An Italian Single-Center Genomic Surveillance Study: Two-Year Analysis of SARS-CoV-2 Spike Protein Mutations
Abstract
1. Introduction
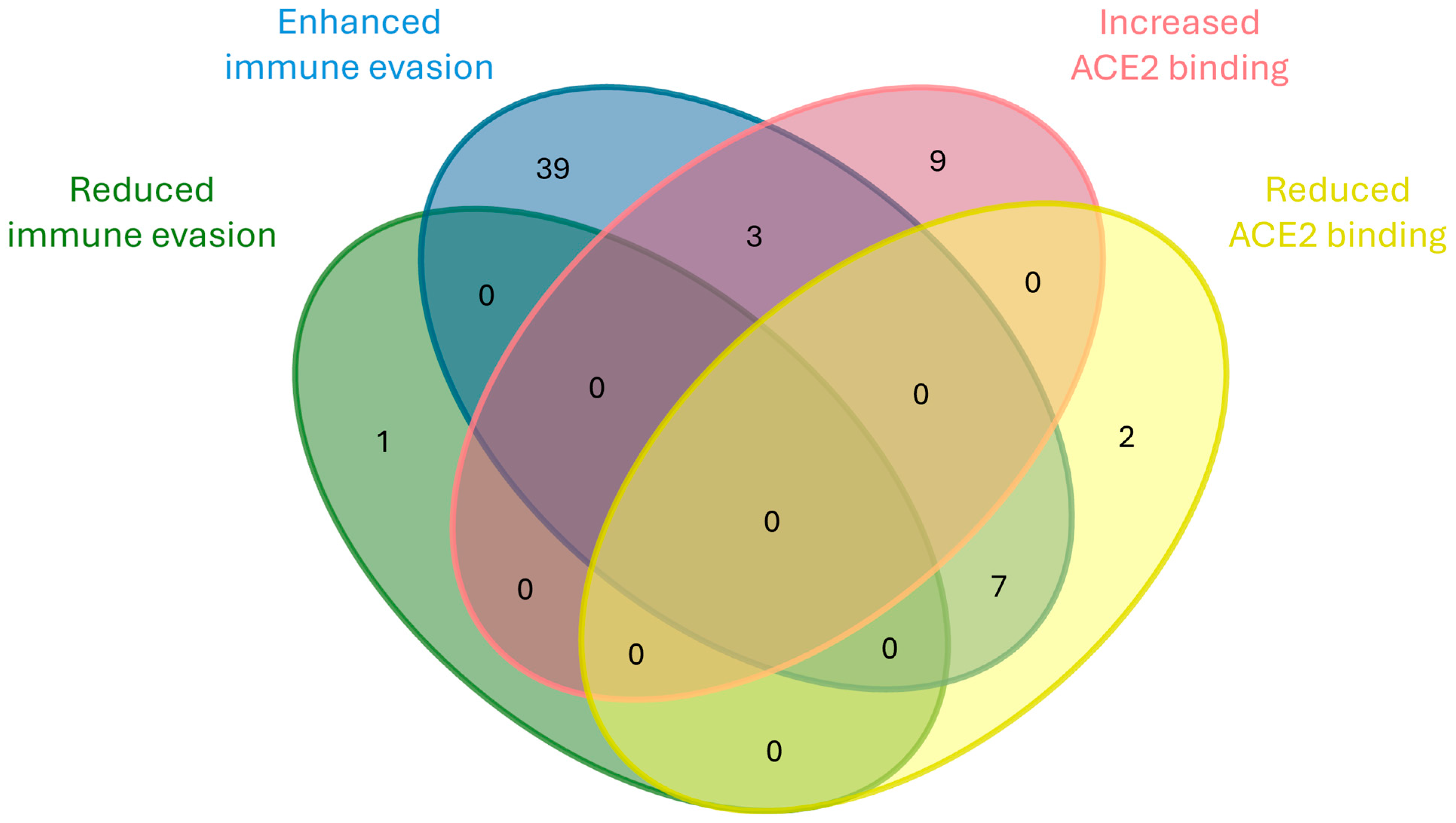
2. Results and Discussion
2.1. Fixed Mutations
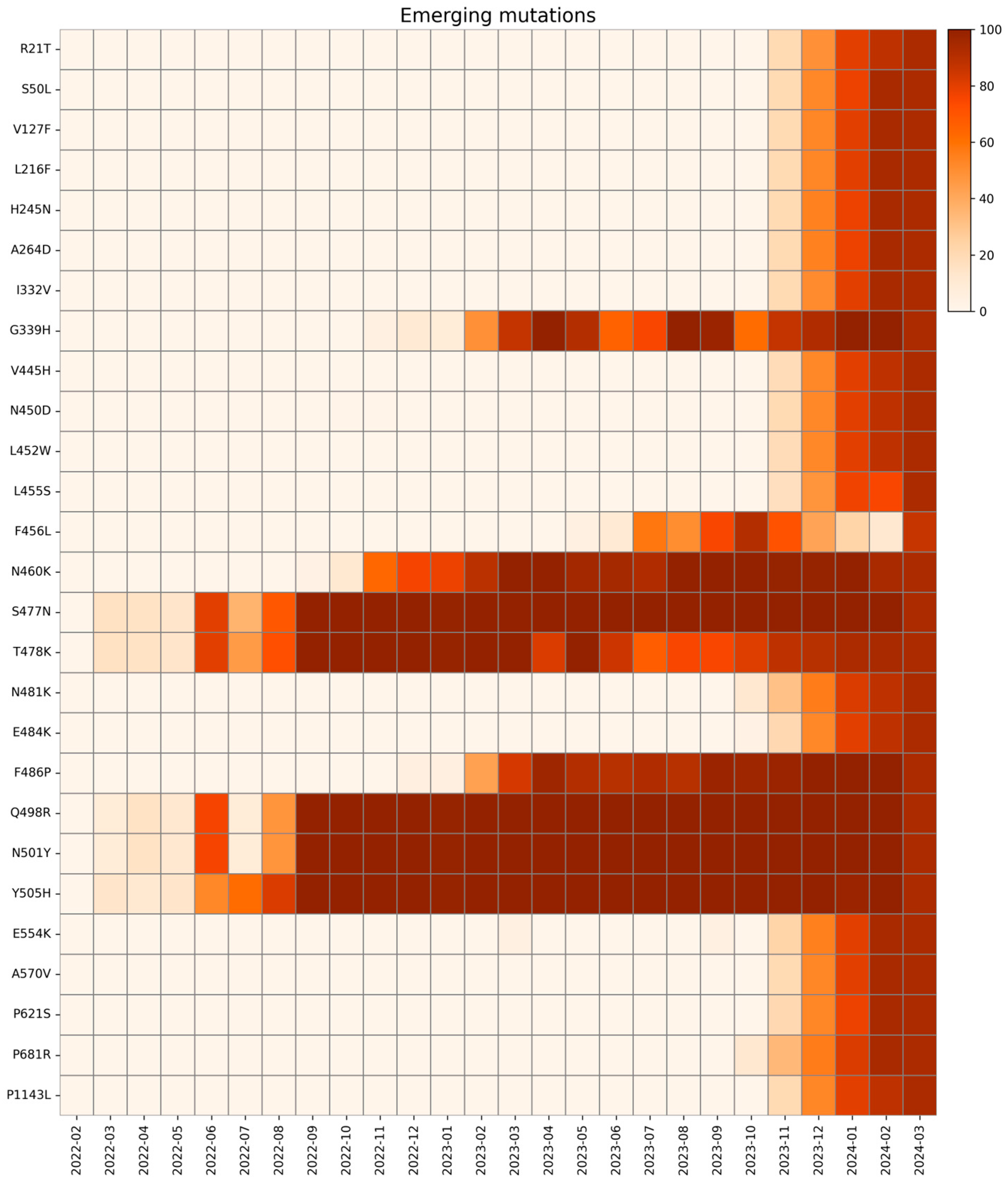
2.2. Emerging Mutations
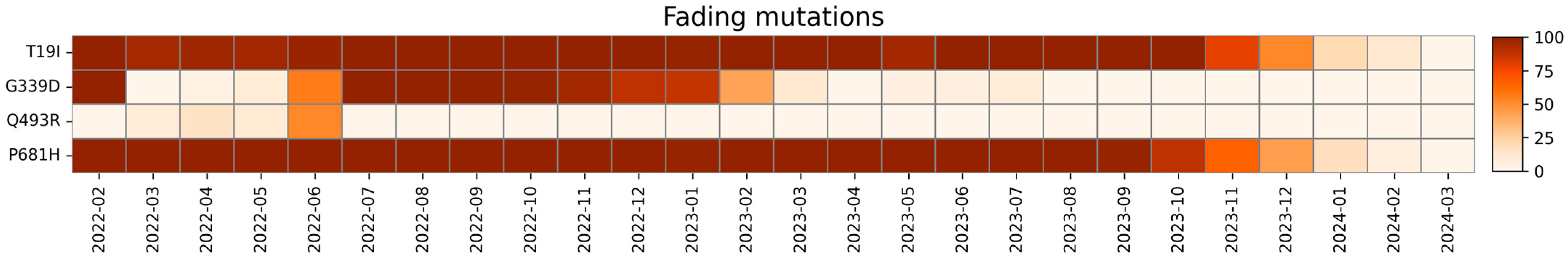
2.3. Fading Mutations
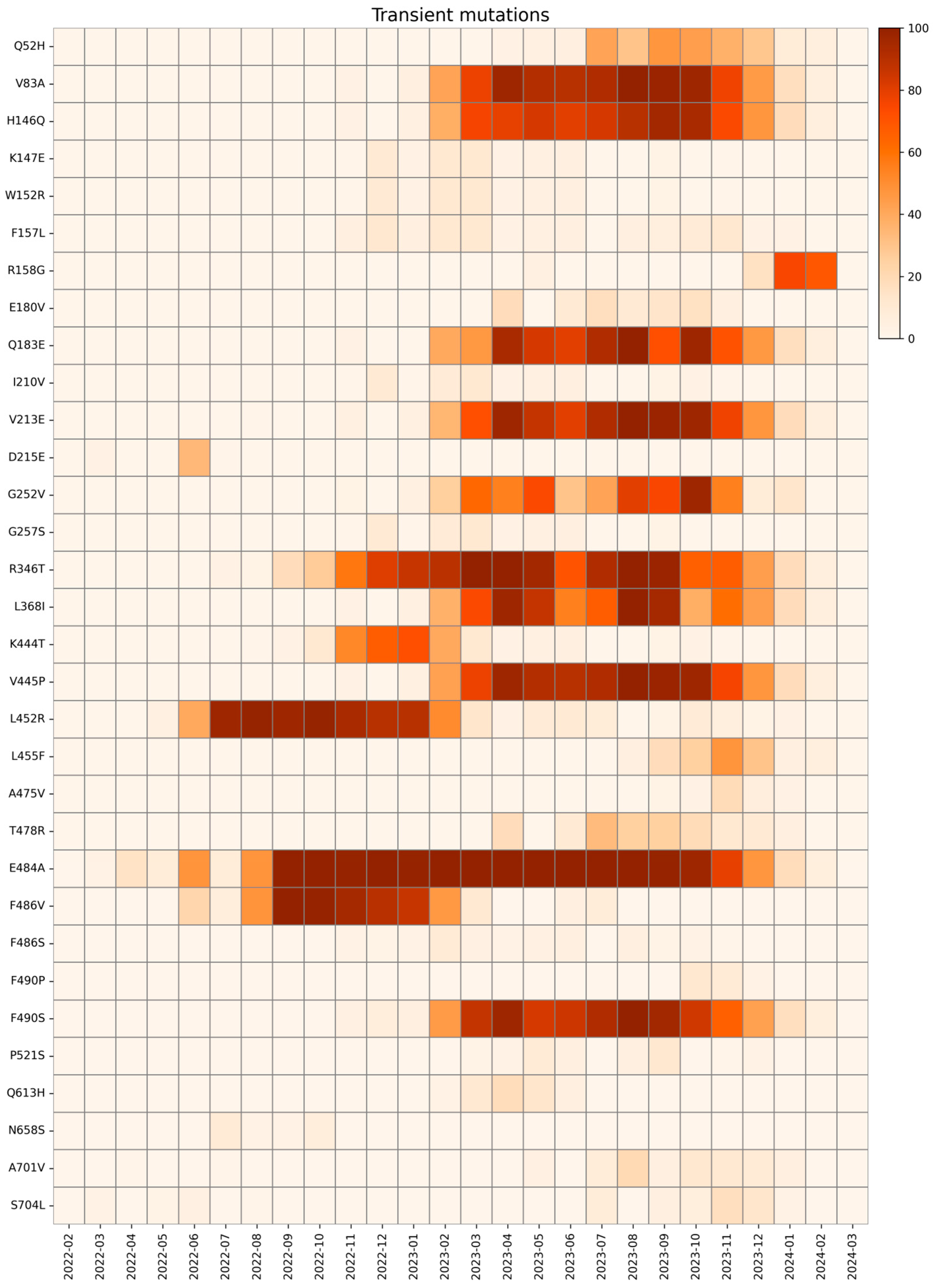
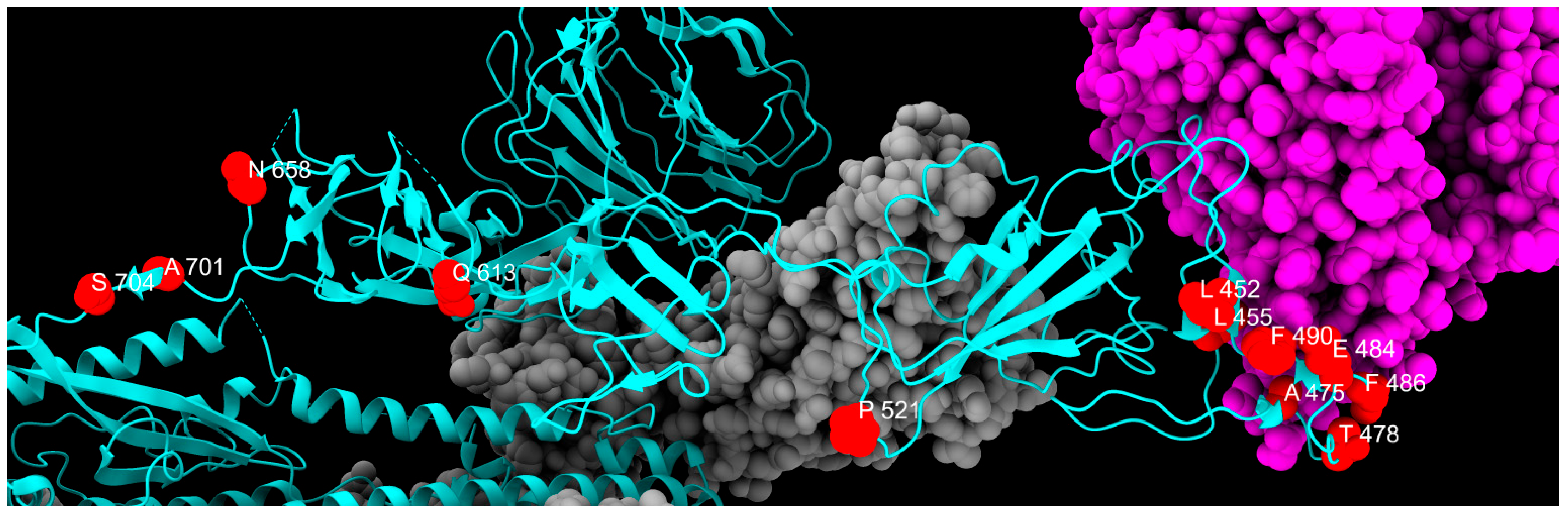
2.4. Transient Mutations
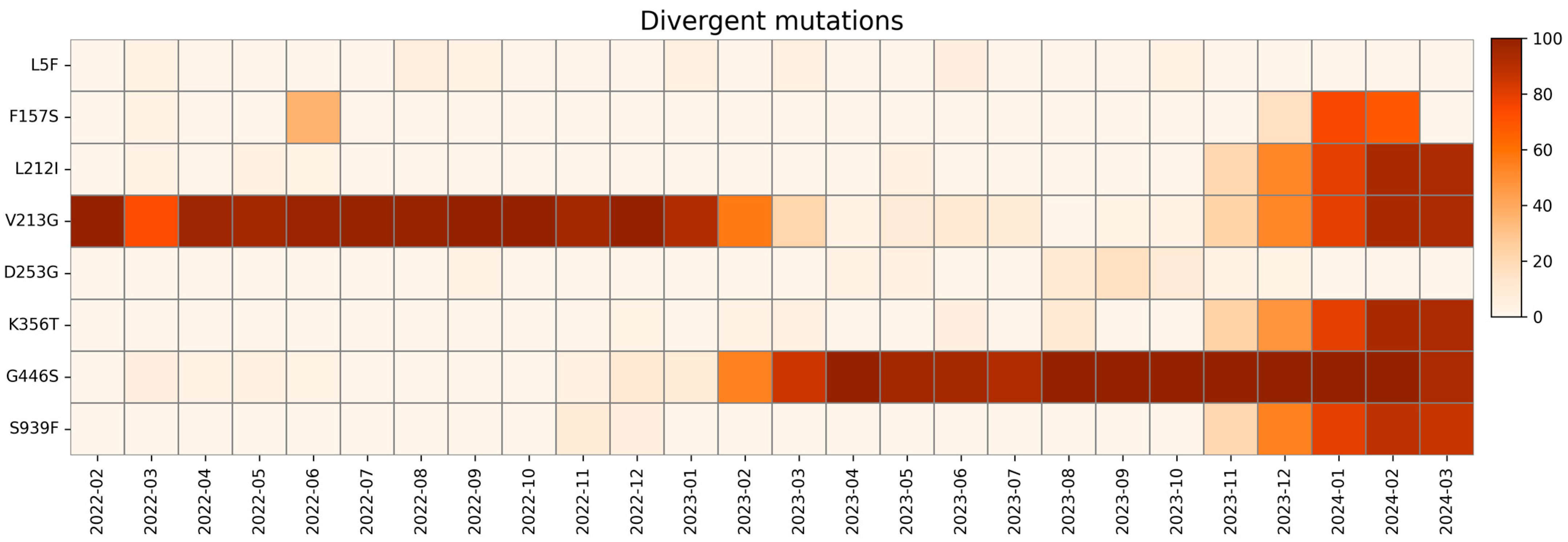
2.5. Divergent Mutations
3. Materials and Methods
3.1. Sample Collection and RNA Extraction
3.2. Library Preparation and Sequencing
3.3. Bioinformatic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| WHO | World Health Organization |
| S | Spike |
| E | Envelope |
| M | Membrane |
| N | Nucleocapsid |
| NTD | N-terminal domain |
| RBD | Receptor-binding domain |
| ACE2 | Angiotensin-converting enzyme 2 |
| NSP | Non-structural protein |
| PANGO | Phylogenetic Assignment of Named Global Outbreak |
| VOC | Variant of Concern |
| VOI | Variant of Interest |
| CTD | C-termina domain |
| FP | Fusion peptide |
| FPPR | Fusion-peptide proximal region |
| HR | Heptad repeat |
| CH | Central helix |
| CD | Connector domain |
| TM | Transmembrane segment |
| mAb | Monoclonal antibody |
| NGS | Next Generation Sequencing |
References
- Lazarevic, I.; Pravica, V.; Miljanovic, D.; Cupic, M. Immune Evasion of SARS-CoV-2 Emerging Variants: What Have We Learnt So Far? Viruses 2021, 13, 1192. [Google Scholar] [CrossRef]
- Flores-Vega, V.R.; Monroy-Molina, J.V.; Jiménez-Hernández, L.E.; Torres, A.G.; Santos-Preciado, J.I.; Rosales-Reyes, R. SARS-CoV-2: Evolution and Emergence of New Viral Variants. Viruses 2022, 14, 653. [Google Scholar] [CrossRef]
- De Castro, E.; Hulo, C.; Masson, P.; Auchincloss, A.; Bridge, A.; Le Mercier, P. ViralZone 2024 Provides Higher-Resolution Images and Advanced Virus-Specific Resources. Nucleic Acids Res. 2024, 52, D817–D821. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.d.I. Insights into SARS-CoV-2 Genome, Structure, Evolution, Pathogenesis and Therapies: Structural Genomics Approach. Biochim. Et Biophys. Acta (BBA) Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Moeller, N.H.; Shi, K.; Demir, Ö.; Belica, C.; Banerjee, S.; Yin, L.; Durfee, C.; Amaro, R.E.; Aihara, H. Structure and Dynamics of SARS-CoV-2 Proofreading Exoribonuclease ExoN. Proc. Natl. Acad. Sci. USA 2022, 119, e2106379119. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- Obermeyer, F.; Jankowiak, M.; Barkas, N.; Schaffner, S.F.; Pyle, J.D.; Yurkovetskiy, L.; Bosso, M.; Park, D.J.; Babadi, M.; MacInnis, B.L.; et al. Analysis of 6.4 Million SARS-CoV-2 Genomes Identifies Mutations Associated with Fitness. Science 2022, 376, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.; Choudhary, M.C.; Regan, J.; Sparks, J.A.; Padera, R.F.; Qiu, X.; Solomon, I.H.; Kuo, H.-H.; Boucau, J.; Bowman, K.; et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 2020, 383, 2291–2293. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and Functional Properties of SARS-CoV-2 Spike Protein: Potential Antivirus Drug Development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Samrat, S.K.; Tharappel, A.M.; Li, Z.; Li, H. Prospect of SARS-CoV-2 Spike Protein: Potential Role in Vaccine and Therapeutic Development. Virus Res. 2020, 288, 198141. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, T.; Cai, Y.; Chen, B. Structure of SARS-CoV-2 Spike Protein. Curr. Opin. Virol. 2021, 50, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Magazine, N.; Zhang, T.; Wu, Y.; McGee, M.C.; Veggiani, G.; Huang, W. Mutations and Evolution of the SARS-CoV-2 Spike Protein. Viruses 2022, 14, 640. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Kannan, S.R.; Spratt, A.N.; Sharma, K.; Chand, H.S.; Byrareddy, S.N.; Singh, K. Omicron SARS-CoV-2 Variant: Unique Features and Their Impact on Pre-Existing Antibodies. J. Autoimmun. 2022, 126, 102779. [Google Scholar] [CrossRef]
- Shen, L.; Triche, T.J.; Dien Bard, J.; Biegel, J.A.; Judkins, A.R.; Gai, X. Spike Protein NTD Mutation G142D in SARS-CoV-2 Delta VOC Lineages Is Associated with Frequent Back Mutations, Increased Viral Loads, and Immune Evasion. medRxiv 2021. [Google Scholar] [CrossRef]
- Shiraz, R.; Tripathi, S. Enhanced Recombination among Omicron Subvariants of SARS-CoV-2 Contributes to Viral Immune Escape. J. Med. Virol. 2023, 95, e28519. [Google Scholar] [CrossRef]
