Functional Express Proteomics for Search and Identification of Differentially Regulated Proteins Involved in the Reaction of Wheat (Triticum aestivum L.) to Nanopriming by Gold Nanoparticles
Abstract
1. Introduction
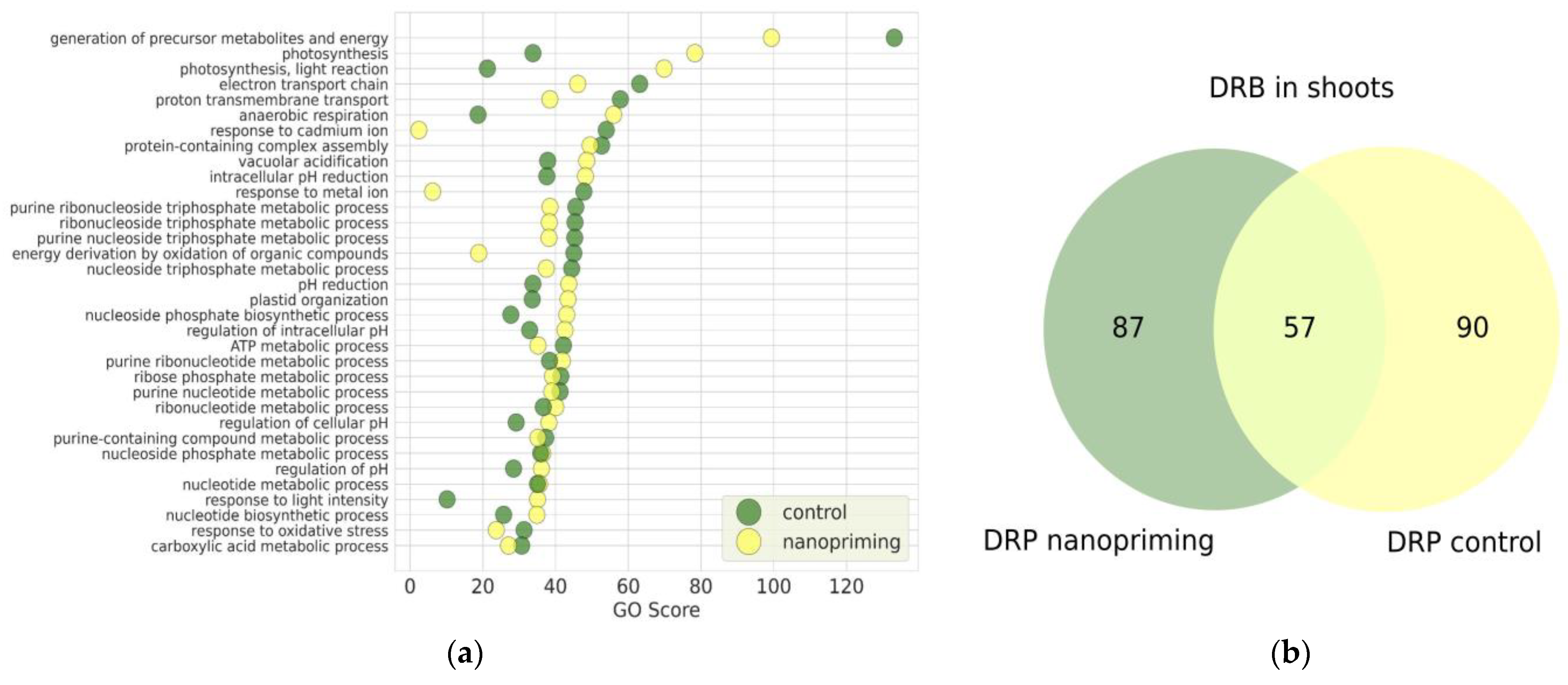
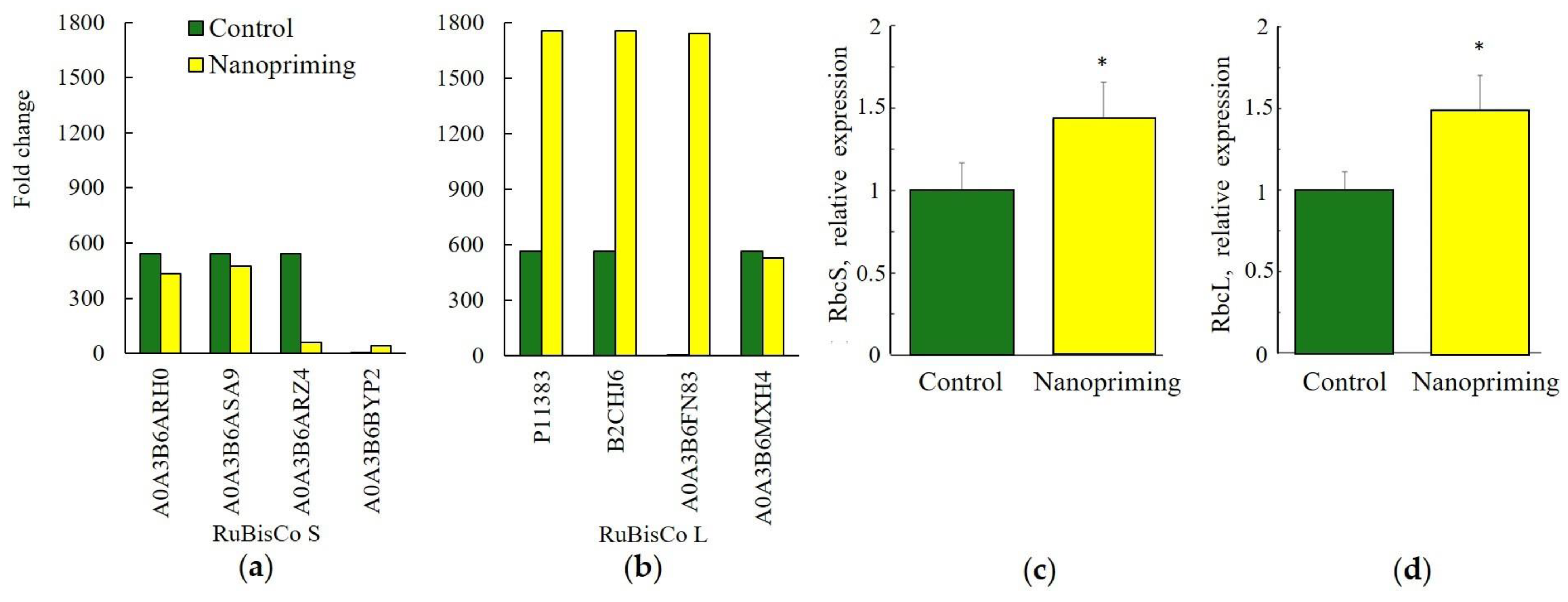
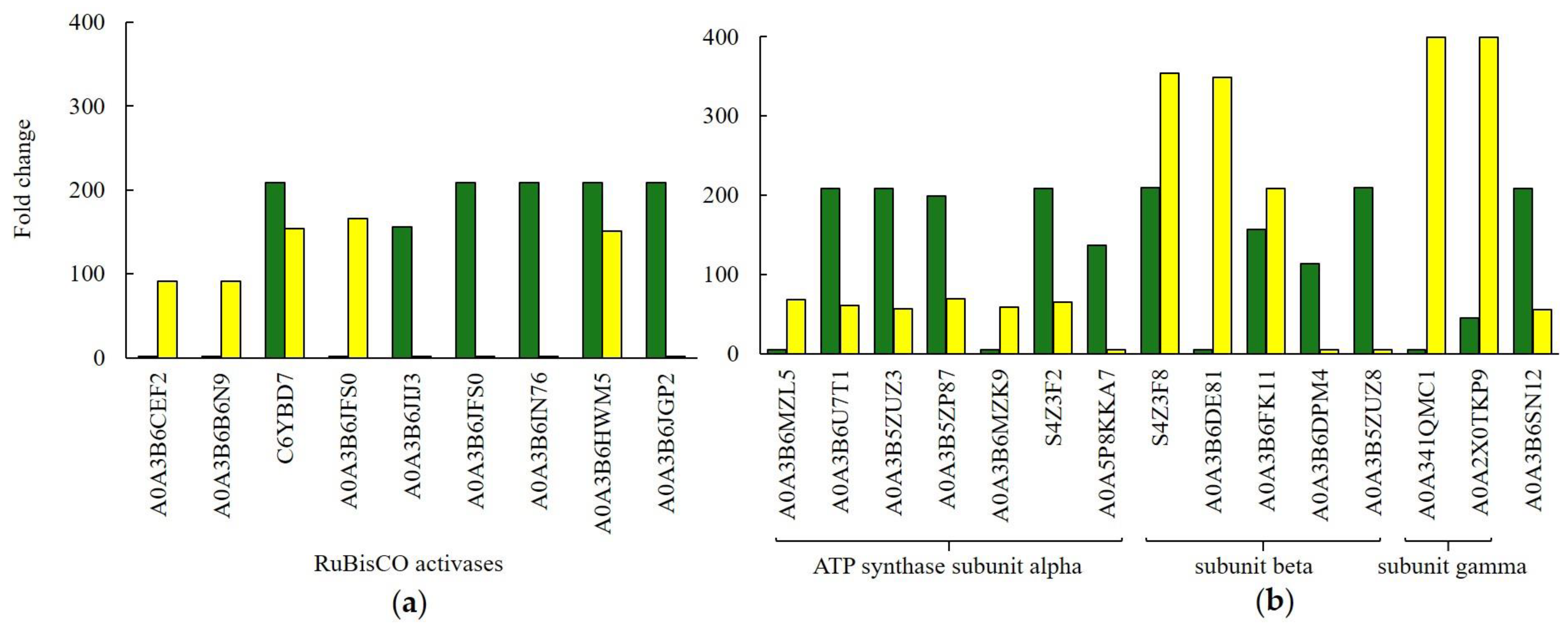
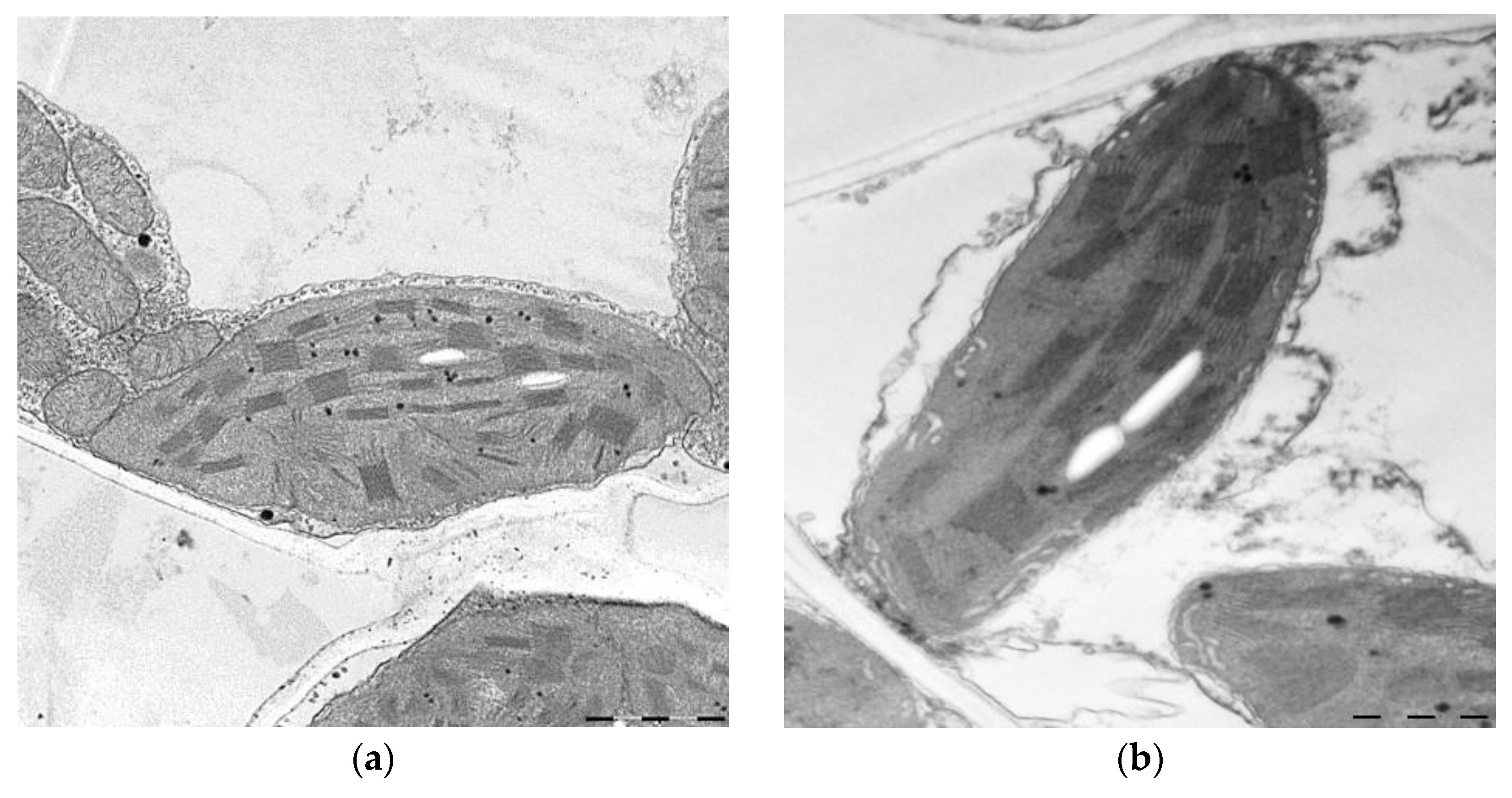
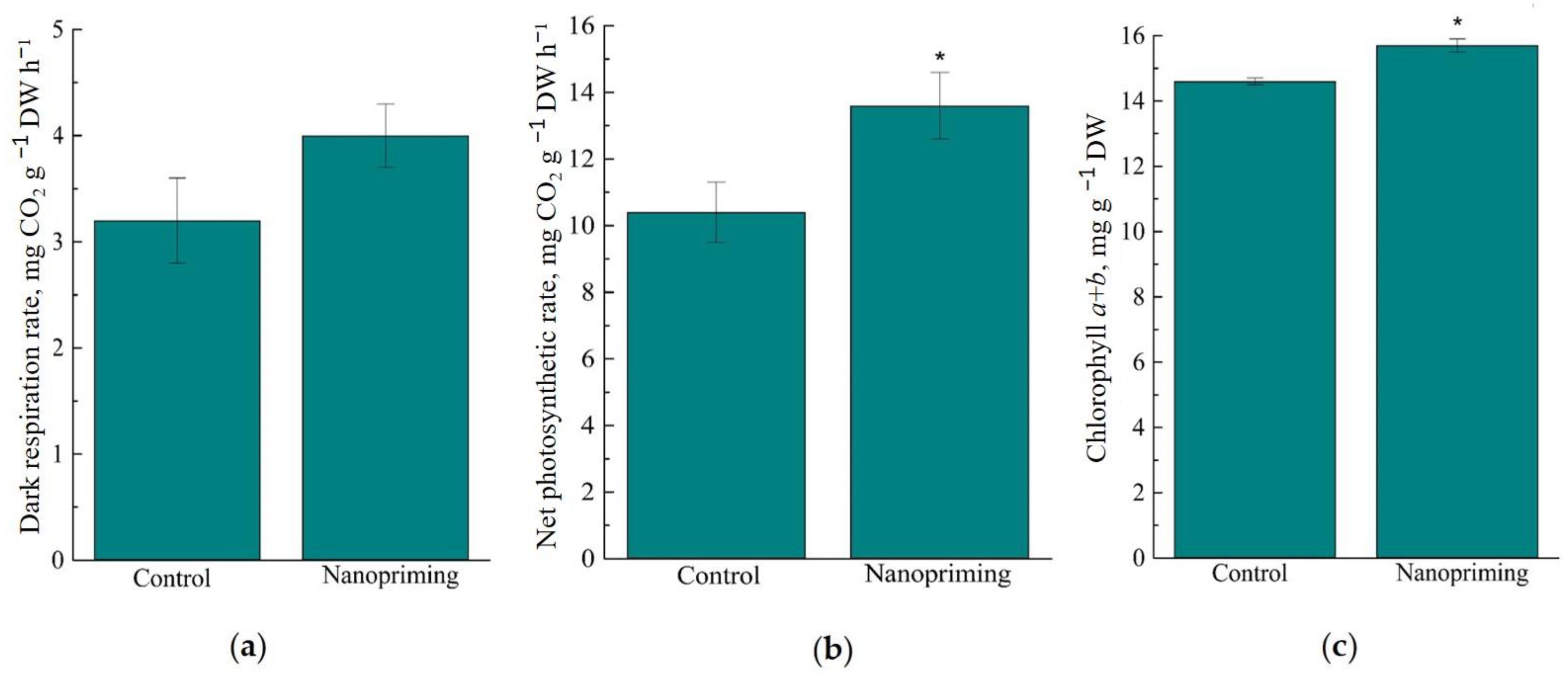
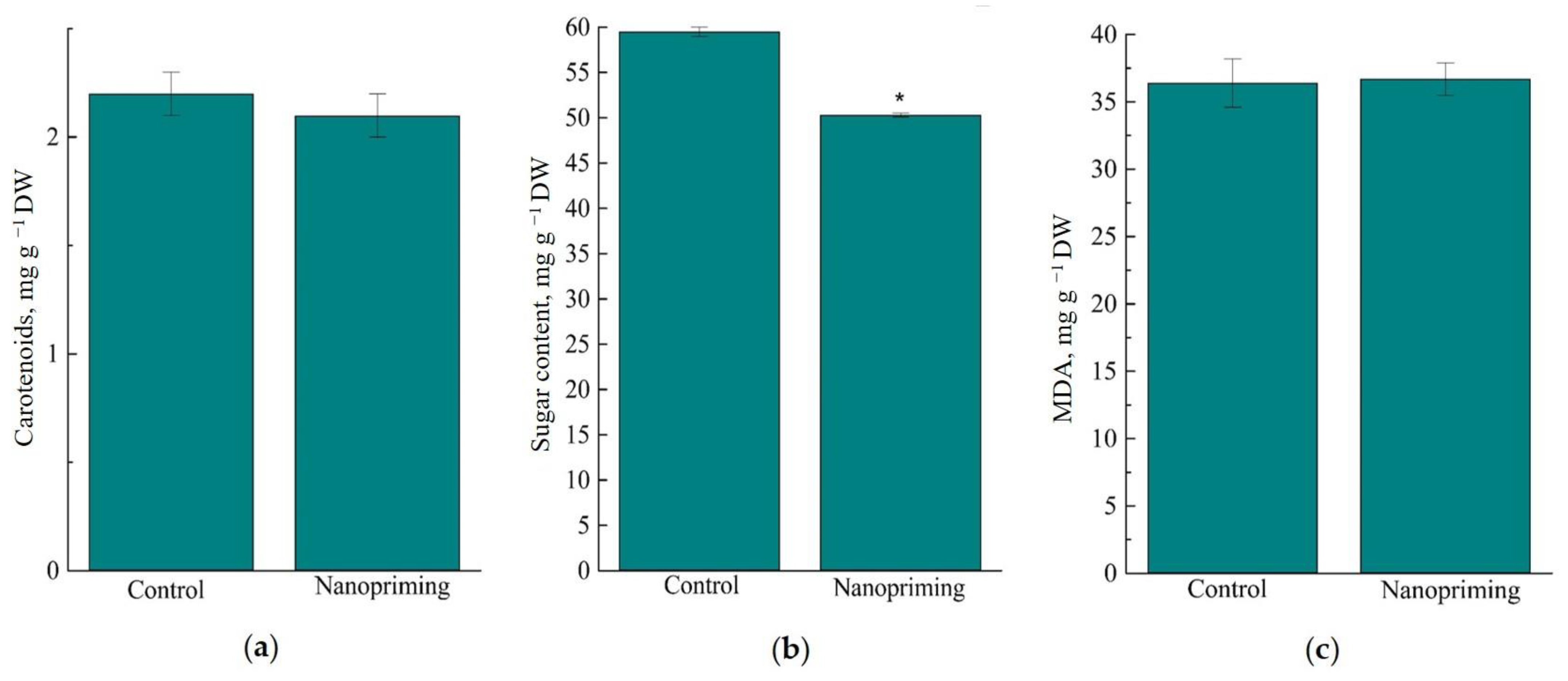
2. Results
3. Discussion
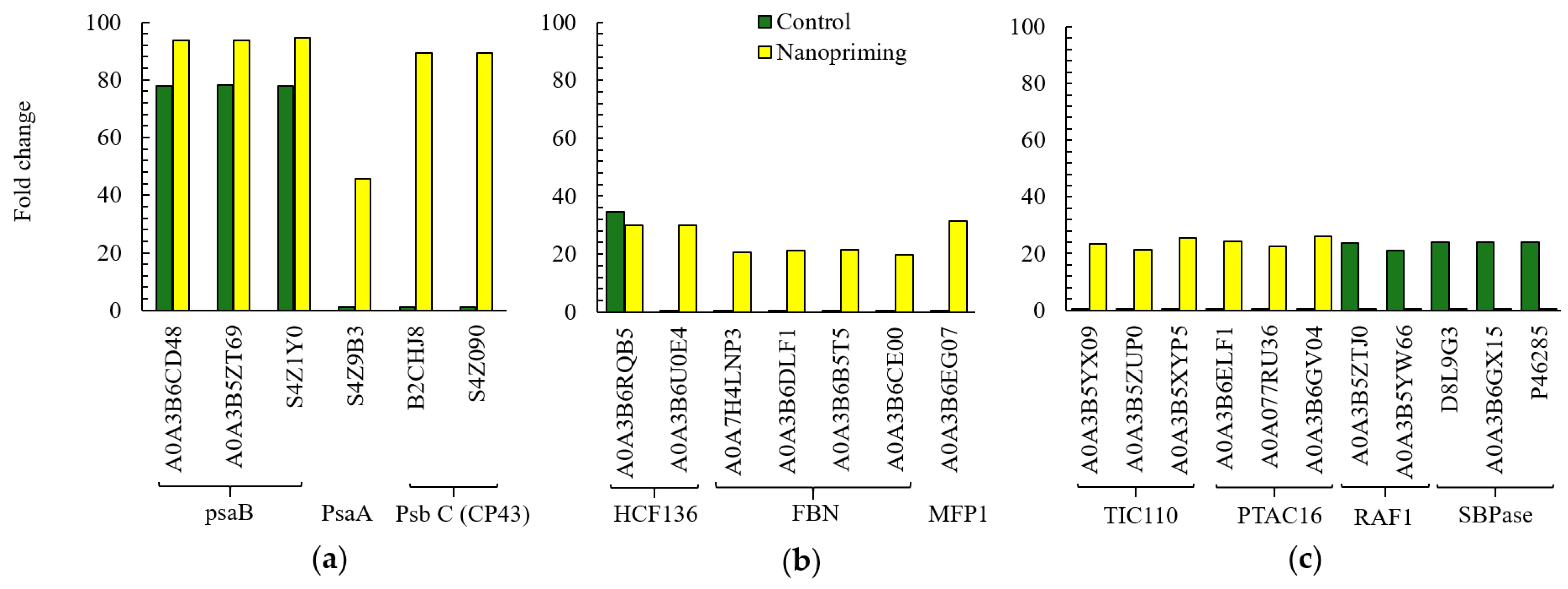
3.1. Photosynthesis
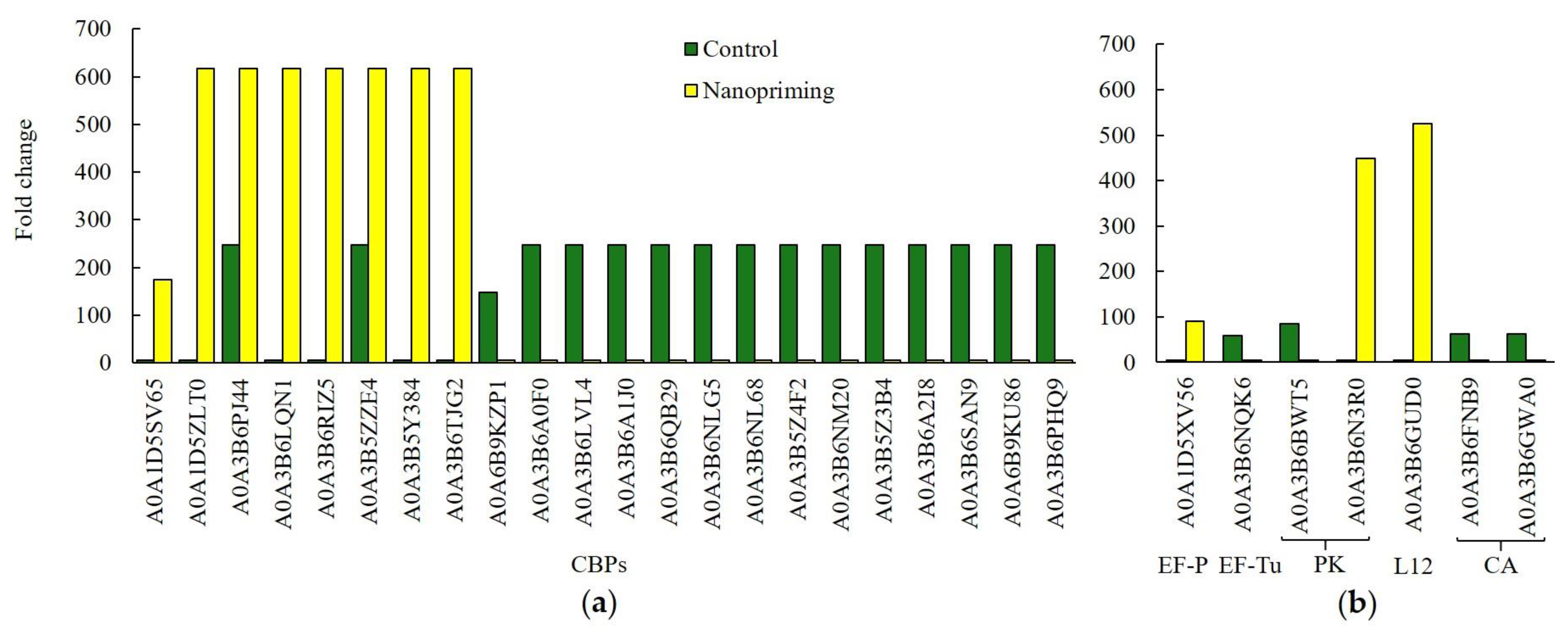
3.2. Generation of Precursor Metabolites and Energy
3.3. Transmembrane Transport
3.4. Oxidative Stress
4. Materials and Methods
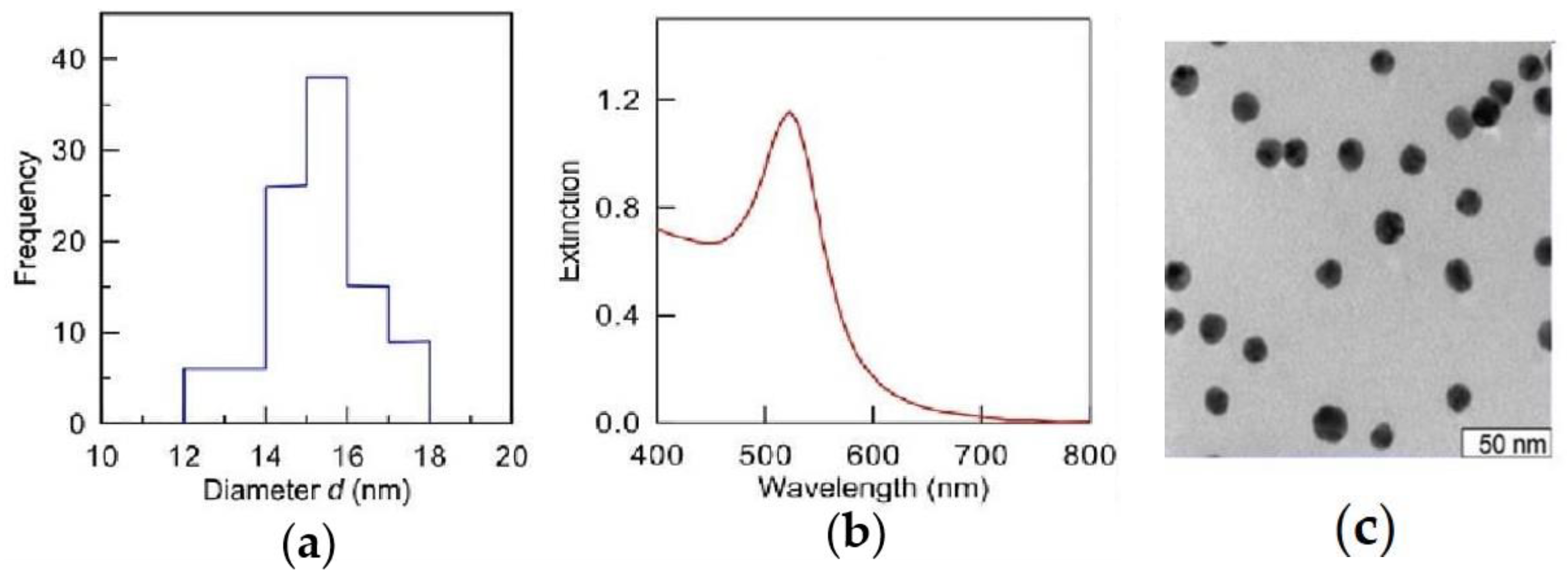
4.1. The Synthesis and Description of Au-NPs
4.2. Plant Material and Growth Conditions
4.3. Protein Extraction and Enzymatic Hydrolysis for Proteomic Analysis
4.4. HPLC-MS1 Analysis
4.5. Proteomic Data Processing
4.6. Ultrastructure of Leaf Cells
4.7. CO2-Gas Exchange
4.8. Photosynthetic Pigments Content
4.9. Soluble Sugars (Glucose, Fructose, and Sucrose) Content
4.10. Malondialdehyde (MDA) Content
4.11. Extraction Soluble Proteins and Activity of Antioxidant Enzymes
4.12. RNA Isolation and RT-PCR (qRT-PCR)
4.13. Gold Content
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Au-NP | gold nanoparticle |
| DRPs | differentially regulated proteins |
| PSA | photosynthetic apparatus |
| RuBisCo | ribulose-1,5-bisphosphate carboxylase/oxygenase |
| AOS | antioxidant system |
| GO | gene ontology |
| psaB | photosystem I P700 chlorophyll a apoprotein A2 |
| PsaA | photosystem I P700 chlorophyll a apoprotein A1 |
| FBN | fibrillins |
| MFP1 | MAR-binding filament-like protein 1 |
| HCF136 | Photosynthesis system II assembly factor Ycf48/Hcf136 |
| CBPs | chlorophyll a-b binding proteins |
| TIC110 | translocon at the inner envelope membrane of chloroplasts 110 |
| PTAC16 | plastid transcriptionally active 16 proteins |
| RAF1 | Rubisco accumulation factor 1 C-terminal domain-containing protein 1 |
| EF-P | elongation factor P |
| PGK | phosphoglycerate kinase |
| MDA | malondialdehyde |
| SOD | superoxide dismutase |
| APX | ascorbate peroxidase |
| CAT | catalase |
| POD | class III peroxidases |
| DW | dry weight |
| LHC | light-harvesting complexes |
| GLO | Glycolate oxidase |
| CDSP32 | thioredoxin domain-containing protein |
