Make Acetylcholine Great Again! Australian Skinks Evolved Multiple Neurotoxin-Proof Nicotinic Acetylcholine Receptors in Defiance of Snake Venom
Abstract
1. Introduction
2. Results
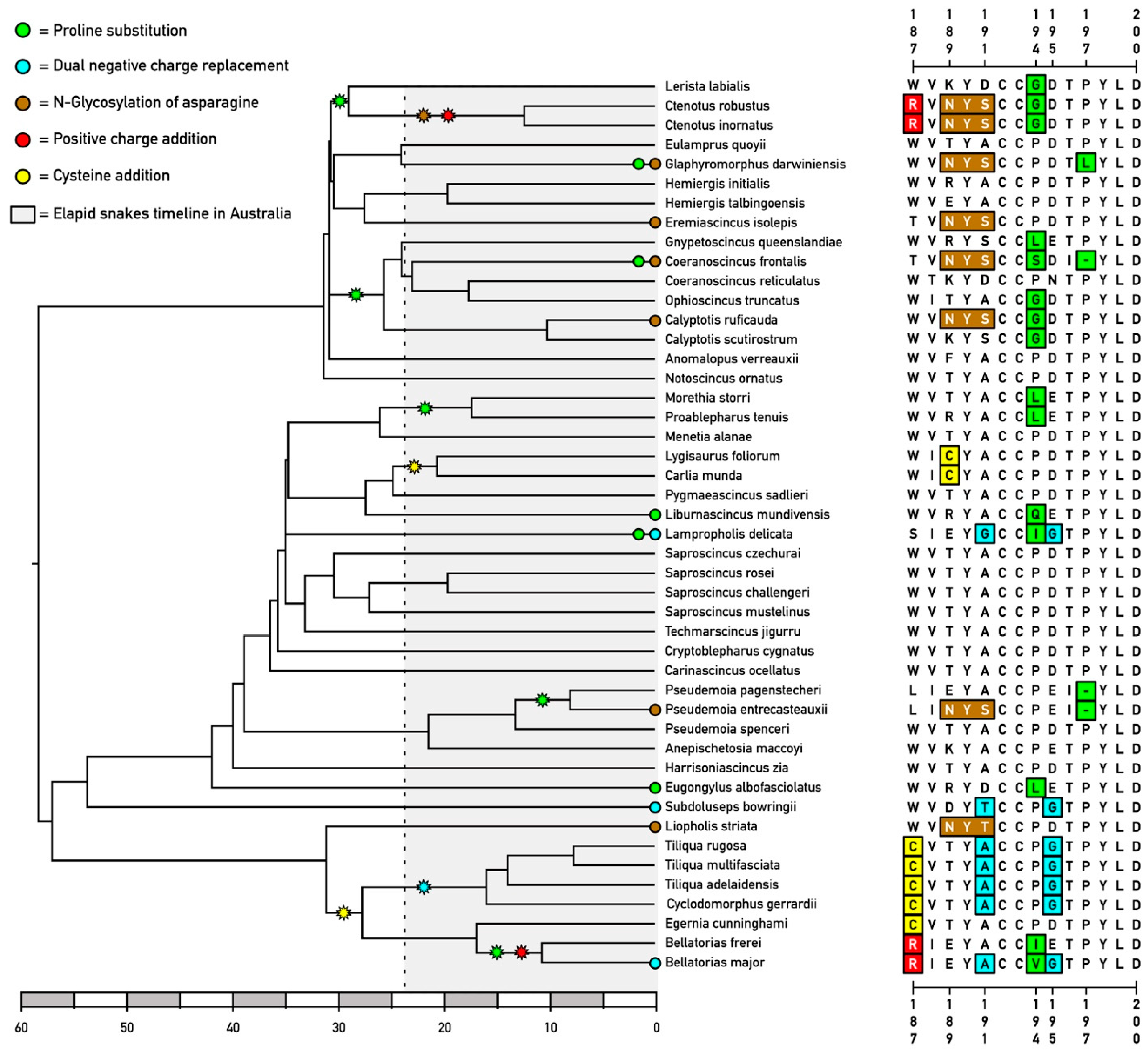
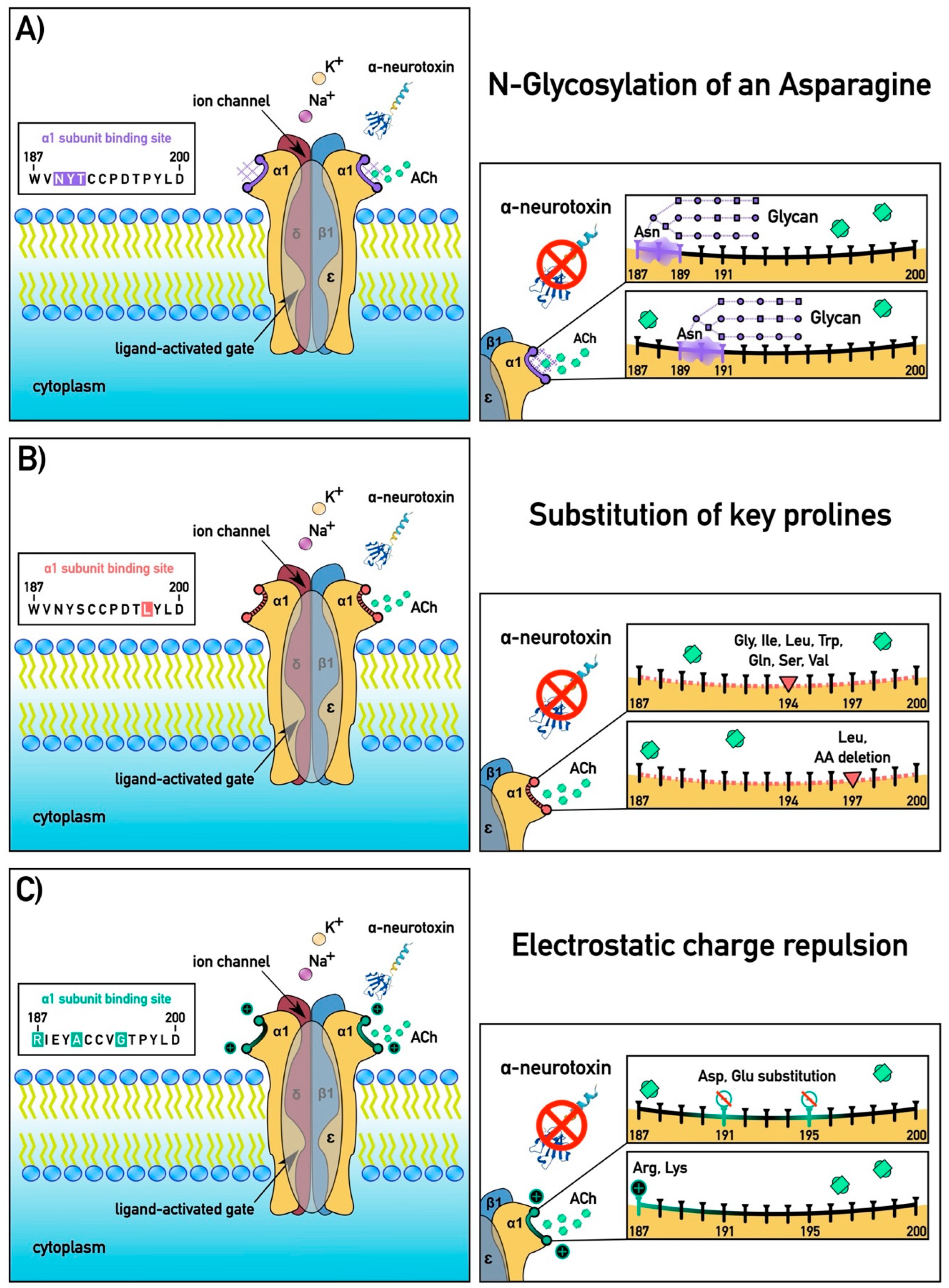
2.1. Convergent Molecular Adaptations in Skink nAChR α1 Subunits
2.2. Comparison with Other Vertebrates
2.3. Signs of Selection Analyses Reveal Complex Evolutionary History with Instances of Positive Selection
2.4. Receptor Binding Assays
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Species Selection
4.2. DNA Extraction and nAChR α1 Gene Sequencing
- Samples were amplified using following primers designed for the nAChR orthosteric using our Tiliqua rugosa genomic sequence:
- ○
- Forward 5′ TGAGTAACTTCATGGAGAGCGG 3′.
- ○
- Reverse 5′ TGTGGGCAGATAAAACACTAAGCC 3′.
- PCR reaction contents were as follows:
- ○
- 25 μL of Taq PCR master mix.
- ○
- 3 μL of each primer (10 μM).
- ○
- 500 ng of DNA.
- ○
- PCR water to adjust to the 50 μL total PCR reaction volume.
- The PCR reaction conditions were as follows:
- ○
- Initial denaturation at 95 °C for 3 min (all subsequent denaturation steps were at 95 °C for 30 s).
- ○
- Annealing was at 55 °C for 30 s.
- ○
- Extension was at 72 °C for 1 min.
- ○
- The PCR steps of denaturation, annealing, and extension were repeated for 35 cycles.