- Zheng, B.; Xiao, Y.; Tong, B.; Mao, Y.; Ge, R.; Tian, F.; Dong, X.; Zheng, P. S373P Mutation Stabilizes the Receptor-Binding Domain of the Spike Protein in Omicron and Promotes Binding. JACS Au 2023, 3, 1902–1910. [Google Scholar] [CrossRef]
- Zeng, X.-T.; Yu, X.-X.; Cheng, W. The Interactions of ZDHHC5/GOLGA7 with SARS-CoV-2 Spike (S) Protein and Their Effects on S Protein’s Subcellular Localization, Palmitoylation and Pseudovirus Entry. Virol. J. 2021, 18, 257. [Google Scholar] [CrossRef]
- Cao, Y.; Yisimayi, A.; Jian, F.; Song, W.; Xiao, T.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X.; et al. BA.2.12.1, BA.4 and BA.5 Escape Antibodies Elicited by Omicron Infection. Nature 2022, 608, 593–602. [Google Scholar] [CrossRef]
- Li, L.; Liao, H.; Meng, Y.; Li, W.; Han, P.; Liu, K.; Wang, Q.; Li, D.; Zhang, Y.; Wang, L.; et al. Structural Basis of Human ACE2 Higher Binding Affinity to Currently Circulating Omicron SARS-CoV-2 Sub-Variants BA.2 and BA.1.1. Cell 2022, 185, 2952–2960.e10. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Chan, J.F.-W.; Liu, H.; Liu, Y.; Chai, Y.; Shi, J.; Shuai, H.; Hou, Y.; Huang, X.; Yuen, T.T.-T.; et al. Spike Mutations Contributing to the Altered Entry Preference of SARS-CoV-2 Omicron BA.1 and BA.2. Emerg. Microbes Infect. 2022, 11, 2275–2287. [Google Scholar] [CrossRef] [PubMed]
- Pondé, R.A.A. Physicochemical Effect of the N501Y, E484K/Q, K417N/T, L452R and T478K Mutations on the SARS-CoV-2 Spike Protein RBD and Its Influence on Agent Fitness and on Attributes Developed by Emerging Variants of Concern. Virology 2022, 572, 44–54. [Google Scholar] [CrossRef]
- Kullappan, M.; Mary, U.; Ambrose, J.M.; Veeraraghavan, V.P.; Surapaneni, K.M. Elucidating the Role of N440K Mutation in SARS-CoV-2 Spike—ACE-2 Binding Affinity and COVID-19 Severity by Virtual Screening, Molecular Docking and Dynamics Approach. J. Biomol. Struct. Dyn. 2023, 41, 912–929. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Thao, T.T.N.; Hoffmann, D.; Taddeo, A.; Ebert, N.; Labroussaa, F.; Pohlmann, A.; King, J.; Steiner, S.; Kelly, J.N.; et al. SARS-CoV-2 Spike D614G Change Enhances Replication and Transmission. Nature 2021, 592, 122–127. [Google Scholar] [CrossRef]
- Qu, P.; Evans, J.P.; Kurhade, C.; Zeng, C.; Zheng, Y.-M.; Xu, K.; Shi, P.-Y.; Xie, X.; Liu, S.-L. Determinants and Mechanisms of the Low Fusogenicity and High Dependence on Endosomal Entry of Omicron Subvariants. mBio 2023, 14, e0317622. [Google Scholar] [CrossRef]
- Lubinski, B.; Jaimes, J.A.; Whittaker, G.R. Intrinsic Furin-Mediated Cleavability of the Spike S1/S2 Site from SARS-CoV-2 Variant B.1.1.529 (Omicron). bioRxiv 2022. [Google Scholar] [CrossRef]
- Shishir, T.A.; Jannat, T.; Naser, I. Bin An In-Silico Study of the Mutation-Associated Effects on the Spike Protein of SARS-CoV-2, Omicron Variant. PLoS ONE 2022, 17, e0266844. [Google Scholar] [CrossRef]
- Maaroufi, H. The N764K and N856K Mutations in SARS-CoV-2 Omicron BA.1 S Protein Generate Potential Cleavage Sites for SKI-1/S1P Protease. bioRxiv 2022. [Google Scholar] [CrossRef]
- Elko, E.A.; Mead, H.L.; Nelson, G.A.; Zaia, J.A.; Ladner, J.T.; Altin, J.A. Recurrent SARS-CoV-2 Mutations at Spike D796 Evade Antibodies from Pre-Omicron Convalescent and Vaccinated Subjects. Microbiol. Spectr. 2024, 12, e0329123. [Google Scholar] [CrossRef]
- Cheng, Y.; Zheng, D.; Zhang, D.; Guo, D.; Wang, Y.; Liu, W.; Liang, L.; Hu, J.; Luo, T. Molecular Recognition of SARS-CoV-2 Spike Protein with Three Essential Partners: Exploring Possible Immune Escape Mechanisms of Viral Mutants. J. Mol. Model. 2023, 29, 109. [Google Scholar] [CrossRef]
- Gobeil, S.M.-C.; Henderson, R.; Stalls, V.; Janowska, K.; Huang, X.; May, A.; Speakman, M.; Beaudoin, E.; Manne, K.; Li, D.; et al. Structural Diversity of the SARS-CoV-2 Omicron Spike. Mol. Cell 2022, 82, 2050–2068.e6. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.J.C.; Verma, A.K.; Odle, A.; Lei, R.; Meyerholz, D.K.; Matreyek, K.A.; Perlman, S.; Wong, L.-Y.R.; Wu, N.C. Evidence of Antigenic Drift in the Fusion Machinery Core of SARS-CoV-2 Spike. Proc. Natl. Acad. Sci. USA 2024, 121, e2317222121. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Brown, J.C.; Zhou, J.; Thakur, N.; Sukhova, K.; Kugathasan, R.; Yan, A.W.; Furnon, W.; De Lorenzo, G.; Reuss, D.; et al. The Altered Entry Pathway and Antigenic Distance of the SARS-CoV-2 Omicron Variant Map to Separate 2 Domains of Spike Protein. bioRxiv 2022. [Google Scholar] [CrossRef]
- Yang, S.; Yu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Chen, X.; Xu, Y.; Wang, P.; Wang, J.; Yu, L.; et al. Antigenicity and Infectivity Characterisation of SARS-CoV-2 BA.2.86. Lancet Infect. Dis. 2023, 23, e457–e459. [Google Scholar] [CrossRef]