References
- Kumari, A.; Gupta, A.K.; Sharma, S.; Jadon, V.S.; Sharma, V.; Chun, S.C.; Sivanesan, I. Nanoparticles as a Tool for Alleviating Plant Stress: Mechanisms, Implications, and Challenges. Plants 2024, 13, 1528. [Google Scholar] [CrossRef] [PubMed]
- Swain, R.; Behera, M.; Sahoo, S.; Rout, G.R. Nanoscience in Plant Stress Mitigation: A Comprehensive Review. BioNanoScience 2024, 15, 24. [Google Scholar] [CrossRef]
- Azameti, M.K.; Imoro, A.-W.M. Nanotechnology: A promising field in enhancing abiotic stress tolerance in plants. Crop Des. 2023, 2, 100037. [Google Scholar] [CrossRef]
- Fincheira, P.; Hoffmann, N.; Tortella, G.; Ruiz, A.; Cornejo, P.; Diez, M.C.; Seabra, A.B.; Benavides-Mendoza, A.; Rubilar, O. Eco-Efficient Systems Based on Nanocarriers for the Controlled Release of Fertilizers and Pesticides: Toward Smart Agriculture. Nanomaterials 2023, 13, 1978. [Google Scholar] [CrossRef]
- Rajput, P.; Singh, A.; Agrawal, S.; Ghazaryan, K.; Rajput, V.D.; Movsesyan, H.; Mandzhieva, S.; Minkina, T.; Alexiou, A. Effects of environmental metal and metalloid pollutants on plants and human health: Exploring nano-remediation approach. Stress Biol. 2024, 4, 27. [Google Scholar] [CrossRef]
- Balusamy, S.R.; Joshi, A.S.; Perumalsamy, H.; Mijakovic, I.; Singh, P. Advancing sustainable agriculture: A critical review of smart and eco-friendly nanomaterial applications. J. Nanobiotechnol. 2023, 21, 372. [Google Scholar] [CrossRef]
- Francis, D.V.; Abdalla, A.K.; Mahakham, W.; Sarmah, A.K.; Ahmed, Z.F.R. Interaction of plants and metal nanoparticles: Exploring its molecular mechanisms for sustainable agriculture and crop improvement. Environ. Int. 2024, 190, 108859. [Google Scholar] [CrossRef]
- Yadav, N.; Bora, S.; Devi, B.; Upadhyay, C.; Singh, P. Nanoparticle-mediated defense priming: A review of strategies for enhancing plant resilience against biotic and abiotic stresses. Plant Physiol. Biochem. 2024, 213, 108796. [Google Scholar] [CrossRef]
- Nile, S.H.; Thiruvengadam, M.; Wang, Y.; Samynathan, R.; Shariati, M.A.; Rebezov, M.; Nile, A.; Sun, M.; Venkidasamy, B.; Xiao, J.; et al. Nano-priming as emerging seed priming technology for sustainable agriculture-recent developments and future perspectives. J. Nanobiotechnol. 2022, 20, 254. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Bai, T.; Wei, H.; Gardea-Torresdey, J.L.; Keller, A.; White, J.C. Nanobiotechnology-based strategies for enhanced crop stress resilience. Nat. Food 2022, 3, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, L.; Zhang, Q.; Wei, J.; Zhao, X.; Zheng, Z.; Chen, B.; Xu, Z. Nanopriming boost seed vigor: Deeper insights into the effect mechanism. Plant Physiol. Biochem. 2024, 214, 108895. [Google Scholar] [CrossRef]
