q, R100W, L103V, R120H, E123V, E125K, Y127C, R145H, R147Q, G148S, M151I, P153L, G163D, G163S, R182Q, G189D, R197C, R201W, A205D, D211N, R212H, L214F, A226T, D239G, R246C, R246P, P254S, V264I, G265S, G281C |
| Endocrine | R194S, R197S, Q223P, P248A |
| Genitourinary | P112S, V158M, L180F, K202R, R261Q, H266N, F277L |
| Gynecologic | A5D, A5T, R17W, K21N, A38T, D50N, P65L, P68L, P94L, G114D, R118Q, M151I, P153Q, T192I, R207H, D211Y, E293K |
| Head and Neck | G265S |
| Hematologic/Blood | R17Q, T124I, S172R, H266Y |
| Musculoskeletal | R24Q |
| Neurologic | R118Q, P130S, M164I, D175N, L180F, V264A |
| Respiratory/ Thoracic | A52S, S77L, L180F, R246H, S282R |
| Skin | M22Q, S48L, R120H, M168I, R201Q, E213K, P248S, R261W, E269K, R272Q, L275I, R280W |
| № | SNP | Predicted Effect | Cancer Type | Conservation of Amino Acid Residue |
|---|---|---|---|---|
| 1 | P94L | High | Endometrioid carcinoma | Non conserved |
| 2 | R100W | High | Stomach carcinoma, breast carcinoma | Non conserved |
| 3 | R120C | High | Breast carcinoma | Non conserved |
| 4 | E125K | High | Upper aerodigestive tract carcinoma | Conserved |
| 5 | Y127C | High | Large intestine adenocarcinoma | Conserved |
| 6 | R145H | High | Stomach carcinoma | Conserved |
| 7 | P153L | High | Stomach carcinoma | Conserved |
| 8 | V158M | High | Kidney carcinoma, clear cell renal cell carcinoma | Conserved |
| 9 | G163S | High | Large intestine adenocarcinoma | Conserved |
| 10 | R182Q | High | Large intestine adenocarcinoma | Conserved |
| 11 | R197C | High | Esophagus adenocarcinoma | Non conserved |
| 12 | R207H | High | Endometrioid carcinoma, uterine adenomas and adenocarcinomas | Non conserved |
| 13 | R261Q | High | Urinary tract carcinoma | Non conserved |
| 14 | R261W | High | Malignant melanoma | Non conserved |
| 15 | E269K | High | Malignant melanoma | Non conserved |
| 16 | R280W | High | Cutaneous melanoma | Non conserved |
| 17 | G281C | High | Stomach adenocarcinoma | Non conserved |
| 18 | E293K | High | Cervix carcinoma, squamous cell carcinoma | Non conserved |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kladova, O.A.; Kuznetsova, A.A. The Link Between Human Alkyladenine DNA Glycosylase and Cancer Development. Int. J. Mol. Sci. 2025, 26, 7647. https://doi.org/10.3390/ijms26157647
Kladova OA, Kuznetsova AA. The Link Between Human Alkyladenine DNA Glycosylase and Cancer Development. International Journal of Molecular Sciences. 2025; 26(15):7647. https://doi.org/10.3390/ijms26157647
Chicago/Turabian StyleKladova, Olga A., and Aleksandra A. Kuznetsova. 2025. "The Link Between Human Alkyladenine DNA Glycosylase and Cancer Development" International Journal of Molecular Sciences 26, no. 15: 7647. https://doi.org/10.3390/ijms26157647
APA StyleKladova, O. A., & Kuznetsova, A. A. (2025). The Link Between Human Alkyladenine DNA Glycosylase and Cancer Development. International Journal of Molecular Sciences, 26(15), 7647. https://doi.org/10.3390/ijms26157647