- Yang, H.; Guo, H.; Wang, A.; Cao, L.; Fan, Q.; Jiang, J.; Wang, M.; Lin, L.; Ge, X.; Wang, H.; et al. Structural Basis for the Evolution and Antibody Evasion of SARS-CoV-2 BA.2.86 and JN.1 Subvariants. Nat. Commun. 2024, 15, 7715. [Google Scholar] [CrossRef]
- Qu, P.; Xu, K.; Faraone, J.N.; Goodarzi, N.; Zheng, Y.-M.; Carlin, C.; Bednash, J.S.; Horowitz, J.C.; Mallampalli, R.K.; Saif, L.J.; et al. Immune Evasion, Infectivity, and Fusogenicity of SARS-CoV-2 BA.2.86 and FLip Variants. Cell 2024, 187, 585–595.e6. [Google Scholar] [CrossRef]
- Yajima, H.; Anraku, Y.; Kaku, Y.; Kimura, K.T.; Plianchaisuk, A.; Okumura, K.; Nakada-Nakura, Y.; Atarashi, Y.; Hemmi, T.; Kuroda, D.; et al. Structural Basis for Receptor-Binding Domain Mobility of the Spike in SARS-CoV-2 BA.2.86 and JN.1. Nat. Commun. 2024, 15, 8574. [Google Scholar] [CrossRef] [PubMed]
- Qu, P.; Faraone, J.N.; Evans, J.P.; Zheng, Y.-M.; Carlin, C.; Anghelina, M.; Stevens, P.; Fernandez, S.; Jones, D.; Panchal, A.R.; et al. Enhanced Evasion of Neutralizing Antibody Response by Omicron XBB.1.5, CH.1.1, and CA.3.1 Variants. Cell Rep. 2023, 42, 112443. [Google Scholar] [CrossRef]
- Zhang, L.; Kempf, A.; Nehlmeier, I.; Cossmann, A.; Richter, A.; Bdeir, N.; Graichen, L.; Moldenhauer, A.-S.; Dopfer-Jablonka, A.; Stankov, M.V.; et al. SARS-CoV-2 BA.2.86 Enters Lung Cells and Evades Neutralizing Antibodies with High Efficiency. Cell 2024, 187, 596–608.e17. [Google Scholar] [CrossRef] [PubMed]
- Kannan, S.R.; Spratt, A.N.; Cohen, A.R.; Naqvi, S.H.; Chand, H.S.; Quinn, T.P.; Lorson, C.L.; Byrareddy, S.N.; Singh, K. Evolutionary Analysis of the Delta and Delta Plus Variants of the SARS-CoV-2 Viruses. J. Autoimmun. 2021, 124, 102715. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, L.; Liang, Z.; Nie, J.; Wu, J.; Li, Q.; Ding, R.; Zhang, Y.; Chen, G.; Wang, Y.; et al. Infectivity and Antigenicity of Pseudoviruses with High-Frequency Mutations of SARS-CoV-2 Identified in Portugal. Arch. Virol. 2022, 167, 459–470. [Google Scholar] [CrossRef]
- Cao, Y.; Jian, F.; Wang, J.; Yu, Y.; Song, W.; Yisimayi, A.; Wang, J.; An, R.; Chen, X.; Zhang, N.; et al. Imprinted SARS-CoV-2 Humoral Immunity Induces Convergent Omicron RBD Evolution. Nature 2022, 614, 521–529. [Google Scholar] [CrossRef]
- Mohapatra, R.K.; Mahal, A.; Mishra, S.; Kandi, V.; Obaidullah, W.J. SARS-CoV-2 Variants BA.2.86 and EG.5.1 Alongside Scrub Typhus and Nipah in India During the Ongoing Cricket World Cup 2023: Threat Perceptions and Countermeasures. Cureus 2023, 15, e48895. [Google Scholar] [CrossRef]
- Pavia, G.; Quirino, A.; Marascio, N.; Veneziano, C.; Longhini, F.; Bruni, A.; Garofalo, E.; Pantanella, M.; Manno, M.; Gigliotti, S.; et al. Persistence of SARS-CoV-2 Infection and Viral Intra- and Inter-host Evolution in COVID-19 Hospitalized Patients. J. Med. Virol. 2024, 96, e29708. [Google Scholar] [CrossRef]
- Lu, Y.; Ao, D.; He, X.; Wei, X. The Rising SARS-CoV-2 JN.1 Variant: Evolution, Infectivity, Immune Escape, and Response Strategies. MedComm 2024, 5, e675. [Google Scholar] [CrossRef]
- Yang, S.; Yu, Y.; Xu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Wang, P.; Wang, J.; Liu, J.; Yu, L.; et al. Fast Evolution of SARS-CoV-2 BA.2.86 to JN.1 under Heavy Immune Pressure. Lancet Infect. Dis. 2024, 24, e70–e72. [Google Scholar] [CrossRef]
- Jian, F.; Feng, L.; Yang, S.; Yu, Y.; Wang, L.; Song, W.; Yisimayi, A.; Chen, X.; Xu, Y.; Wang, P.; et al. Convergent Evolution of SARS-CoV-2 XBB Lineages on Receptor-Binding Domain 455–456 Synergistically Enhances Antibody Evasion and ACE2 Binding. PLOS Pathog. 2023, 19, e1011868. [Google Scholar] [CrossRef]
- Focosi, D.; Spezia, P.G.; Gueli, F.; Maggi, F. The Era of the FLips: How Spike Mutations L455F and F456L (and A475V) Are Shaping SARS-CoV-2 Evolution. Viruses 2023, 16, 3. [Google Scholar] [CrossRef]
- Mondeali, M.; Etemadi, A.; Barkhordari, K.; Mobini Kesheh, M.; Shavandi, S.; Bahavar, A.; Tabatabaie, F.H.; Mahmoudi Gomari, M.; Modarressi, M.H. The Role of S477N Mutation in the Molecular Behavior of SARS-CoV-2 Spike Protein: An In-silico Perspective. J. Cell. Biochem. 2023, 124, 308–319. [Google Scholar] [CrossRef]
- Jhun, H.; Park, H.-Y.; Hisham, Y.; Song, C.-S.; Kim, S. SARS-CoV-2 Delta (B.1.617.2) Variant: A Unique T478K Mutation in Receptor Binding Motif (RBM) of Spike Gene. Immune Netw. 2021, 21, e32. [Google Scholar] [CrossRef]
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S.; et al. SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms 2021, 9, 1542. [Google Scholar] [CrossRef]