- ○
- Final extension at 72 °C for 10 min.
4.3. Identification of Putative Resistance Mutations
4.4. Data Analysis and Ancestral State Reconstruction
4.5. Receptor Binding Assays
4.5.1. Mimotope Design and Preparation
- Post-synthesis uncontrollable thiol oxidation was prevented by the synthetic peptides having a serine doublet in place of the cysteine-doublet as per previous work [83].
- Mimotopes were dissolved in 100% dimethyl sulfoxide (DMSO) followed by 1:10 dilution with double-deionised water in order to make 50 µg/mL working stocks.
- All prepared mimotope stock solutions were stored at −20 °C for future use.
4.5.2. Biolayer Interferometry Assay (BLI)
- A validated Octet HTX biolayer interferometry assay was used to measure neurotoxin-receptor binding affinities as per previously published protocols, as were data acquisition, processing, and statistical analyses [2,26,32,33,34,35,44,46,52,54,88]. It must be noted that the mimotopes only approximate the orthosteric site and cannot capture full conformational or functional receptor dynamics. Acanthophis wellsi venom (received under UQ Animal Ethics Approval 15 March 2021/AE000075) was used, as it has been previously shown to be a model species for potent broadly acting alpha-neurotoxic Australian elapid snake venom [26]. Blank sensors were used as negative controls.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekara, U.; Harris, R.J.; Fry, B.G. The target selects the toxin: Specific amino acids in snake-prey nicotinic acetylcholine receptors that are selectively bound by king cobra venoms. Toxins 2022, 14, 528. [Google Scholar] [CrossRef] [PubMed]
- Barchan, D.; Kachalsky, S.; Neumann, D.; Vogel, Z.; Ovadia, M.; Kochva, E.; Fuchs, S. How the mongoose can fight the snake—The binding-site of the mongoose acetylcholine-receptor. Proc. Natl. Acad. Sci. USA 1992, 89, 7717–7721. [Google Scholar] [CrossRef] [PubMed]
- Barchan, D.; Ovadia, M.; Kochva, E.; Fuchs, S. The binding-site of the nicotinic acetylcholine-receptor in animal species resistant to alpha-bungarotoxin. Biochemistry 1995, 34, 9172–9176. [Google Scholar] [CrossRef]
- Geffeney, S.L.; Fujimoto, E.; Brodie, E.D.; Brodie, E.D.; Ruben, P.C. Evolutionary diversification of TTX-resistant sodium channels in a predator-prey interaction. Nature 2005, 434, 759–763. [Google Scholar] [CrossRef]
- Jansa, S.A.; Voss, R.S. Adaptive evolution of the venom-targeted vWF protein in opossums that eat pitvipers. PLoS ONE 2011, 6, e20997. [Google Scholar] [CrossRef]
- Tarvin, R.D.; Borghese, C.M.; Sachs, W.; Santos, J.C.; Lu, Y.; O’Connell, L.A.; Cannatella, D.C.; Harris, R.A.; Zakon, H.H. Interacting amino acid replacements allow poison frogs to evolve epibatidine resistance. Science 2017, 357, 1261–1266. [Google Scholar] [CrossRef]
- Karageorgi, M.; Groen, S.C.; Sumbul, F.; Pelaez, J.N.; Verster, K.I.; Aguilar, J.M.; Hastings, A.P.; Bernstein, S.L.; Matsunaga, T.; Astourian, M.; et al. Genome editing retraces the evolution of toxin resistance in the Monarch Butterfly. Nature 2019, 574, 409–412. [Google Scholar] [CrossRef]
- van Thiel, J.; Khan, M.A.; Wouters, R.M.; Harris, R.J.; Casewell, N.R.; Fry, B.G.; Kini, R.M.; Mackessy, S.P.; Vonk, F.J.; Wuster, W.; et al. Convergent evolution of toxin resistance in animals. Biol. Rev. Camb. Philos. Soc. 2022, 97, 1823–1843. [Google Scholar] [CrossRef]
- Solé, R. Revisiting Leigh Van Valen’s “A New Evolutionary Law” (1973). Biol. Theory 2022, 17, 120–125. [Google Scholar] [CrossRef]
- Holding, M.L.; Biardi, J.E.; Gibbs, H.L. Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proceedings. Biol. Sci. R. Soc. 2016, 283, 20152841. [Google Scholar] [CrossRef]
- Holding, M.L.; Drabeck, D.H.; Jansa, S.A.; Gibbs, H.L. Venom resistance as a model for understanding the molecular basis of complex coevolutionary adaptations. Integr. Comp. Biol. 2016, 56, 1032–1043. [Google Scholar] [CrossRef]