- Dykman, L.; Khlebtsov, N. Gold Nanoparticles in Biomedical Applications, 1st ed.; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar] [CrossRef]
- Dykman, L.A. Gold nanoparticles for preparation of antibodies and vaccines against infectious diseases. Expert Rev. Vaccines 2020, 19, 465–477. [Google Scholar] [CrossRef]
- Alabdallah, N.M. Do gold nanoparticles consistently benefit crop plants under both non-stressed and abiotic stress conditions? Nanotechnol. Rev. 2025, 14, 20250153. [Google Scholar] [CrossRef]
- Kusainova, T.T.; Emekeeva, D.D.; Kazakova, E.M.; Gorshkov, V.A.; Kjeldsen, F.; Kuskov, M.L.; Zhigach, A.N.; Olkhovskaya, I.P.; Bogoslovskaya, O.A.; Glushchenko, N.N.; et al. Ultra-Fast Mass Spectrometry in Plant Biochemistry: Response of Winter Wheat Proteomics to Pre-Sowing Treatment with Iron Compounds. Biochemistry 2023, 88, 1390–1403. [Google Scholar] [CrossRef]
- Tiwari, M.; Krishnamurthy, S.; Shukla, D.; Kiiskila, J.; Jain, A.; Datta, R.; Sharma, N.; Sahi, S.V. Comparative transcriptome and proteome analysis to reveal the biosynthesis of gold nanoparticles in Arabidopsis. Sci. Rep. 2016, 6, 21733. [Google Scholar] [CrossRef]
- Hossain, Z.; Yasmeen, F.; Komatsu, S. Nanoparticles: Synthesis, Morphophysiological Effects, and Proteomic Responses of Crop Plants. Int. J. Mol. Sci. 2020, 21, 3056. [Google Scholar] [CrossRef]
- Venzhik, Y.; Deryabin, A.; Zhukova, K. Au-Based Nanoparticles Enhance Low Temperature Tolerance in Wheat by Regulating Some Physiological Parameters and Gene Expression. Plants 2024, 13, 1261. [Google Scholar] [CrossRef]
- Ivanov, M.V.; Bubis, J.A.; Gorshkov, V.; Tarasova, I.A.; Levitsky, L.I.; Lobas, A.A.; Solovyeva, E.M.; Pridatchenko, M.L.; Kjeldsen, F.; Gorshkov, M.V. DirectMS1: MS/MS-Free Identification of 1000 Proteins of Cellular Proteomes in 5 Minutes. Anal. Chem. 2020, 92, 4326–4333. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, M.V.; Bubis, J.A.; Gorshkov, V.; Abdrakhimov, D.A.; Kjeldsen, F.; Gorshkov, M.V. Boosting MS1-only Proteomics with Machine Learning Allows 2000 Protein Identifications in Single-Shot Human Proteome Analysis Using 5 min HPLC Gradient. J. Proteome Res. 2021, 20, 1864–1873. