Supervised Machine-Based Learning and Computational Analysis to Reveal Unique Molecular Signatures Associated with Wound Healing and Fibrotic Outcomes to Lens Injury
Abstract
1. Introduction
2. Results
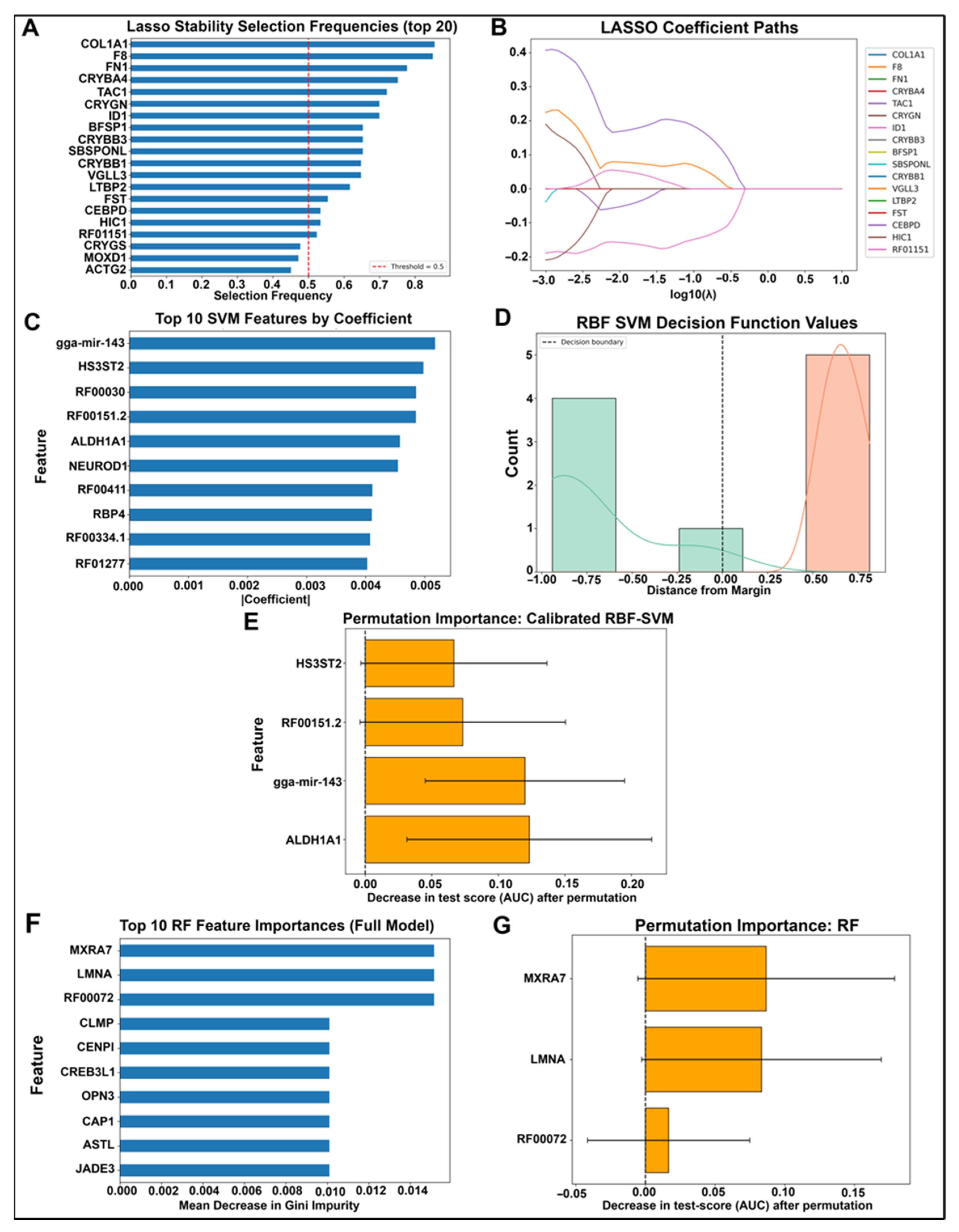
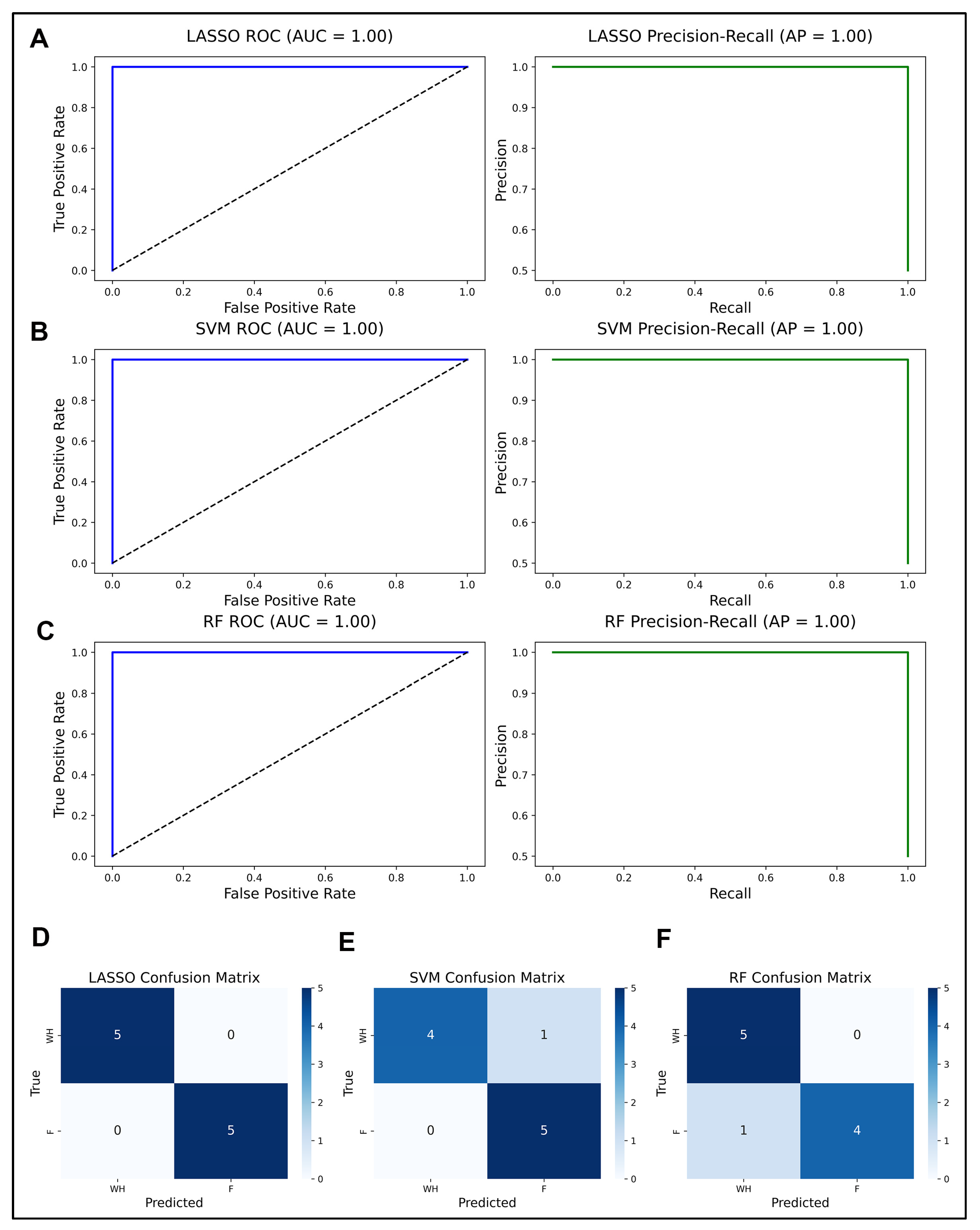
2.1. Model Training and Validation Strategy for Identifying Unique Molecular Signatures Associated with Wound Healing vs. Fibrosis
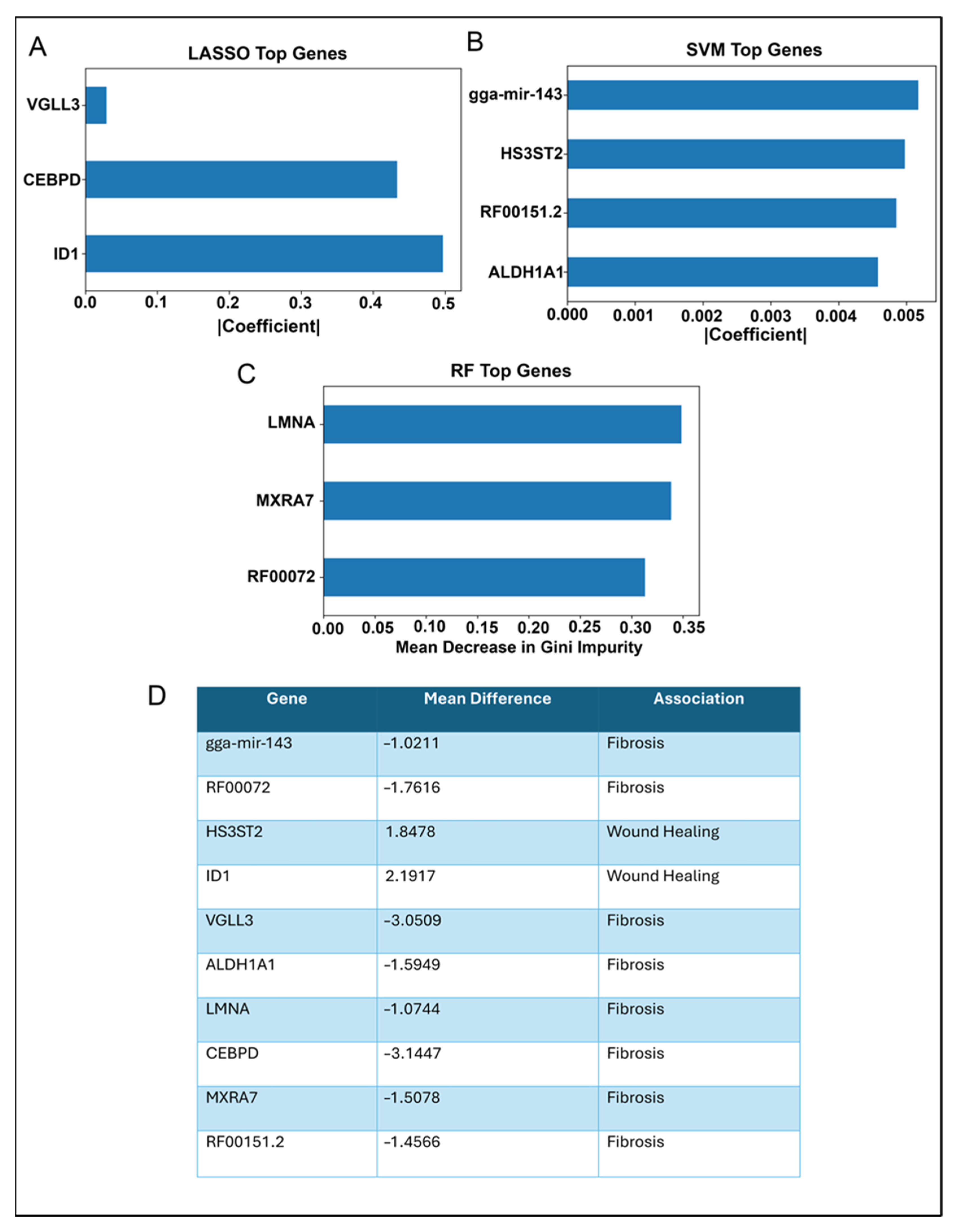
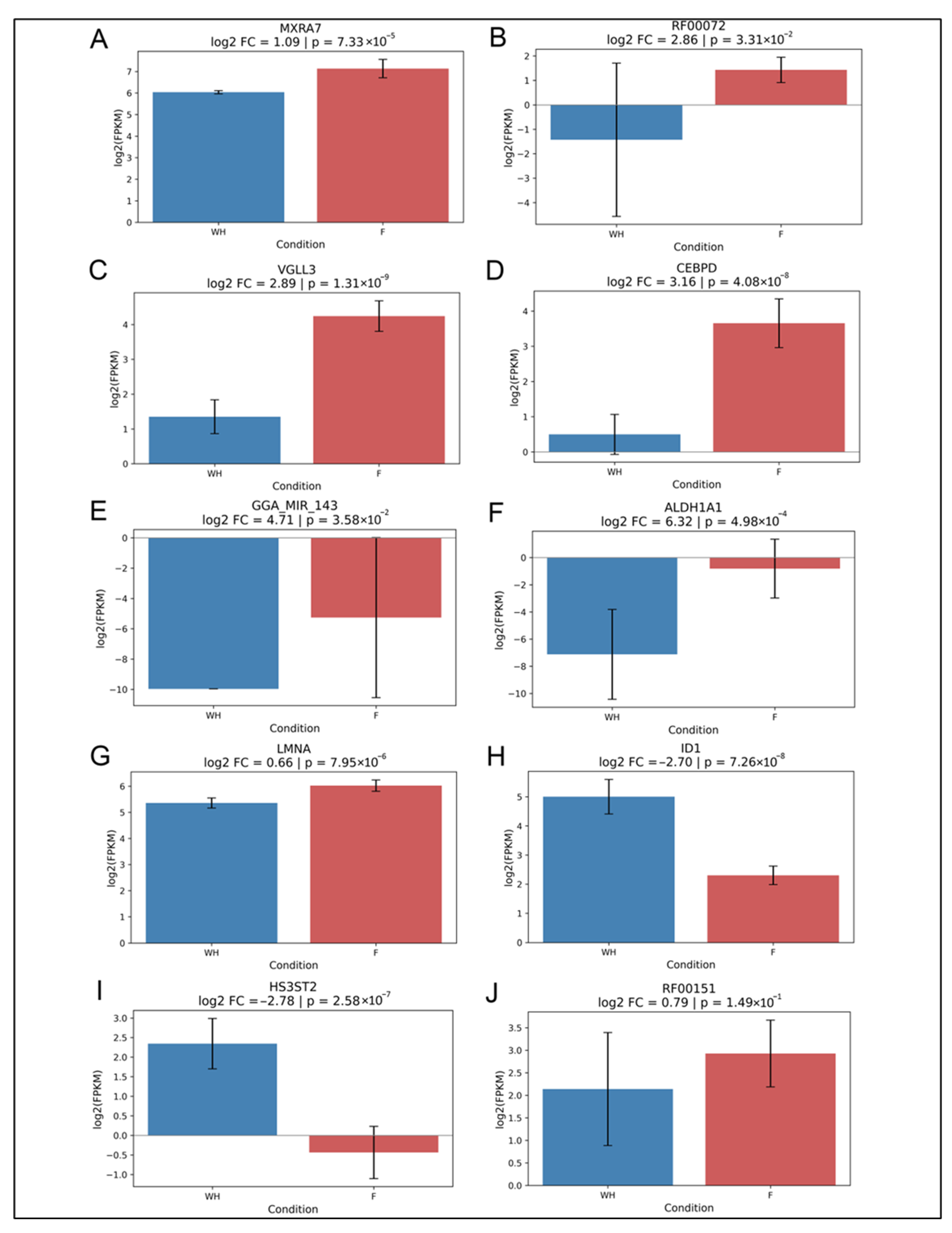
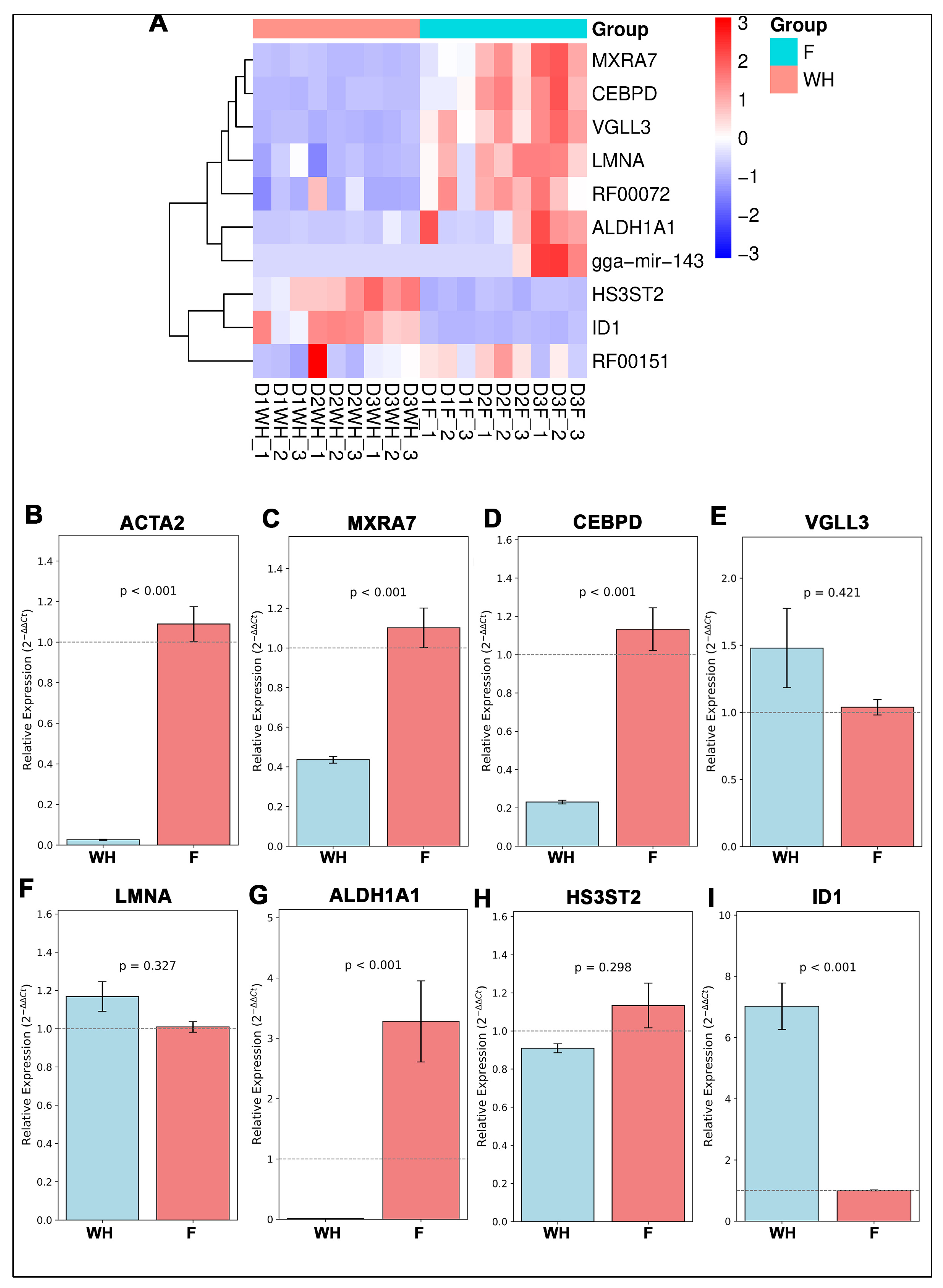
2.2. Validation of Potential Gene Candidates Associated with Wound Healing vs. Fibrosis
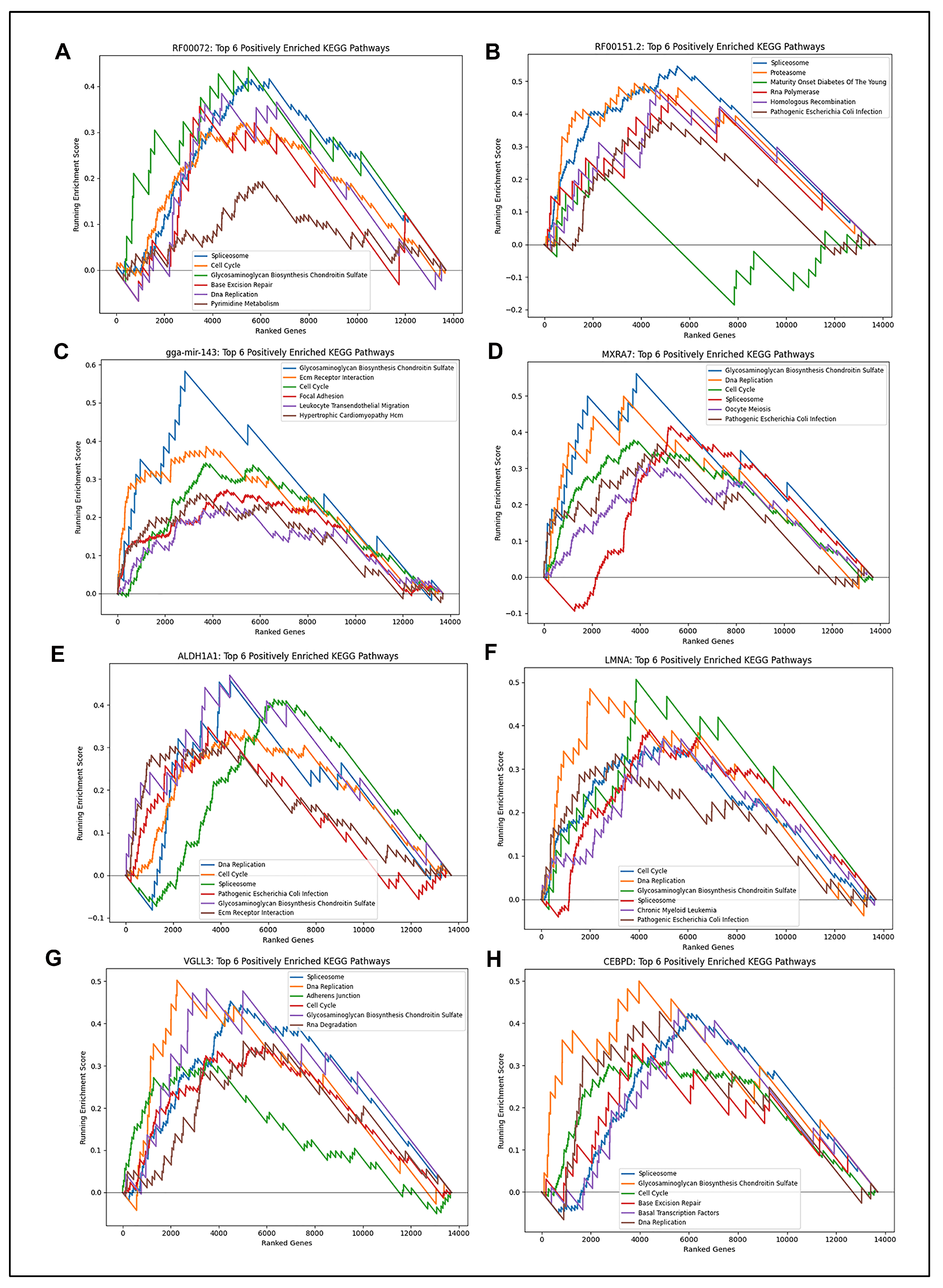
2.3. GSEA KEGG Pathway Analysis Reveals Potential Biological Function Associated with Candidate Biomarkers
3. Discussion
Limitations
4. Materials and Methods
4.1. Animals
4.2. Preparation, Imaging, and Treatment of Ex Vivo Wounded Lens Epithelial Explants
4.3. RNA Sequencing and Bioinformatics
4.4. Model Training and Validation
4.5. GSEA Analysis
4.6. RT-PCR Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PCO | Posterior Capsule Opacification |