- Biardi, J.; Coss, R.; Smith, D. California ground squirrel (Spermophilus beecheyi) blood sera inhibits crotalid venom proteolytic activity. Toxicon 2000, 38, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wuster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. Biol. Sci. R. Soc. 2009, 276, 2443–2449. [Google Scholar] [CrossRef] [PubMed]
- Aird, S.D.; Aggarwal, S.; Villar-Briones, A.; Tin, M.M.; Terada, K.; Mikheyev, A.S. Snake venoms are integrated systems, but abundant venom proteins evolve more rapidly. BMC Genom. 2015, 16, 647. [Google Scholar] [CrossRef]
- Arbuckle, K.; Rodriguez de la Vega, R.C.; Casewell, N.R. Coevolution takes the sting out of it: Evolutionary biology and mechanisms of toxin resistance in animals. Toxicon 2017, 140, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, H.L.; Sanz, L.; Perez, A.; Ochoa, A.; Hassinger, A.T.B.; Holding, M.L.; Calvete, J.J. The molecular basis of venom resistance in a rattlesnake-squirrel predator-prey system. Mol. Ecol. 2020, 29, 2871–2888. [Google Scholar] [CrossRef]
- Holding, M.L.; Putman, B.J.; Kong, L.M.; Smith, J.E.; Clark, R.W. Physiological stress integrates resistance to rattlesnake venom and the onset of risky foraging in California ground squirrels. Toxins 2020, 12, 617. [Google Scholar] [CrossRef]
- Ochoa, A.; Hassinger, A.T.B.; Holding, M.L.; Gibbs, H.L. Genetic characterization of potential venom resistance proteins in California ground squirrels (Otospermophilus beecheyi) using transcriptome analyses. J. Exp. Zool. B Mol. Dev. Evol. 2023, 340, 259–269. [Google Scholar] [CrossRef]
- Pomento, A.M.; Perry, B.W.; Denton, R.D.; Gibbs, H.L.; Holding, M.L. No safety in the trees: Local and species-level adaptation of an arboreal squirrel to the venom of sympatric rattlesnakes. Toxicon 2016, 118, 149–155. [Google Scholar] [CrossRef]
- Robinson, K.E.; Holding, M.L.; Whitford, M.D.; Saviola, A.J.; Yates, J.R., 3rd; Clark, R.W. Phenotypic and functional variation in venom and venom resistance of two sympatric rattlesnakes and their prey. J. Evol. Biol. 2021, 34, 1447–1465. [Google Scholar] [CrossRef] [PubMed]
- Schield, D.R.; Perry, B.W.; Adams, R.H.; Holding, M.L.; Nikolakis, Z.L.; Gopalan, S.S.; Smith, C.F.; Parker, J.M.; Meik, J.M.; DeGiorgio, M.; et al. The roles of balancing selection and recombination in the evolution of rattlesnake venom. Nat. Ecol. Evol. 2022, 6, 1367–1380. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.S.; Holding, M.L.; Haynes, L.M.; Ginsburg, D. Tandem duplication of serpin genes yields functional variation and snake venom inhibitors. bioRxiv 2025, 631777. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.C.; Pichyangkul, S.; Garcia, V.E. The resistance of three species of warm-blooded animals to Western Diamondback Rattlesnake (Crotalus atrox) venom. Toxicon 1979, 17, 601–607. [Google Scholar] [CrossRef]
- Biardi, J.E.; Coss, R.G. Rock squirrel (Spermophilus variegatus) blood sera affects proteolytic and hemolytic activities of rattlesnake venoms. Toxicon 2011, 57, 323–331. [Google Scholar] [CrossRef]
- Chandrasekara, U.; Mancuso, M.; Seneci, L.; Bourke, L.; Trembath, D.F.; Sumner, J.; Zdenek, C.N.; Fry, B.G. A Russian doll of resistance: Nested gains and losses of venom immunity in varanid lizards. Int. J. Mol. Sci. 2024, 25, 2628. [Google Scholar] [CrossRef]
- Khan, M.A.; Dashevsky, D.; Kerkkamp, H.; Kordis, D.; de Bakker, M.A.G.; Wouters, R.; van Thiel, J.; Op den Brouw, B.; Vonk, F.; Kini, R.M.; et al. Widespread evolution of molecular resistance to snake venom alpha-neurotoxins in vertebrates. Toxins 2020, 12, 638. [Google Scholar] [CrossRef]
- Fry, B.G.; Wuster, W.; Kini, R.M.; Brusic, V.; Khan, A.; Venkataraman, D.; Rooney, A.P. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 2003, 57, 110–129. [Google Scholar] [CrossRef]