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, M.V.; Bubis, J.A.; Gorshkov, V.; Tarasova, I.A.; Levitsky, L.I.; Solovyeva, E.M.; Lipatova, A.V.; Kjeldsen, F.; Gorshkov, M.V. DirectMS1Quant: Ultrafast Quantitative Proteomics with MS/MS-Free Mass Spectrometry. Anal. Chem. 2022, 94, 13068–13075. [Google Scholar] [CrossRef] [PubMed]
- Green, B.R.; Pichersky, E.; Kloppstech, K. Chlorophyll a/b-binding proteins: An extended family. Trends Biochem. Sci. 1991, 16, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Tyutereva, E.V.; Dmitrieva, V.A.; Voitsekhovskaja, O.V. Chlorophyll b as a source of signals steering plant development. Agric. Biol. 2017, 52, 843–855. [Google Scholar] [CrossRef]
- Mao, Y.; Catherall, E.; Diaz-Ramos, A.; Greiff, G.R.L.; Azinas, S.; Gunn, L.; McCormick, A.J. The small subunit of Rubisco and its potential as an engineering target. J. Exp. Bot. 2023, 74, 543–561. [Google Scholar] [CrossRef]
- Ng, J.; Guo, Z.; Mueller-Cajar, O. Rubisco activase requires residues in the large subunit N terminus to remodel inhibited plant Rubisco. J. Biol. Chem. 2020, 295, 16427–16435. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.; Cui, L.; Meng, S.; Ye, N.; Peng, X. Catalytic and functional aspects of different isozymes of glycolate oxidase in rice. BMC Plant Biol. 2017, 17, 135. [Google Scholar] [CrossRef]
- Cui, L.L.; Lu, Y.S.; Li, Y.; Yang, C.; Peng, X.X. Overexpression of Glycolate Oxidase Confers Improved Photosynthesis under High Light and High Temperature in Rice. Front. Plant Sci. 2016, 7, 1165. [Google Scholar] [CrossRef]
- Lu, Y.; Li, Y.; Yang, Q.; Zhang, Z.; Chen, Y.; Zhang, S.; Peng, X.X. Suppression of glycolate oxidase causes glyoxylate accumulation that inhibits photosynthesis through deactivating Rubisco in rice. Physiol. Plant 2014, 150, 463–476. [Google Scholar] [CrossRef]
- Satti, S.H.; Raja, N.I.; Ikram, M.; Oraby, H.F.; Mashwani, Z.U.; Mohamed, A.H.; Singh, A.; Omar, A.A. Plant-Based Titanium Dioxide Nanoparticles Trigger Biochemical and Proteome Modifications in Triticum aestivum L. under Biotic Stress of Puccinia striiformis. Molecules 2022, 27, 4274. [Google Scholar] [CrossRef]