| ML | Machine Learning |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| SVM | Support Vector Machine |
| RF | Random Forest |
| ECM | Extracellular Matrix |
| SVM-RFE | Support Vector Machine Recursive Feature Elimination |
| PCA | Principal Component Analysis |
| WH | Wound Healing |
| F | Fibrosis |
| GSEA | Gene Set Enrichment Analysis |
References
- Zhao, X.; Chen, J.; Sun, H.; Zhang, Y.; Zou, D. New insights into fibrosis from the ECM degradation perspective: The macrophage-MMP-ECM interaction. Cell Biosci. 2022, 12, 117. [Google Scholar]
- Mayorca-Guiliani, A.; Leeming, D.; Henriksen, K.; Mortensen, J.; Nielsen, S.; Anstee, Q.; Sanyal, A.; Karsdal, M.; Schuppan, D. ECM formation and degradation during fibrosis, repair, and regeneration. npj Metab. Health Dis. 2025, 3, 25. [Google Scholar] [CrossRef]
- Ghazal, R.; Wang, M.; Liu, D.; Tschumperlin, D.J.; Pereira, N.L. Cardiac Fibrosis in the Multi-Omics Era: Implications for Heart Failure. Circ. Res. 2025, 136, 773–802. [Google Scholar] [CrossRef]