- Sunagar, K.; Jackson, T.N.; Undheim, E.A.; Ali, S.A.; Antunes, A.; Fry, B.G. Three-fingered RAVERs: Rapid Accumulation of Variations in Exposed Residues of snake venom toxins. Toxins 2013, 5, 2172–2208. [Google Scholar] [CrossRef]
- Utkin, Y.; Sunagar, K.; Jackson, T.N.W.; Reeks, T.; Fry, B.G. Three-Finger Toxins (3FTxs). In Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery; Fry, B.G., Ed.; Oxford University Press: New York, NY, USA, 2015; pp. 215–227. [Google Scholar]
- Barber, C.M.; Isbister, G.K.; Hodgson, W.C. Alpha neurotoxins. Toxicon 2013, 66, 47–58. [Google Scholar] [CrossRef]
- Harris, R.J.; Youngman, N.J.; Zdenek, C.N.; Huynh, T.M.; Nouwens, A.; Hodgson, W.C.; Harrich, D.; Dunstan, N.; Portes-Junior, J.A.; Fry, B.G. Assessing the binding of venoms from aquatic elapids to the nicotinic acetylcholine receptor orthosteric site of different prey models. Int. J. Mol. Sci. 2020, 21, 7377. [Google Scholar] [CrossRef]
- Harris, R.J.; Zdenek, C.N.; Debono, J.; Harrich, D.; Fry, B.G. Evolutionary interpretations of nicotinic acetylcholine receptor targeting venom effects by a clade of Asian Viperidae snakes. Neurotox. Res. 2020, 38, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.J.; Zdenek, C.N.; Harrich, D.; Frank, N.; Fry, B.G. An appetite for destruction: Detecting prey-selective binding of alpha-neurotoxins in the venom of Afro-Asian elapids. Toxins 2020, 12, 205. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.J.; Zdenek, C.N.; Nouwens, A.; Sweeney, C.; Dunstan, N.; Fry, B.G. A symmetry or asymmetry: Functional and compositional comparison of venom from the left and right glands of the Indochinese spitting cobra (Naja siamensis). Toxicon X 2020, 7, 100050. [Google Scholar] [CrossRef] [PubMed]
- Kachalsky, S.G.; Jensen, B.S.; Barchan, D.; Fuchs, S. Two subsites in the binding domain of the acetylcholine receptor: An aromatic subsite and a proline subsite. Proc. Natl. Acad. Sci. USA 1995, 92, 10801–10805. [Google Scholar] [CrossRef]
- Hart, A.J.; Isbister, G.K.; Hodgson, W.C. In vitro neurotoxic effects of Pseudechis spp. venoms: A comparison of avian and murine skeletal muscle preparations. Toxicon 2013, 63, 112–115. [Google Scholar] [CrossRef]
- Hart, A.J.; Isbister, G.K.; O’Donnell, P.; Williamson, N.A.; Hodgson, W.C. Species differences in the neuromuscular activity of post-synaptic neurotoxins from two Australian black snakes (Pseudechis porphyriacus and Pseudechis colletti). Toxicol. Lett. 2013, 219, 262–268. [Google Scholar] [CrossRef]
- Heyborne, W.H.; Mackessy, S.P. Identification and characterization of a taxon-specific three-finger toxin from the venom of the Green Vinesnake (Oxybelis fulgidus; family Colubridae). Biochimie 2013, 95, 1923–1932. [Google Scholar] [CrossRef]
- Mackessy, S.P.; Sixberry, N.A.; Heyborne, W.H.; Fritts, T. Venom of the Brown Treesnake, Boiga irregularis: Ontogenetic shifts and taxa-specific toxicity. Toxicon 2006, 47, 537–548. [Google Scholar] [CrossRef]
- Modahl, C.M.; Mrinalini; Frietze, S.; Mackessy, S.P. Adaptive evolution of distinct prey-specific toxin genes in rear-fanged snake venom. Proceedings. Biol. Sci. R. Soc. 2018, 285, 20181003. [Google Scholar] [CrossRef]
- Pawlak, J.; Mackessy, S.P.; Fry, B.G.; Bhatia, M.; Mourier, G.; Fruchart-Gaillard, C.; Servent, D.; Menez, R.; Stura, E.; Menez, A.; et al. Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (Mangrove Catsnake) with bird-specific activity. J. Biol. Chem. 2006, 281, 29030–29041. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, J.; Mackessy, S.P.; Sixberry, N.M.; Stura, E.A.; Le Du, M.H.; Menez, R.; Foo, C.S.; Menez, A.; Nirthanan, S.; Kini, R.M. Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity. FASEB J. 2009, 23, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Zdenek, C.N.; Harris, R.J.; Kuruppu, S.; Youngman, N.J.; Dobson, J.S.; Debono, J.; Khan, M.; Smith, I.; Yarski, M.; Harrich, D.; et al. A taxon-specific and high-throughput method for measuring ligand binding to nicotinic acetylcholine receptors. Toxins 2019, 11, 600. [Google Scholar] [CrossRef] [PubMed]