- Schröder, S.; Richter, A.; Veith, T.; Emanuel, J.; Gudermann, L.; Friedmann, K.; Jeworowski, L.M.; Mühlemann, B.; Jones, T.C.; Müller, M.A.; et al. Characterization of Intrinsic and Effective Fitness Changes Caused by Temporarily Fixed Mutations in the SARS-CoV-2 Spike E484 Epitope and Identification of an Epistatic Precondition for the Evolution of E484A in Variant Omicron. Virol. J. 2023, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Shi, K.; Geng, Q.; Ye, G.; Aihara, H.; Li, F. Structural Basis for Mouse Receptor Recognition by SARS-CoV-2 Omicron Variant. Proc. Natl. Acad. Sci. USA 2022, 119, e2206509119. [Google Scholar] [CrossRef] [PubMed]
- Khairnar, K.; Tomar, S.S. COVID-19 Genome Surveillance: A Geographical Landscape and Mutational Mapping of SARS-CoV-2 Variants in Central India over Two Years. Virus Res. 2024, 344, 199365. [Google Scholar] [CrossRef]
- Zahradník, J.; Marciano, S.; Shemesh, M.; Zoler, E.; Harari, D.; Chiaravalli, J.; Meyer, B.; Rudich, Y.; Li, C.; Marton, I.; et al. SARS-CoV-2 Variant Prediction and Antiviral Drug Design Are Enabled by RBD in Vitro Evolution. Nat. Microbiol. 2021, 6, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Lupala, C.S.; Ye, Y.; Chen, H.; Su, X.-D.; Liu, H. Mutations on RBD of SARS-CoV-2 Omicron Variant Result in Stronger Binding to Human ACE2 Receptor. Biochem. Biophys. Res. Commun. 2022, 590, 34–41. [Google Scholar] [CrossRef]
- Zhou, D.; Supasa, P.; Liu, C.; Dijokaite-Guraliuc, A.; Duyvesteyn, H.M.E.; Selvaraj, M.; Mentzer, A.J.; Das, R.; Dejnirattisai, W.; Temperton, N.; et al. The SARS-CoV-2 Neutralizing Antibody Response to SD1 and Its Evasion by BA.2.86. Nat. Commun. 2024, 15, 2734. [Google Scholar] [CrossRef]
- Ray, D.; Le, L.; Andricioaei, I. Distant Residues Modulate Conformational Opening in SARS-CoV-2 Spike Protein. Proc. Natl. Acad. Sci. USA 2021, 118, e2100943118. [Google Scholar] [CrossRef]
- Liu, P.; Yue, C.; Meng, B.; Xiao, T.; Yang, S.; Liu, S.; Jian, F.; Zhu, Q.; Yu, Y.; Ren, Y.; et al. Spike N354 Glycosylation Augments SARS-CoV-2 Fitness for Human Adaptation through Structural Plasticity. Natl. Sci. Rev. 2024, 11, nwae206. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, P.; Wang, N.; Wang, L.; Fan, K.; Zhu, Q.; Wang, K.; Chen, R.; Feng, R.; Jia, Z.; et al. Structural and Functional Characterizations of Infectivity and Immune Evasion of SARS-CoV-2 Omicron. Cell 2022, 185, 860–871.e13. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced Fusogenicity and Pathogenicity of SARS-CoV-2 Delta P681R Mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef]
- Donzelli, S.; Spinella, F.; di Domenico, E.G.; Pontone, M.; Cavallo, I.; Orlandi, G.; Iannazzo, S.; Ricciuto, G.M.; ISG Virology Covid Team; Pellini, R.; et al. Evidence of a SARS-CoV-2 Double Spike Mutation D614G/S939F Potentially Affecting Immune Response of Infected Subjects. Comput. Struct. Biotechnol. J. 2022, 20, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, Y.; Liu, L.; Schwanz, L.T.; Li, Z.; Nair, M.S.; Ho, J.; Zhang, R.M.; Iketani, S.; Yu, J.; et al. Antigenicity and Receptor Affinity of SARS-CoV-2 BA.2.86 Spike. Nature 2023, 624, 639–644. [Google Scholar] [CrossRef]
- Dadonaite, B.; Crawford, K.H.D.; Radford, C.E.; Farrell, A.G.; Yu, T.C.; Hannon, W.W.; Zhou, P.; Andrabi, R.; Burton, D.R.; Liu, L.; et al. A Pseudovirus System Enables Deep Mutational Scanning of the Full SARS-CoV-2 Spike. Cell 2023, 186, 1263–1278.e20. [Google Scholar] [CrossRef] [PubMed]
- Dadonaite, B.; Brown, J.; McMahon, T.E.; Farrell, A.G.; Figgins, M.D.; Asarnow, D.; Stewart, C.; Lee, J.; Logue, J.; Bedford, T.; et al. Spike Deep Mutational Scanning Helps Predict Success of SARS-CoV-2 Clades. Nature 2024, 631, 617–626. [Google Scholar] [CrossRef]
- Subramoney, K.; Mtileni, N.; Davis, A.; Giandhari, J.; Tegally, H.; Wilkinson, E.; Naidoo, Y.; Ramphal, Y.; Pillay, S.; Ramphal, U.; et al. SARS-CoV-2 Spike Protein Diversity at an Intra-Host Level, among SARS-CoV-2 Infected Individuals in South Africa, 2020 to 2022. PLoS ONE 2023, 18, e0286373. [Google Scholar] [CrossRef]
- Pastorio, C.; Zech, F.; Noettger, S.; Jung, C.; Jacob, T.; Sanderson, T.; Sparrer, K.M.J.; Kirchhoff, F. Determinants of Spike Infectivity, Processing, and Neutralization in SARS-CoV-2 Omicron Subvariants BA.1 and BA.2. Cell Host Microbe 2022, 30, 1255–1268.e5. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron Escapes the Majority of Existing SARS-CoV-2 Neutralizing Antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef]