- Yang, F.; Hong, F.; You, W.; Liu, C.; Gao, F.; Wu, C.; Yang, P. Influences of nano-anatase TiO2 on the nitrogen metabolism of growing spinach. Biol. Trace Elem. Res. 2006, 110, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Ford, K.L.; Cassin, A.; Bacic, A. Quantitative proteomic analysis of wheat cultivars with differing drought stress tolerance. Front. Plant Sci. 2011, 2, 44. [Google Scholar] [CrossRef] [PubMed]
- Plochinger, M.; Schwenkert, S.; von Sydow, L.; Schroder, W.P.; Meurer, J. Functional Update of the Auxiliary Proteins PsbW, PsbY, HCF136, PsbN, TerC and ALB3 in Maintenance and Assembly of PSII. Front. Plant Sci. 2016, 7, 423. [Google Scholar] [CrossRef] [PubMed]
- El-Sappah, A.H.; Li, J.; Yan, K.; Zhu, C.; Huang, Q.; Zhu, Y.; Chen, Y.; El-Tarabily, K.A.; AbuQamar, S.F. Fibrillin gene family and its role in plant growth, development, and abiotic stress. Front. Plant Sci. 2024, 15, 1453974. [Google Scholar] [CrossRef]
- Meier, I.; Phelan, T.; Gruissem, W.; Spiker, S.; Schneider, D. MFP1, a novel plant filament-like protein with affinity for matrix attachment region DNA. Plant Cell 1996, 8, 2105–2115. [Google Scholar] [CrossRef][Green Version]
- Akiyama, K.; Kondo, K.; Inabe, K.; Murakami, S.; Wakabayashi, K.I.; Hisabori, T. The beta-hairpin region of the cyanobacterial F(1)-ATPase gamma-subunit plays a regulatory role in the enzyme activity. Biochem. J. 2019, 476, 1771–1780. [Google Scholar] [CrossRef]
- Richardson, L.G.L.; Singhal, R.; Schnell, D.J. The integration of chloroplast protein targeting with plant developmental and stress responses. BMC Biol. 2017, 15, 118. [Google Scholar] [CrossRef]
- Kovacheva, S.; Bedard, J.; Patel, R.; Dudley, P.; Twell, D.; Rios, G.; Koncz, C.; Jarvis, P. In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 2005, 41, 412–428. [Google Scholar] [CrossRef]
- Hao, Q.; Shang, W.; Zhang, C.; Chen, H.; Chen, L.; Yuan, S.; Chen, S.; Zhang, X.; Zhou, X. Identification and Comparative Analysis of CBS Domain-Containing Proteins in Soybean (Glycine max) and the Primary Function of GmCBS21 in Enhanced Tolerance to Low Nitrogen Stress. Int. J. Mol. Sci. 2016, 17, 620. [Google Scholar] [CrossRef] [PubMed]
- Tomar, S.; Subba, A.; Bala, M.; Singh, A.K.; Pareek, A.; Singla-Pareek, S.L. Genetic Conservation of CBS Domain Containing Protein Family in Oryza Species and Their Association with Abiotic Stress Responses. Int. J. Mol. Sci. 2022, 23, 1687. [Google Scholar] [CrossRef] [PubMed]