- Asher, O.; Jensen, B.S.; Lupu-Meiri, M.; Oron, Y.; Fuchs, S. The mongoose acetylcholine receptor α-subunit: Analysis of glycosylation and α-bungarotoxin binding. FEBS Lett. 1998, 426, 212–216. [Google Scholar] [CrossRef]
- Chandrasekara, U.; Broussard, E.M.; Rokyta, D.R.; Fry, B.G. High-voltage toxin’roll: Electrostatic charge repulsion as a dynamic venom resistance trait in pythonid snakes. Toxins 2024, 16, 176. [Google Scholar] [CrossRef]
- Chandrasekara, U.; Mancuso, M.; Sumner, J.; Edwards, D.; Zdenek, C.N.; Fry, B.G. Sugar-coated survival: N-glycosylation as a unique bearded dragon venom resistance trait within Australian agamid lizards. Comp. Biochem. Physiol. Toxicol. Pharmacol. CBP 2024, 282, 109929. [Google Scholar] [CrossRef]
- Dashevsky, D.; Harris, R.J.; Zdenek, C.N.; Benard-Valle, M.; Alagon, A.; Portes-Junior, J.A.; Tanaka-Azevedo, A.M.; Grego, K.F.; Sant’Anna, S.S.; Frank, N.; et al. Red-on-yellow queen: Bio-layer interferometry reveals functional diversity within Micrurus venoms and toxin resistance in prey species. J. Mol. Evol. 2024, 92, 317–328. [Google Scholar] [CrossRef]
- Dellisanti, C.D.; Yao, Y.; Stroud, J.C.; Wang, Z.Z.; Chen, L. Structural determinants for alpha-neurotoxin sensitivity in muscle nAChR and their implications for the gating mechanism. Channels 2007, 1, 234–237. [Google Scholar] [CrossRef]
- Drabeck, D.H.; Dean, A.M.; Jansa, S.A. Why the honey badger don’t care: Convergent evolution of venom-targeted nicotinic acetylcholine receptors in mammals that survive venomous snake bites. Toxicon 2015, 99, 68–72. [Google Scholar] [CrossRef]
- Drabeck, D.H.; Holt, J.; McGaugh, S.E. Widespread convergent evolution of alpha-neurotoxin resistance in African mammals. Biol. Lett. 2022, 18, 20220361. [Google Scholar] [CrossRef]
- Harris, R.J.; Nekaris, K.A.; Fry, B.G. Monkeying around with venom: An increased resistance to alpha-neurotoxins supports an evolutionary arms race between Afro-Asian primates and sympatric cobras. BMC Biol. 2021, 19, 253. [Google Scholar] [CrossRef]
- Harris, R.J.; Fry, B.G. Electrostatic resistance to alpha-neurotoxins conferred by charge reversal mutations in nicotinic acetylcholine receptors. Proc. Biol. Sci. R. Soc. 2021, 288, 20202703. [Google Scholar] [CrossRef]
- Jones, L.; Harris, R.J.; Fry, B.G. Not goanna get me: Mutations in the savannah monitor lizard (Varanus exanthematicus) nicotinic acetylcholine receptor confer reduced susceptibility to sympatric cobra venoms. Neurotox. Res. 2021, 39, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, M.; Zaman, S.; Maddock, S.T.; Kamei, R.G.; Salazar-Valenzuela, D.; Wilkinson, M.; Roelants, K.; Fry, B.G. Resistance Is Not Futile: Widespread convergent evolution of resistance to alpha-neurotoxic snake venoms in caecilians (Amphibia: Gymnophiona). Int. J. Mol. Sci. 2023, 24, 11353. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Teng, J.; Worrell, B.T.; Noviello, C.M.; Lee, M.; Karlin, A.; Stowell, M.H.B.; Hibbs, R.E. Structure of the native muscle-type nicotinic receptor and inhibition by snake venom toxins. Neuron 2020, 106, 952–962. [Google Scholar] [CrossRef]
- Takacs, Z.; Wilhelmsen, K.C.; Sorota, S. Snake α-neurotoxin binding site on the Egyptian Cobra (Naja haje) nicotinic acetylcholine receptor is conserved. Mol. Biol. Evol. 2001, 18, 1800–1809. [Google Scholar] [CrossRef]
- Takacs, Z.; Wilhelmsen, K.C.; Sorota, S. Cobra (Naja spp.) nicotinic acetylcholine receptor exhibits resistance to Erabu sea snake (Laticauda semifasciata) short-chain alpha-neurotoxin. J. Mol. Evol. 2004, 58, 516–526. [Google Scholar] [CrossRef]