- Huang, R.; Fu, P.; Ma, L. Kidney fibrosis: From mechanisms to therapeutic medicines. Signal Transduct. Target. Ther. 2023, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Liu, Y. Fibroblast activation and heterogeneity in fibrotic disease. Nat. Rev. Nephrol. 2025. [Google Scholar] [CrossRef] [PubMed]
- Cakir, S.N.; de Castro Brás, L.E. Injury-specific inflammation leads to organ-specific fibrosis. Am. J. Physiol. Heart Circ. Physiol. 2020, 319, H610–H612. [Google Scholar] [CrossRef]
- Gilgenkrantz, H.; Sayegh, R.A.; Lotersztajn, S. Immunoregulation of Liver Fibrosis: New Opportunities for Antifibrotic Therapy. Annu. Rev. Pharmacol. Toxicol. 2025, 65, 281–299. [Google Scholar] [CrossRef]
- Eldred, J.A.; Dawes, L.J.; Wormstone, I.M. The lens as a model for fibrotic disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 1301–1319. [Google Scholar] [CrossRef] [PubMed]
- Shinde, A.V.; Humeres, C.; Frangogiannis, N.G. The role of α-smooth muscle actin in fibroblast-mediated matrix contraction and remodeling. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 298–309. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123 Pt 24, 4195–4200. [Google Scholar] [CrossRef]