- Shikha, S.; Jogi, M.K.; Jha, R.; Kumar, R.A.; Sah, T.; Singh, P.; Sagar, R.; Kumar, A.; Marwal, R.; Ponnusamy, K.; et al. Genome Sequencing of SARS-CoV-2 Omicron Variants in Delhi Reveals Alterations in Immunogenic Regions in Spike Glycoprotein. Front. Immunol. 2023, 14, 1209513. [Google Scholar] [CrossRef] [PubMed]
- Philip, A.M.; Ahmed, W.S.; Biswas, K.H. Reversal of the Unique Q493R Mutation Increases the Affinity of Omicron S1-RBD for ACE2. Comput. Struct. Biotechnol. J. 2023, 21, 1966–1977. [Google Scholar] [CrossRef]
- Bansal, K.; Kumar, S. Mutational Cascade of SARS-CoV-2 Leading to Evolution and Emergence of Omicron Variant. Virus Res. 2022, 315, 198765. [Google Scholar] [CrossRef] [PubMed]
- Guigon, A.; Faure, E.; Lemaire, C.; Chopin, M.-C.; Tinez, C.; Assaf, A.; Lazrek, M.; Hober, D.; Bocket, L.; Engelmann, I.; et al. Emergence of Q493R Mutation in SARS-CoV-2 Spike Protein during Bamlanivimab/Etesevimab Treatment and Resistance to Viral Clearance. J. Infect. 2022, 84, 248–288. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, A.; Abubaker, J.; Al-Mulla, F. Structural Modelling of SARS-CoV-2 Alpha Variant (B.1.1.7) Suggests Enhanced Furin Binding and Infectivity. Virus Res. 2021, 303, 198522. [Google Scholar] [CrossRef]
- Lista, M.J.; Winstone, H.; Wilson, H.D.; Dyer, A.; Pickering, S.; Galao, R.P.; De Lorenzo, G.; Cowton, V.M.; Furnon, W.; Suarez, N.; et al. The P681H Mutation in the Spike Glycoprotein of the Alpha Variant of SARS-CoV-2 Escapes IFITM Restriction and Is Necessary for Type I Interferon Resistance. J. Virol. 2022, 96, e0125022. [Google Scholar] [CrossRef]
- Benlarbi, M.; Ding, S.; Bélanger, É.; Tauzin, A.; Poujol, R.; Medjahed, H.; El Ferri, O.; Bo, Y.; Bourassa, C.; Hussin, J.; et al. Temperature-Dependent Spike-ACE2 Interaction of Omicron Subvariants Is Associated with Viral Transmission. mBio 2024, 15, e0090724. [Google Scholar] [CrossRef]
- Tamura, T.; Ito, J.; Uriu, K.; Zahradnik, J.; Kida, I.; Anraku, Y.; Nasser, H.; Shofa, M.; Oda, Y.; Lytras, S.; et al. Virological Characteristics of the SARS-CoV-2 XBB Variant Derived from Recombination of Two Omicron Subvariants. Nat. Commun. 2023, 14, 2800. [Google Scholar] [CrossRef]
- Feng, Y.; Yi, J.; Yang, L.; Wang, Y.; Wen, J.; Zhao, W.; Kim, P.; Zhou, X. COV2Var, a Function Annotation Database of SARS-CoV-2 Genetic Variation. Nucleic Acids Res. 2024, 52, D701–D713. [Google Scholar] [CrossRef]
- Zappa, M.; Verdecchia, P.; Angeli, F. Severe Acute Respiratory Syndrome Coronavirus 2 Evolution: How Mutations Affect XBB.1.5 Variant. Eur. J. Intern. Med. 2023, 112, 128–132. [Google Scholar] [CrossRef]
- Wang, Q.; Iketani, S.; Li, Z.; Guo, Y.; Yeh, A.Y.; Liu, M.; Yu, J.; Sheng, Z.; Huang, Y.; Liu, L.; et al. Antigenic Characterization of the SARS-CoV-2 Omicron Subvariant BA.2.75. Cell Host Microbe 2022, 30, 1512–1517.e4. [Google Scholar] [CrossRef]
- Wang, Q.; Iketani, S.; Li, Z.; Liu, L.; Guo, Y.; Huang, Y.; Bowen, A.D.; Liu, M.; Wang, M.; Yu, J.; et al. Alarming Antibody Evasion Properties of Rising SARS-CoV-2 BQ and XBB Subvariants. Cell 2023, 186, 279–286.e8. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, Y.; Yang, C.; Ren, W.; Qiu, C.; Fan, S.; Ding, Q.; Lan, J. Receptor Binding Mechanism and Immune Evasion Capacity of SARS-CoV-2 BQ.1.1 Lineage. Virology 2024, 600, 110241. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Nehlmeier, I.; Kempf, A.; Cossmann, A.; Schulz, S.R.; Dopfer-Jablonka, A.; Baier, E.; Tampe, B.; Moerer, O.; Dickel, S.; et al. Lung Cell Entry, Cell–Cell Fusion Capacity, and Neutralisation Sensitivity of Omicron Sublineage BA.2.75. Lancet Infect. Dis. 2022, 22, 1537–1538. [Google Scholar] [CrossRef] [PubMed]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; et al. SARS-CoV-2 Spike L452R Variant Evades Cellular Immunity and Increases Infectivity. Cell Host Microbe 2021, 29, 1124–1136.e11. [Google Scholar] [CrossRef]
- Peng, D.; He, C.; Chen, Z.; Lei, H.; Huang, X.; Ye, C.; Wang, B.; Hao, Y.; Du, X.; Lu, S.; et al. XBB.1.16-RBD-based Trimeric Protein Vaccine Can Effectively Inhibit XBB.1.16-included XBB Subvariant Infection. MedComm 2024, 5, e687. [Google Scholar] [CrossRef] [PubMed]
- Muecksch, F.; Weisblum, Y.; Barnes, C.O.; Schmidt, F.; Schaefer-Babajew, D.; Wang, Z.; Lorenzi, J.C.C.; Flyak, A.I.; DeLaitsch, A.T.; Huey-Tubman, K.E.; et al. Affinity Maturation of SARS-CoV-2 Neutralizing Antibodies Confers Potency, Breadth, and Resilience to Viral Escape Mutations. Immunity 2021, 54, 1853–1868.e7. [Google Scholar] [CrossRef]