- Gavel, Y.; von Heijne, G. Sequence differences between glycosylated and non-glycosylated Asn-X Thr/Ser acceptor sites: Implications for protein engineering. Protein Eng. 1990, 3, 433–442. [Google Scholar] [CrossRef]
- Mellquist, J.; Kasturi, L.; Spitalnik, S.; Shakin-Eshleman, S. The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 1998, 37, 6833–6837. [Google Scholar] [CrossRef]
- Ohtsubo, K.; Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Chapple, D.G.; Roll, U.; Böhm, M.; Aguilar, R.; Amey, A.P.; Austin, C.C.; Baling, M.; Barley, A.J.; Bates, M.F.; Bauer, A.M.; et al. Conservation status of the world’s skinks (Scincidae): Taxonomic and geographic patterns in extinction risk. Biol. Conserv. 2021, 257, 109101. [Google Scholar] [CrossRef]
- Wilson, S.; Swan, G. A Complete Guide to Reptiles of Australia (Seventh Edition); Reed New Holland: Sydney, Australia, 2025. [Google Scholar]
- Youngman, N.J.; Llinas, J.; Fry, B.G. Evidence for resistance to coagulotoxic effects of Australian elapid snake venoms by sympatric prey (blue tongue skinks) but not by predators (monitor lizards). Toxins 2021, 13, 590. [Google Scholar] [CrossRef] [PubMed]
- Minton, S.A.; Minton, M.R. Toxicity of some Australian snake venoms for potential prey species of reptiles and amphibians. Toxicon 1981, 19, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Wickramaratna, J.C.; Hodgson, W.C.; Alewood, P.F.; Kini, R.M.; Ho, H.; Wuster, W. Electrospray liquid chromatography/mass spectrometry fingerprinting of Acanthophis (death adder) venoms: Taxonomic and toxinological implications. Rapid Commun. Mass Spectrom. 2002, 16, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Wang, C.R.; Ellis, S.; Pukala, T.L.; Sumner, J.; Murphy, K.; Dunstan, N.; Isbister, G.K. The venom proteome of the ecologically divergent Australian elapid, Southern Death Adder Acanthophis antarcticus. Toxins 2025, 17, 352. [Google Scholar] [CrossRef]
- Lee, M.S.; Sanders, K.L.; King, B.; Palci, A. Diversification rates and phenotypic evolution in venomous snakes (Elapidae). R. Soc. Open Sci. 2016, 3, 150277. [Google Scholar] [CrossRef]
- Kumar, S.; Suleski, M.; Craig, J.M.; Kasprowicz, A.E.; Sanderford, M.; Li, M.; Stecher, G.; Hedges, S.B. TimeTree 5: An expanded resource for species divergence times. Mol. Biol. Evol. 2022, 39, msac174. [Google Scholar] [CrossRef]
- Brennan, I.G.; Chapple, D.G.; Keogh, J.S.; Donnellan, S. Evolutionary bursts drive morphological novelty in the world’s largest skinks. Curr. Biol. 2024, 34, 3905–3916. [Google Scholar] [CrossRef]
- Chapple, D.G.; Chapple, S.N.J.; Smith, S.A.; Shea, G.M.; Brennan, I.G.; Sadlier, R.A. Phylogenetic relationships in the Eugongylini (Squamata: Scincidae): Generic limits and biogeography. Aust. J. Zool. 2023, 70, 165–203. [Google Scholar] [CrossRef]
- Gardner, M.G.; Hugall, A.F.; Donnellan, S.C.; Hutchinson, M.N.; Foster, R. Molecular systematics of social skinks: Phylogeny and taxonomy of the Egernia group (Reptilia: Scincidae). Zool. J. Linn. Soc 2008, 154, 781–794. [Google Scholar] [CrossRef][Green Version]
- Haines, M.L.; Moussalli, A.; Stuart-Fox, D.; Clemann, N.; Melville, J. Phylogenetic evidence of historic mitochondrial introgression and cryptic diversity in the genus Pseudemoia (Squamata: Scincidae). Mol. Phylogenetics Evol. 2014, 81, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Oliver, P.M.; Hugall, A.F. Phylogenetic evidence for mid-Cenozoic turnover of a diverse continental biota. Nat. Ecol. Evol. 2017, 1, 1896–1902. [Google Scholar] [CrossRef] [PubMed]
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Rabosky, D.L.; Donnellan, S.C.; Talaba, A.L.; Lovette, I.J. Exceptional among-lineage variation in diversification rates during the radiation of Australia’s most diverse vertebrate clade. Proc. Biol. Sci. R. Soc. 2007, 274, 2915–2923. [Google Scholar] [CrossRef]