- Wynn, T.A. Cellular and molecular mechanisms of fibrosis. J. Pathol. 2008, 214, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.X.; Sun, L.; Zhang, X.J.; Chen, J.L.; Zhou, Y.; Ji, X.M.; Meng, P.P.; Wu, T.; Wang, X.B.; Hou, Y.X. Machine learning-based models for advanced fibrosis in non-alcoholic steatohepatitis patients: A cohort study. World J. Gastroenterol. 2025, 31, 101383. [Google Scholar] [CrossRef]
- Bai, X.; Pu, C.; Zhen, W.; Huang, Y.; Zhang, Q.; Li, Z.; Zhang, Y.; Xu, R.; Yao, Z.; Wu, W.; et al. Identifying liver cirrhosis in patients with chronic hepatitis B: An interpretable machine learning algorithm based on LSM. Ann. Med. 2025, 57, 2477294. [Google Scholar] [CrossRef]
- Wu, L.; Liu, Y.; Zhang, Y.; Xu, R.; Bi, K.; Li, J.; Wang, J.; Liu, Y.; Guo, W.; Wang, Q.; et al. Identification of PANoptosis-related genes for idiopathic pulmonary fibrosis by machine learning and molecular subtype analysis. Sci. Rep. 2024, 14, 24068. [Google Scholar] [CrossRef]