- Broin, M.; Cuine, S.; Eymery, F.; Rey, P. The plastidic 2-cysteine peroxiredoxin is a target for a thioredoxin involved in the protection of the photosynthetic apparatus against oxidative damage. Plant Cell 2002, 14, 1417–1432. [Google Scholar] [CrossRef]
- Kohzuma, K.; Froehlich, J.E.; Davis, G.A.; Temple, J.A.; Minhas, D.; Dhingra, A.; Cruz, J.A.; Kramer, D.M. The Role of Light-Dark Regulation of the Chloroplast ATP Synthase. Front. Plant Sci. 2017, 8, 1248. [Google Scholar] [CrossRef]
- Dykman, L.A.; Khlebtsov, N.G. Methods for chemical synthesis of colloidal gold. Russ. Chem. Rev. 2019, 88, 229. [Google Scholar] [CrossRef]
- Keiblinger, K.M.; Wilhartitz, I.C.; Schneider, T.; Roschitzki, B.; Schmid, E.; Eberl, L.; Riedel, K.; Zechmeister-Boltenstern, S. Soil metaproteomics—Comparative evaluation of protein extraction protocols. Soil. Biol. Biochem. 2012, 54, 14–24. [Google Scholar] [CrossRef]
- Wessel, D.; Flugge, U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Hulstaert, N.; Shofstahl, J.; Sachsenberg, T.; Walzer, M.; Barsnes, H.; Martens, L.; Perez-Riverol, Y. ThermoRawFileParser: Modular, Scalable, and Cross-Platform RAW File Conversion. J. Proteome Res. 2020, 19, 537–542. [Google Scholar] [CrossRef]
- Abdrakhimov, D.A.; Bubis, J.A.; Gorshkov, V.; Kjeldsen, F.; Gorshkov, M.V.; Ivanov, M.V. Biosaur: An open-source Python software for liquid chromatography-mass spectrometry peptide feature detection with ion mobility support. Rapid Commun. Mass Spectrom. 2021, 39, e9045. [Google Scholar] [CrossRef]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 2010, 604, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pirmoradian, M.; Zubarev, R.; Kall, L. Covariation of Peptide Abundances Accurately Reflects Protein Concentration Differences. Mol. Cell Proteom. 2017, 16, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kazakova, E.M.; Solovyeva, E.M.; Levitsky, L.I.; Bubis, J.A.; Emekeeva, D.D.; Antonets, A.A.; Nazarov, A.A.; Gorshkov, M.V.; Tarasova, I.A. Proteomics-based scoring of cellular response to stimuli for improved characterization of signaling pathway activity. Proteomics 2023, 23, e2200275. [Google Scholar] [CrossRef]
- Kulmanov, M.; Guzmán-Vega, F.J.; Duek Roggli, P.; Lane, L.; Arold, S.T.; Hoehndorf, R. Protein function prediction as approximate semantic entailment. Nat. Mach. Intell. 2024, 6, 220–228. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramirez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, D.D.; Bensch, K.; Barrnett, R.J. Cytochemistry and electron microscopy. The preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. J. Cell Biol. 1963, 17, 19–58. [Google Scholar] [CrossRef]