- Kumar, P.; Zhang, X.; Shaha, R.; Kschischo, M.; Dobbelstein, M. Identification of Antibody-Resistant SARS-CoV-2 Mutants via N4-Hydroxycytidine Mutagenesis. Antivir. Res. 2024, 231, 106006. [Google Scholar] [CrossRef]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef] [PubMed]
- Faraone, J.N.; Qu, P.; Goodarzi, N.; Zheng, Y.-M.; Carlin, C.; Saif, L.J.; Oltz, E.M.; Xu, K.; Jones, D.; Gumina, R.J.; et al. Immune Evasion and Membrane Fusion of SARS-CoV-2 XBB Subvariants EG.5.1 and XBB.2.3. Emerg. Microbes Infect. 2023, 12, 2270069. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Egri, S.; Kurhade, C.; Diaz-Salinas, M.A.; Jaimes, J.A.; Nyalile, T.; Xie, X.; Choudhary, M.C.; Dauphin, A.; Li, J.Z.; et al. S:D614G and S:H655Y Are Gateway Mutations That Act Epistatically to Promote SARS-CoV-2 Variant Fitness. bioRxiv 2023. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Z.; Ho, J.; Guo, Y.; Yeh, A.Y.; Mohri, H.; Liu, M.; Wang, M.; Yu, J.; Shah, J.G.; et al. Resistance of SARS-CoV-2 Omicron Subvariant BA.4.6 to Antibody Neutralisation. Lancet Infect. Dis. 2022, 22, 1666–1668. [Google Scholar] [CrossRef]
- Kimura, I.; Yamasoba, D.; Tamura, T.; Nao, N.; Suzuki, T.; Oda, Y.; Mitoma, S.; Ito, J.; Nasser, H.; Zahradnik, J.; et al. Virological Characteristics of the SARS-CoV-2 Omicron BA.2 Subvariants, Including BA.4 and BA.5. Cell 2022, 185, 3992–4007.e16. [Google Scholar] [CrossRef]
- Chen, C.; Nadeau, S.; Yared, M.; Voinov, P.; Xie, N.; Roemer, C.; Stadler, T. CoV-Spectrum: Analysis of Globally Shared SARS-CoV-2 Data to Identify and Characterize New Variants. Bioinformatics 2022, 38, 1735–1737. [Google Scholar] [CrossRef]
- Zhang, Z.; Wan, X.; Li, X.; Wan, C. Effects of a Shift of the Signal Peptide Cleavage Site in Signal Peptide Variant on the Synthesis and Secretion of SARS-CoV-2 Spike Protein. Molecules 2022, 27, 6688. [Google Scholar] [CrossRef] [PubMed]
- Aljindan, R.Y.; Al-Subaie, A.M.; Al-Ohali, A.I.; Kumar D, T.; Doss C, G.P.; Kamaraj, B. Investigation of Nonsynonymous Mutations in the Spike Protein of SARS-CoV-2 and Its Interaction with the ACE2 Receptor by Molecular Docking and MM/GBSA Approach. Comput. Biol. Med. 2021, 135, 104654. [Google Scholar] [CrossRef]
- Nersisyan, S.; Zhiyanov, A.; Zakharova, M.; Ishina, I.; Kurbatskaia, I.; Mamedov, A.; Galatenko, A.; Shkurnikov, M.; Gabibov, A.; Tonevitsky, A. Alterations in SARS-CoV-2 Omicron and Delta Peptides Presentation by HLA Molecules. PeerJ 2022, 10, e13354. [Google Scholar] [CrossRef]
- Sun, L.; Wang, L.; Zhang, C.; Xiao, Y.; Zhang, L.; Zhao, Z.; Ren, L.; Peng, J. Rapid Detection of Predominant SARS-CoV-2 Variants Using Multiplex High-Resolution Melting Analysis. Microbiol. Spectr. 2023, 11, e0005523. [Google Scholar] [CrossRef] [PubMed]
- West, A.P.; Wertheim, J.O.; Wang, J.C.; Vasylyeva, T.I.; Havens, J.L.; Chowdhury, M.A.; Gonzalez, E.; Fang, C.E.; Di Lonardo, S.S.; Hughes, S.; et al. Detection and Characterization of the SARS-CoV-2 Lineage B.1.526 in New York. Nat. Commun. 2021, 12, 4886. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Cai, S.; Fang, X.; Wang, X.; Zhang, Z.; Damiani, M.; Hudlerova, C.; Rosa, A.; Hope, J.; Cook, N.J.; et al. Multiplexed Detection of Viral Antigen and RNA Using Nanopore Sensing and Encoded Molecular Probes. Nat. Commun. 2023, 14, 7362. [Google Scholar] [CrossRef]
- Motozono, C.; Toyoda, M.; Tan, T.S.; Hamana, H.; Goto, Y.; Aritsu, Y.; Miyashita, Y.; Oshiumi, H.; Nakamura, K.; Okada, S.; et al. The SARS-CoV-2 Omicron BA.1 Spike G446S Mutation Potentiates Antiviral T-Cell Recognition. Nat. Commun. 2022, 13, 5440. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Hwang, M.; Cornelius, L.; Navarathna, D.H.; Chatterjee, P.; Jinadatha, C. Evolution of a Distinct SARS-CoV-2 Lineage Identified during an Investigation of a Hospital Outbreak. Viruses 2024, 16, 337. [Google Scholar] [CrossRef]
- Diani, E.; Silvagni, D.; Lotti, V.; Lagni, A.; Baggio, L.; Medaina, N.; Biban, P.; Gibellini, D. Evaluation of Saliva and Nasopharyngeal Swab Sampling for Genomic Detection of SARS-CoV-2 in Children Accessing a Pediatric Emergency Department during the Second Pandemic Wave. Front. Microbiol. 2023, 14, 1163438. [Google Scholar] [CrossRef]
- Tonon, E.; Cecchetto, R.; Diani, E.; Medaina, N.; Turri, G.; Lagni, A.; Lotti, V.; Gibellini, D. Surfing the Waves of SARS-CoV-2: Analysis of Viral Genome Variants Using an NGS Survey in Verona, Italy. Microorganisms 2024, 12, 846. [Google Scholar] [CrossRef]