- Li, X.; Huang, Y.; Ye, N.; He, J. Analysis of immune-related genes in idiopathic pulmonary fibrosis based on bioinformatics and experimental verification. Ann. Palliat. Med. 2021, 10, 11598–11614. [Google Scholar] [CrossRef]
- Wang, S.; Lv, T.; Chen, Q.; Yang, Y.; Xu, L.; Zhang, X.; Wang, E.; Hu, X.; Liu, Y. Transcriptome sequencing and lncRNA-miRNA-mRNA network construction in cardiac fibrosis and heart failure. Bioengineered 2022, 13, 7118–7133. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Lv, G.; Liu, X.; Xiao, Y.; Tan, Y.; Zhu, Y. Machine learning selection of basement membrane-associated genes and development of a predictive model for kidney fibrosis. Sci. Rep. 2025, 15, 6567. [Google Scholar] [CrossRef]
- Walker, J.L.; Bleaken, B.M.; Wolff, I.M.; Menko, A.S. Establishment of a Clinically Relevant Ex Vivo Mock Cataract Surgery Model for Investigating Epithelial Wound Repair in a Native Microenvironment. J. Vis. Exp. 2015, 5, e52886. [Google Scholar]
- Konopińska, J.; Młynarczyk, M.; Dmuchowska, D.A.; Obuchowska, I. Posterior capsule opacification: A review of experimental studies. J. Clin. Med. 2021, 10, 2847. [Google Scholar] [CrossRef]