- Wellburn, A.R. The Spectral Determination of Chlorophylls a and b, as well as Total Carotenoids, Using Various Solvents with Spectrophotometers of Different Resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Nakamura, M. Determination of Fructose in the Presence of a Large Excess of Glucose Part IV A Modified Resorcinol-Thiourea-Hydrochloric Acid Reaction. Agric. Biol. Chem. 1968, 32, 696–700. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Miszalski, Z.; Ślesak, I.; Niewiadomska, E.; Baczek-Kwinta, R.; Lüttge, U.; Ratajczak, R. Subcellular localization and stress responses of superoxide dismutase isoforms from leaves in the C3-CAM intermediate halophyte Mesembryanthemum crystallinum L. Plant Cell Environ. 1998, 21, 169–179. [Google Scholar] [CrossRef]
- Kumar, G.; Knowles, N.R. Changes in Lipid Peroxidation and Lipolytic and Free-Radical Scavenging Enzyme Activities during Aging and Sprouting of Potato (Solanum tuberosum) Seed-Tubers. Plant Physiol. 1993, 102, 115–124. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Hydrogen Peroxide is Scavenged by Ascorbate-specific Peroxidase in Spinach Chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar] [CrossRef]
- Perdomo, J.A.; Buchner, P.; Carmo-Silva, E. The relative abundance of wheat Rubisco activase isoforms is post-transcriptionally regulated. Photosynth. Res. 2021, 148, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]

| Indicator | Control | Nanopriming |
|---|---|---|
| Seed germination, % | 77 ± 2 | 76 ± 2 |
| Leaf length, mm | 136 ± 3 | 154 ± 3 * |
| Area of peroxisome, µm2 | 0.25 ± 0.02 | 0.18 ± 0.02 * |
| Area of mitochondria, µm2 | 0.28 ± 0.01 | 0.16 ± 0.02 * |
| Number of cristae per 1 µm2, pcs | 48 ± 2 | 49 ± 3 |
| Area of chloroplast, µm2 | 5.10 ± 0.1 | 5.70 ± 0.1 * |
| Number of grana per 10 µm2 of chloroplast, pcs | 51 ± 1 | 56 ± 1 * |
| Number of plastoglobules per 10 µm2 of chloroplast, pcs | 37 ± 4 | 36 ± 1 |
| Number of thylakoids in grana, pcs | 7 ± 0.1 | 7 ± 0.2 |
| Area of starch grain, µm2 | 0.04 ± 0.006 | 0.07 ± 0.02 * |
| Area of starch in chloroplast, % | 0.70 ± 0.1 | 1.20 ± 0.1 * |