- Cecchetto, R.; Tonon, E.; Medaina, N.; Turri, G.; Diani, E.; Piccaluga, P.P.; Salomoni, A.; Conti, M.; Tacconelli, E.; Lagni, A.; et al. Detection of SARS-CoV-2 Δ426 ORF8 Deletion Mutant Cluster in NGS Screening. Microorganisms 2023, 11, 2378. [Google Scholar] [CrossRef]
- Bonfield, J.K.; Marshall, J.; Danecek, P.; Li, H.; Ohan, V.; Whitwham, A.; Keane, T.; Davies, R.M. HTSlib: C Library for Reading/Writing High-Throughput Sequencing Data. Gigascience 2021, 10, giab007. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; McCauley, J. GISAID: Global Initiative on Sharing All Influenza Data—From Vision to Reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Piccaluga, P.P.; Di Guardo, A.; Lagni, A.; Lotti, V.; Diani, E.; Navari, M.; Gibellini, D. COVID-19 Vaccine: Between Myth and Truth. Vaccines 2022, 10, 349. [Google Scholar] [CrossRef]
- Monaco, M.G.L.; Spiteri, G.; Caliskan, G.; Lotti, V.; Carta, A.; Gibellini, D.; Verlato, G.; Porru, S. SARS-CoV-2 and Its Variants in Thrice-Infected Health Workers: A Case Series from an Italian University Hospital. Viruses 2022, 14, 2536. [Google Scholar] [CrossRef]

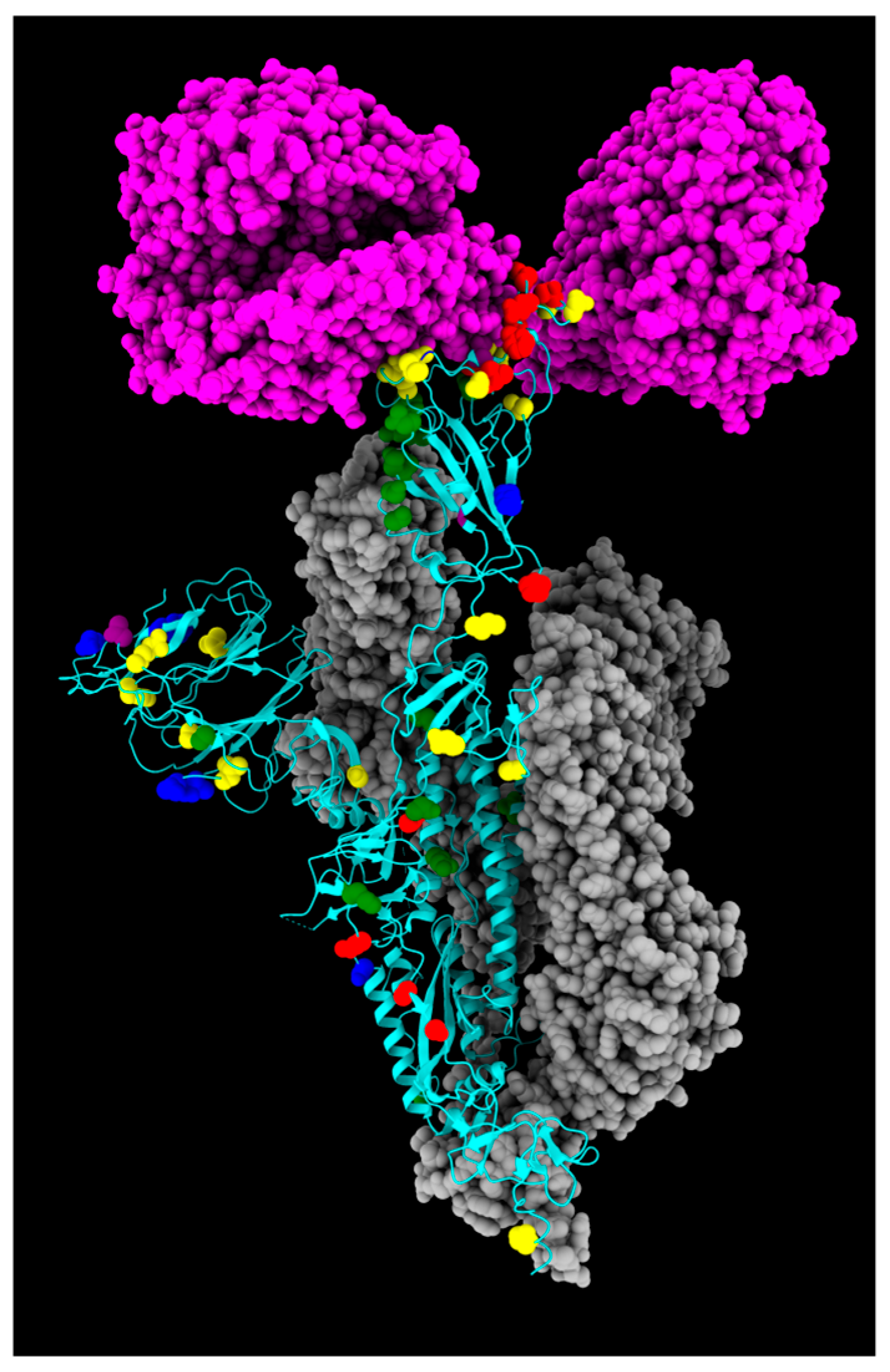
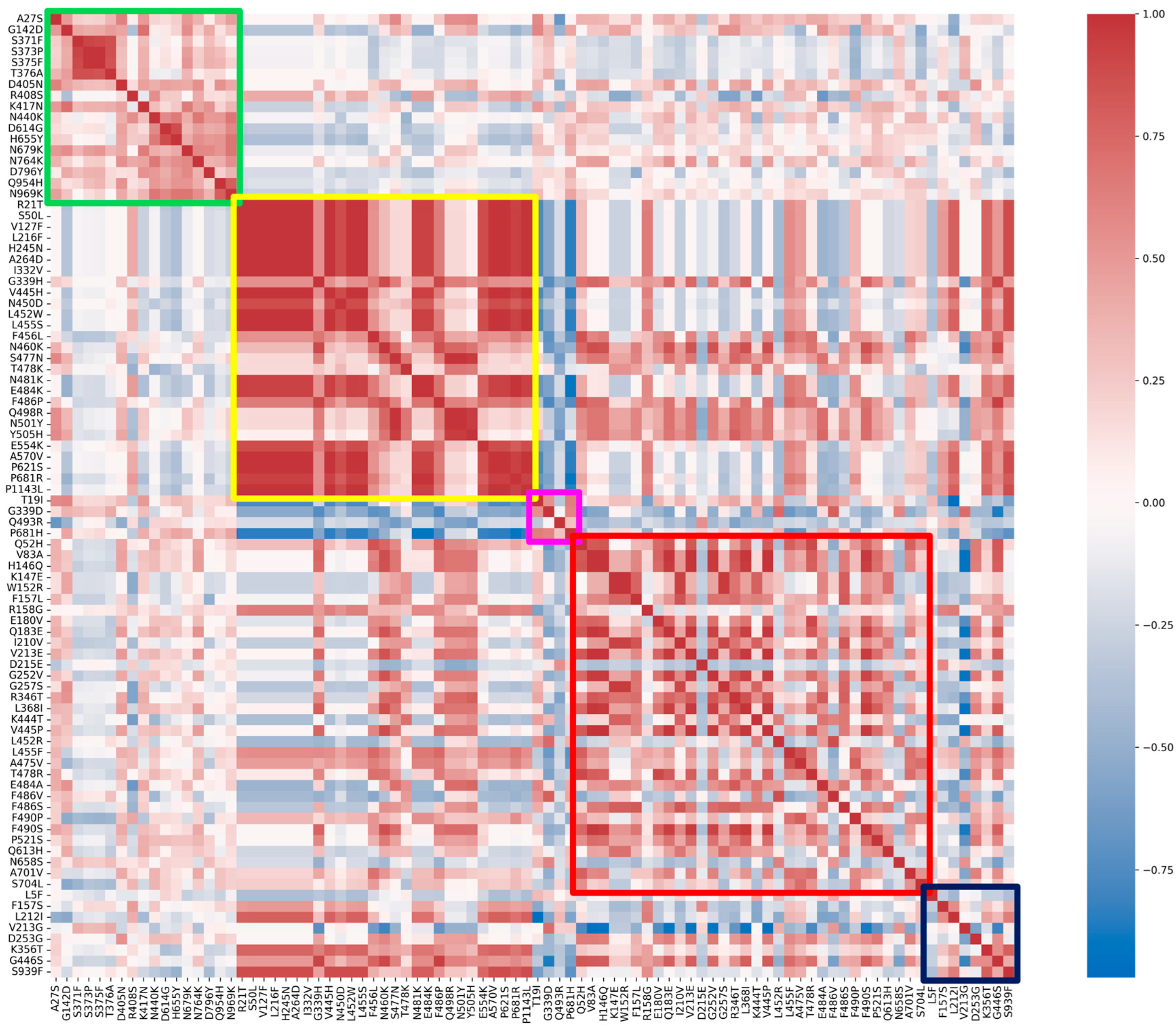
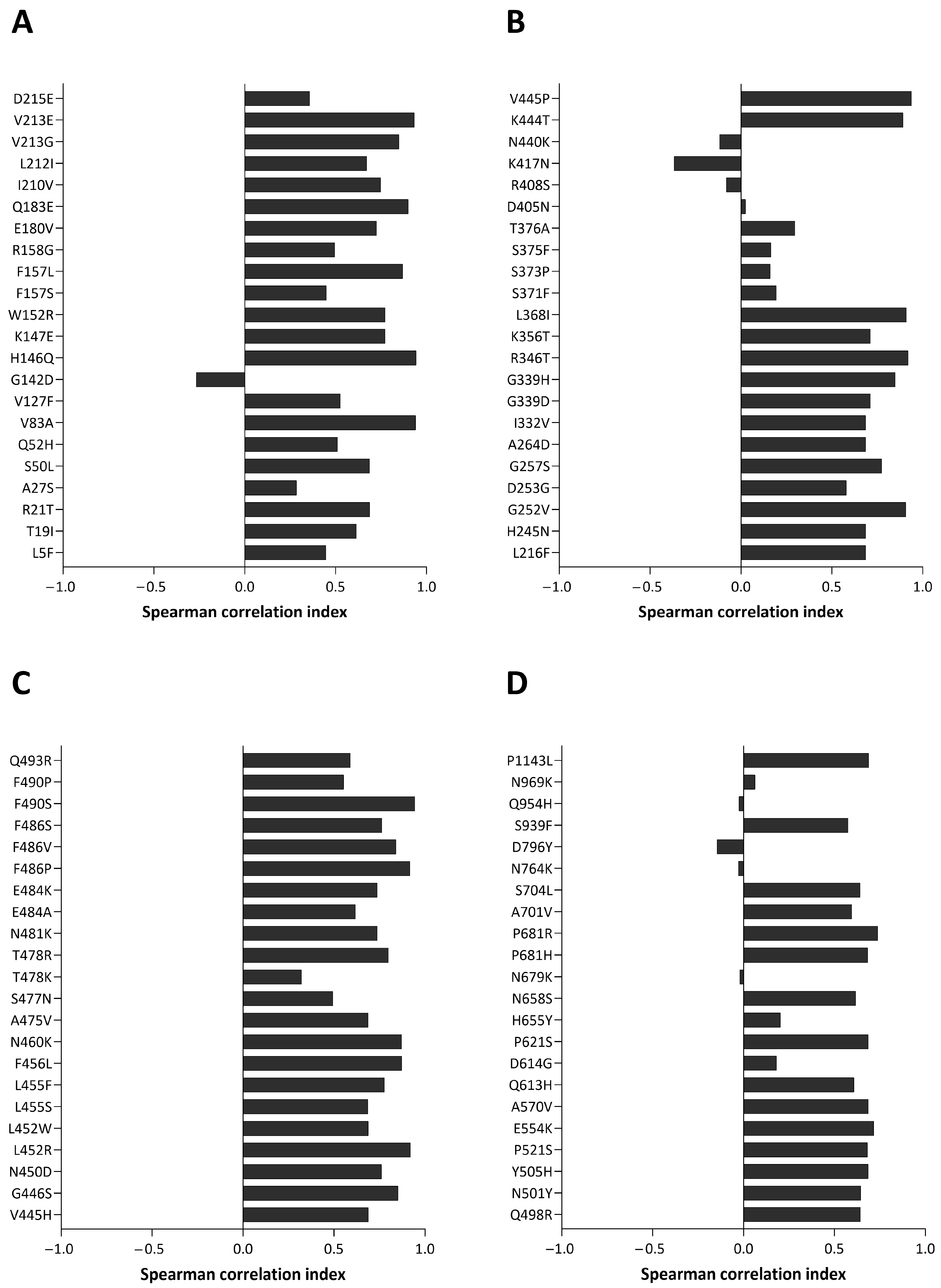
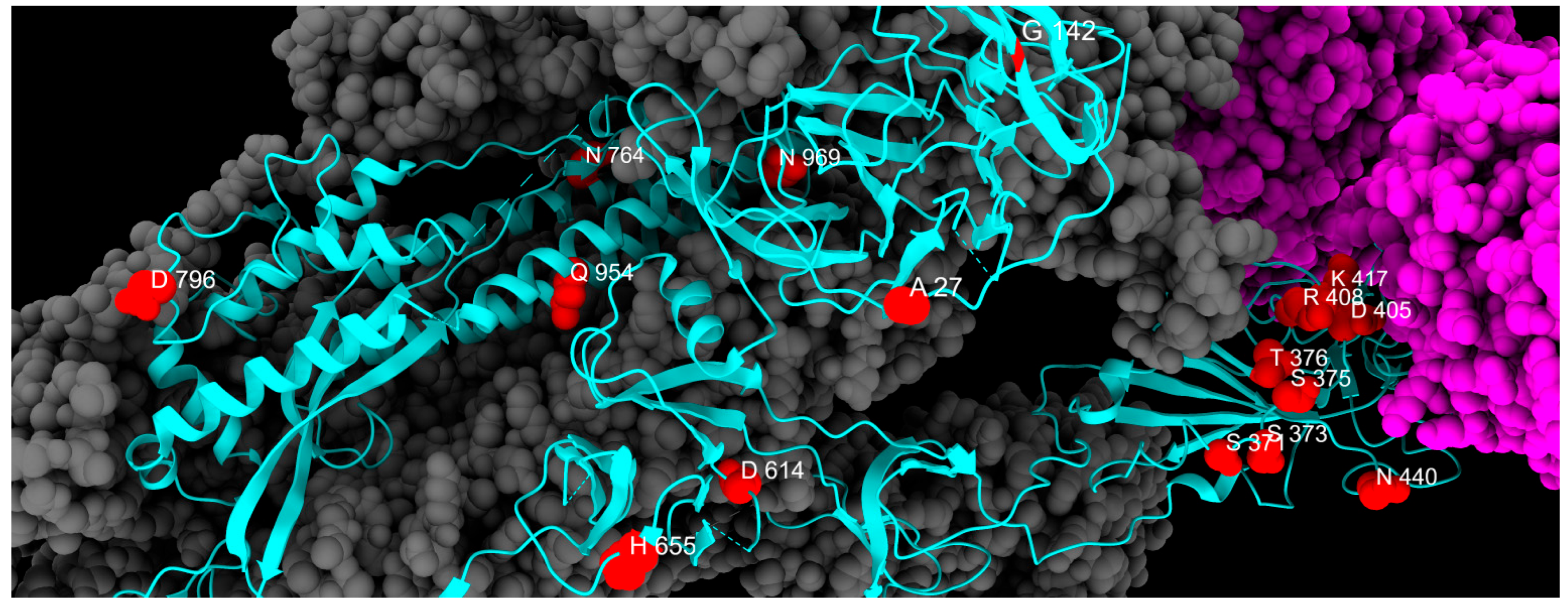



| Fixed Mutations | Wild Type Polarity | Mutated Polarity | Domain | Involvement | First > 90% Prevalence Date |
|---|---|---|---|---|---|
| A27S | Hydrophobic | Polar | NTD | Immune evasion | April 2022 |
| G142D | Glycine | Negative | NTD | Immune evasion and increased viral loads | April 2022 |
| S371F | Polar | Hydrophobic | RBD | Cell entry | April 2022 |
| S373P | Polar | Proline | RBD | Increased ACE2 binding | February 2022 |
| S375F | Polar | Hydrophobic | RBD | Reduced pathogenicity | February 2022 |
| T376A | Polar | Hydrophobic | RBD | Reduced pathogenicity | April 2022 |
| D405N | Negative | Polar | RBD | Immune evasion | April 2022 |
| R408S | Positive | Polar | RBD | Immune evasion and increased ACE2 binding | April 2022 |
| K417N | Positive | Polar | RBD | Immune evasion and reduced ACE2 binding | March 2022 |
| N440K | Polar | Positive | RBD | Immune evasion and reduced ACE2 binding | September 2022 |
| D614G | Negative | Glycine | CTD2 | Increased ACE2 binding and increased replication | April 2020 |
| H655Y | Positive | Hydrophobic | CTD2 | Increased endosomal entry | January 2022 |
| N679K | Polar | Positive | CTD2 | Possible increased furin-mediated cleavage | January 2022 |
| N764K | Polar | Positive | S2 cleavage site | Possible new cleavage site introduction | January 2022 |
| D796Y | Negative | Hydrophobic | S2 cleavage site | Immune evasion | January 2022 |
| Q954H | Polar | Positive | HR1 | Immune evasion | January 2022 |
| N969K | Polar | Positive | HR1 | Increased endosomal entry | January 2022 |
| Emerging Mutations | Wild Type Polarity | Mutated Polarity | Domain | Involvement |
|---|---|---|---|---|
| R21T | Positive | Polar | NTD | Unknown |
| S50L | Polar | Hydrophobic | NTD | Increased lung cell entry |
| V127F | Hydrophobic | Hydrophobic | NTD | Immune evasion |
| L216F | Hydrophobic | Hydrophobic | NTD | Immune evasion |
| H245N | Positive | Polar | NTD | Immune evasion |
| A264D | Hydrophobic | Negative | NTD | Immune evasion |
| I332V | Hydrophobic | Hydrophobic | RBD | Immune evasion |
| G339H | Glycine | Positive | RBD | Immune evasion |
| V445H | Hydrophobic | Positive | RBD | Immune evasion |
| N450D | Polar | Negative | RBD | Immune evasion |
| L452W | Hydrophobic | Hydrophobic | RBD | Immune evasion |
| L455S | Hydrophobic | Polar | RBD | Immune evasion and reduced ACE2 binding |
| F456L | Hydrophobic | Hydrophobic | RBD | Increased ACE2 binding with L455F |
| N460K | Polar | Positive | RBD | Immune evasion |
| S477N | Polar | Polar | RBD | Immune evasion and increased ACE2 binding |
| T478K | Polar | Positive | RBD | Increased ACE2 binding |
| N481K | Polar | Positive | RBD | Immune evasion |
| E484K | Negative | Positive | RBD | Immune evasion |
| F486P | Hydrophobic | Proline | RBD | Immune evasion |
| Q498R | Polar | Positive | RBD | Increased ACE2 binding |
| N501Y | Polar | Hydrophobic | RBD | Immune evasion and increased ACE2 binding |
| Y505H | Hydrophobic | Positive | RBD | Increased ACE2 binding |
| E554K | Negative | Positive | CTD1 | Immune evasion |
| A570V | Hydrophobic | Hydrophobic | CTD1 | Trimer stability |
| P621S | Proline | Polar | CTD2 | Reduced fusogenicity |
| P681R | Proline | Positive | CTD2 | Increased pathogenicity |
| P1143L | Proline | Hydrophobic | S2-CTD | Possible increased cell entry |
| Fading Mutations | Wild Type Polarity | Mutated Polarity | Domain | Involvement |