- Shea, G.M. Nomenclature of supra-generic units within the family Scincidae (Squamata). Zootaxa 2021, 5067, 301–351. [Google Scholar] [CrossRef]
- Skinner, A. Phylogenetic relationships and rate of early diversification of Australian Sphenomorphus group scincids (Scincoidea, Squamata). Biol. J. Linn. Soc. 2007, 92, 347–366. [Google Scholar] [CrossRef]
- Skinner, A.; Hutchinson, M.N.; Lee, M.S. Phylogeny and divergence times of Australian Sphenomorphus group skinks (Scincidae, Squamata). Mol. Phylogenetics Evol. 2013, 69, 906–918. [Google Scholar] [CrossRef]
- Torkkola, J.J.; Worthington-Wilmer, J.; Hutchinson, M.N.; Couper, P.J.; Oliver, P.M. Die on this hill? A new monotypic, microendemic and montane vertebrate genus from the Australian Wet Tropics. Zool. Scr. 2022, 51, 483–497. [Google Scholar] [CrossRef]
- Thorn, K.M.; Hutchinson, M.N.; Lee, M.S.Y.; Brown, N.J.; Camens, A.B.; Worthy, T.H. A new species of Proegernia from the Namba Formation in South Australia and the early evolution and environment of Australian egerniine skinks. R. Soc. Open Sci. 2021, 8, 201686. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A modular system for evolutionary analysis. Version Version 3.81. Available online: http://www.mesquiteproject.org (accessed on 2 March 2025).
- Bracci, L.; Lozzi, L.; Lelli, B.; Pini, A.; Neri, P. Mimotopes of the nicotinic receptor binding site selected by a combinatorial peptide library. Biochemistry 2001, 40, 6611–6619. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Yang, Z.H.; Wong, W.S.W.; Nielsen, R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef]
- Agnarsson, I.; Miller, J.A. Is ACCTRAN better than DELTRAN? Cladistics 2008, 24, 1032–1038. [Google Scholar] [CrossRef]
- Farris, J.S. Methods for computing Wagner trees. Syst. Biol. 1970, 19, 83–92. [Google Scholar] [CrossRef]
- Harris, R.J.; Youngman, N.J.; Chan, W.; Bosmans, F.; Cheney, K.L.; Fry, B.G. Getting stoned: Characterisation of the coagulotoxic and neurotoxic effects of reef stonefish (Synanceia verrucosa) venom. Toxicol. Lett. 2021, 346, 16–22. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chandrasekara, U.; Mancuso, M.; Shea, G.; Jones, L.; Kwiatkowski, J.; Trembath, D.; Chowdhury, A.; Bertozzi, T.; Gardner, M.G.; Hoskin, C.J.; et al. Make Acetylcholine Great Again! Australian Skinks Evolved Multiple Neurotoxin-Proof Nicotinic Acetylcholine Receptors in Defiance of Snake Venom. Int. J. Mol. Sci. 2025, 26, 7510. https://doi.org/10.3390/ijms26157510
Chandrasekara U, Mancuso M, Shea G, Jones L, Kwiatkowski J, Trembath D, Chowdhury A, Bertozzi T, Gardner MG, Hoskin CJ, et al. Make Acetylcholine Great Again! Australian Skinks Evolved Multiple Neurotoxin-Proof Nicotinic Acetylcholine Receptors in Defiance of Snake Venom. International Journal of Molecular Sciences. 2025; 26(15):7510. https://doi.org/10.3390/ijms26157510
Chicago/Turabian StyleChandrasekara, Uthpala, Marco Mancuso, Glenn Shea, Lee Jones, Jacek Kwiatkowski, Dane Trembath, Abhinandan Chowdhury, Terry Bertozzi, Michael G. Gardner, Conrad J. Hoskin, and et al. 2025. "Make Acetylcholine Great Again! Australian Skinks Evolved Multiple Neurotoxin-Proof Nicotinic Acetylcholine Receptors in Defiance of Snake Venom" International Journal of Molecular Sciences 26, no. 15: 7510. https://doi.org/10.3390/ijms26157510
APA StyleChandrasekara, U., Mancuso, M., Shea, G., Jones, L., Kwiatkowski, J., Trembath, D., Chowdhury, A., Bertozzi, T., Gardner, M. G., Hoskin, C. J., Zdenek, C. N., & Fry, B. G. (2025). Make Acetylcholine Great Again! Australian Skinks Evolved Multiple Neurotoxin-Proof Nicotinic Acetylcholine Receptors in Defiance of Snake Venom. International Journal of Molecular Sciences, 26(15), 7510. https://doi.org/10.3390/ijms26157510