| Number of Thylakoids in Grana | Number of Grana in Chloroplast, % | |
|---|---|---|
| Control | Nanopriming | |
| 2–6 | 48 ± 1 | 56 ± 2 * |
| 7–16 | 49 ± 1 | 40 ± 2 * |
| 17–21 | 3 ± 0.6 | 4 ± 1 |
| Organs | Control | Nanopriming |
|---|---|---|
| Seeds | <0.05 | 3.8 ± 0.1 |
| Roots | <0.05 | 1.6 ± 0.1 |
| Leaves | <0.05 | 0.3 ± 0.1 |
| Gene | Primer | Primer Sequences |
|---|---|---|
| TaRP15 | Forward Reverse | TCATTGTGGAGGACTCGTGG GCAGACATAGCCCACACAT |
| rbcS | Forward Reverse | GGATTCGACAACATGCGCCAGG ATATGGCCTGTCGTGAGTGAGC |
| rbcL | Forward Reverse | ACCATTTATGCGCTGGAGAGACC CAAGTAATGCCCCTTGATTTCACC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naraikina, N.; Kussainova, T.; Shelepchikov, A.; Tretyakov, A.; Deryabin, A.; Zhukova, K.; Popov, V.; Tarasova, I.; Dykman, L.; Venzhik, Y. Functional Express Proteomics for Search and Identification of Differentially Regulated Proteins Involved in the Reaction of Wheat (Triticum aestivum L.) to Nanopriming by Gold Nanoparticles. Int. J. Mol. Sci. 2025, 26, 7608. https://doi.org/10.3390/ijms26157608
Naraikina N, Kussainova T, Shelepchikov A, Tretyakov A, Deryabin A, Zhukova K, Popov V, Tarasova I, Dykman L, Venzhik Y. Functional Express Proteomics for Search and Identification of Differentially Regulated Proteins Involved in the Reaction of Wheat (Triticum aestivum L.) to Nanopriming by Gold Nanoparticles. International Journal of Molecular Sciences. 2025; 26(15):7608. https://doi.org/10.3390/ijms26157608
Chicago/Turabian StyleNaraikina, Natalia, Tomiris Kussainova, Andrey Shelepchikov, Alexey Tretyakov, Alexander Deryabin, Kseniya Zhukova, Valery Popov, Irina Tarasova, Lev Dykman, and Yuliya Venzhik. 2025. "Functional Express Proteomics for Search and Identification of Differentially Regulated Proteins Involved in the Reaction of Wheat (Triticum aestivum L.) to Nanopriming by Gold Nanoparticles" International Journal of Molecular Sciences 26, no. 15: 7608. https://doi.org/10.3390/ijms26157608
APA StyleNaraikina, N., Kussainova, T., Shelepchikov, A., Tretyakov, A., Deryabin, A., Zhukova, K., Popov, V., Tarasova, I., Dykman, L., & Venzhik, Y. (2025). Functional Express Proteomics for Search and Identification of Differentially Regulated Proteins Involved in the Reaction of Wheat (Triticum aestivum L.) to Nanopriming by Gold Nanoparticles. International Journal of Molecular Sciences, 26(15), 7608. https://doi.org/10.3390/ijms26157608