- O’Neill, L.M.; Wang, Y.; Duncan, M.K. Modeling cataract surgery in mice. J. Vis. Exp. 2023, 202, e66050. [Google Scholar] [CrossRef] [PubMed]
- Wormstone, I.M.; Eldred, J.A. Experimental models for posterior capsule opacification research. Exp. Eye Res. 2016, 142, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Shirai, K.; Tanaka, S.I.; Lovicu, F.J.; Saika, S. The murine lens: A model to investigate in vivo epithelial–mesenchymal transition. Dev. Dyn. 2018, 247, 340–345. [Google Scholar] [CrossRef]
- Basta, M.D.; Paulson, H.; Walker, J.L. The local wound environment is a key determinant of the outcome of TGFβ signaling on the fibrotic response of CD44+ leader cells in an ex vivo post-cataract-surgery model. Exp. Eye Res. 2021, 213, 108829. [Google Scholar] [CrossRef]
- Bleaken, B.M.; Menko, A.S.; Walker, J.L. Cells activated for wound repair have the potential to direct collective invasion of an epithelium. Mol. Biol. Cell. 2016, 27, 451–465. [Google Scholar] [CrossRef]
- Moon, A.F.; Xu, Y.; Woody, S.M.; Krahn, J.M.; Linhardt, R.J.; Liu, J.; Pedersen, L.C. Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin. Proc. Natl. Acad. Sci. USA 2012, 109, 5265–5270. [Google Scholar] [CrossRef]
- Barbosa, G.O.; Biancardi, M.F.; Carvalho, H.F. Heparan sulfate fine-tunes stromal-epithelial communication in the prostate gland. Dev. Dyn. 2021, 250, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Moon, A.F.; Xu, Y.; Woody, S.M.; Krahn, J.M.; Linhardt, R.J.; Liu, J.; Pedersen, L.C. 3-O-sulfated heparan sulfate binds FGFR2b to enhance FGF10/FGFR2b signaling progenitor proliferation. Proc. Natl. Acad. Sci. USA 2014, 111, 5265–5270. [Google Scholar]
- Higginson, J.R.; Thompson, S.M.; Santos-Silva, A.; Guimond, S.E.; Turnbull, J.E.; Barnett, S.C. Differential sulfation remodelling of heparan sulfate by extracellular 6-O-sulfatases regulates fibroblast growth factor-induced boundary formation by glial cells: Implications for glial cell transplantation. J. Neurosci. 2012, 32, 15902–15912. [Google Scholar] [CrossRef]