|---|---|---|---|---|
| T19I | Polar | Hydrophobic | NTD | Reduced fusogenicity |
| G339D | Glycine | Negative | RBD | Immune evasion |
| Q439R | Polar | Positive | RBD | Increased ACE2 binding |
| P681H | Proline | Positive | CTD2 | Increased furin-mediated cleavage |
| Transient Mutations | Wild Type Polarity | Mutated Polarity | Domain | Involvement |
|---|---|---|---|---|
| Q52H | Polar | Positive | NTD | Unknown |
| V83A | Hydrophobic | Hydrophobic | NTD | Increased fusogenicity |
| H146Q | Positive | Polar | NTD | Unknown |
| K147E | Positive | Negative | NTD | Immune evasion |
| W152R | Hydrophobic | Positive | NTD | Immune evasion |
| F157L | Hydrophobic | Hydrophobic | NTD | Immune evasion |
| R158G | Positive | Glycine | NTD | Immune evasion and increased viral load |
| E180V | Negative | Hydrophobic | NTD | Increased stability |
| Q183E | Polar | Negative | NTD | Immune evasion |
| I210V | Hydrophobic | Hydrophobic | NTD | Immune evasion |
| V213E | Hydrophobic | Negative | NTD | Unknown |
| D215E | Negative | Negative | NTD | Unknown |
| G252V | Glycine | Hydrophobic | NTD | Immune evasion |
| G257S | Glycine | Polar | NTD | Immune evasion |
| R346T | Positive | Polar | RBD | Increased fusogenicity |
| L368I | Hydrophobic | Hydrophobic | RBD | Increased ACE2 binding |
| K444T | Positive | Polar | RBD | Immune evasion |
| V445P | Hydrophobic | Proline | RBD | Immune evasion |
| L452R | Hydrophobic | Positive | RBD | Immune evasion |
| L455F | Hydrophobic | Hydrophobic | RBD | Increased ACE2 binding with F456L |
| A475V | Hydrophobic | Hydrophobic | RBD | Immune evasion |
| T478R | Polar | Positive | RBD | Immune evasion |
| E484A | Negative | Hydrophobic | RBD | Immune evasion and reduced ACE2 binding |
| F486V | Hydrophobic | Hydrophobic | RBD | Immune evasion and reduced ACE2 binding |
| F486S | Hydrophobic | Polar | RBD | Immune evasion and reduced ACE2 binding |
| F490P | Hydrophobic | Proline | RBD | Immune evasion and reduced ACE2 binding |
| F490S | Hydrophobic | Polar | RBD | Immune evasion |
| P521S | Proline | Polar | RBD | Reduced viral infectivity |
| Q613H | Polar | Positive | CTD2 | Protein stabilization |
| N658S | Polar | Polar | CTD2 | Unknown |
| A701V | Hydrophobic | Hydrophobic | S2 cleavage site | Reduced ACE2 binding |
| S704L | Polar | Hydrophobic | S2 cleavage site | Reduced surface expression |
| Divergent Mutations | Wild Type Polarity | Mutated Polarity | Domain | Involvement |
|---|---|---|---|---|
| L5F | Hydrophobic | Hydrophobic | NTD | Enhanced ER targeting and protein processing |
| F157S | Hydrophobic | Hydrophobic | NTD | Increased transmissibility and reduced ACE2 binding |
| L212I | Hydrophobic | Hydrophobic | NTD | Immune evasion |
| V213G | Hydrophobic | Glycine | NTD | Immune evasion and increased infectivity |
| D253G | Negative | Glycine | NTD | Immune evasion |
| K356T | Positive | Polar | RBD | Immune evasion and increased infectivity |
| G446S | Glycine | Polar | RBD | Reduced immune evasion |
| S939F | Polar | Hydrophobic | HR1 | Immune evasion |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cecchetto, R.; Tonon, E.; Palmisano, A.; Lagni, A.; Diani, E.; Lotti, V.; Mantoan, M.; Montesarchio, L.; Palladini, F.; Turri, G.; et al. An Italian Single-Center Genomic Surveillance Study: Two-Year Analysis of SARS-CoV-2 Spike Protein Mutations. Int. J. Mol. Sci. 2025, 26, 7558. https://doi.org/10.3390/ijms26157558
Cecchetto R, Tonon E, Palmisano A, Lagni A, Diani E, Lotti V, Mantoan M, Montesarchio L, Palladini F, Turri G, et al. An Italian Single-Center Genomic Surveillance Study: Two-Year Analysis of SARS-CoV-2 Spike Protein Mutations. International Journal of Molecular Sciences. 2025; 26(15):7558. https://doi.org/10.3390/ijms26157558
Chicago/Turabian StyleCecchetto, Riccardo, Emil Tonon, Asia Palmisano, Anna Lagni, Erica Diani, Virginia Lotti, Marco Mantoan, Livio Montesarchio, Francesca Palladini, Giona Turri, and et al. 2025. "An Italian Single-Center Genomic Surveillance Study: Two-Year Analysis of SARS-CoV-2 Spike Protein Mutations" International Journal of Molecular Sciences 26, no. 15: 7558. https://doi.org/10.3390/ijms26157558
APA StyleCecchetto, R., Tonon, E., Palmisano, A., Lagni, A., Diani, E., Lotti, V., Mantoan, M., Montesarchio, L., Palladini, F., Turri, G., & Gibellini, D. (2025). An Italian Single-Center Genomic Surveillance Study: Two-Year Analysis of SARS-CoV-2 Spike Protein Mutations. International Journal of Molecular Sciences, 26(15), 7558. https://doi.org/10.3390/ijms26157558