- Patel, V.N.; Lombaert, I.M.; Cowherd, S.N.; Shworak, N.W.; Xu, Y.; Liu, J.; Hoffman, M.P. Hs3st3-modified heparan sulfate controls KIT+ progenitor expansion by regulating 3-O-sulfotransferases. Dev. Cell 2014, 29, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.N.; Aure, M.H.; Choi, S.H.; Ball, J.R.; Lane, E.D.; Wang, Z.; Xu, Y.; Zheng, C.; Liu, X.; Martin, D.; et al. Specific 3-O-sulfated heparan sulfate domains regulate salivary gland basement membrane metabolism and epithelial differentiation. Nat. Commun. 2024, 15, 7584. [Google Scholar] [CrossRef]
- Thein, T.; de Melo, J.; Zibetti, C.; Clark, B.S.; Juarez, F.; Blackshaw, S. Control of lens development by Lhx2-regulated neuroretinal FGFs. Development 2016, 143, 3994–4002. [Google Scholar] [CrossRef]
- Lovicu, F.J.; Shin, E.H.; McAvoy, J.W. Fibrosis in the lens. Sprouty regulation of TGFβ-signaling prevents lens EMT leading to cataract. Exp. Eye Res. 2016, 142, 92–101. [Google Scholar] [CrossRef]
- Valcourt, U.; Kowanetz, M.; Niimi, H.; Heldin, C.H.; Moustakas, A. TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol. Biol. Cell. 2005, 16, 1987–2002. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Huang, S.S.; Huang, J.S. Cellular heparan sulfate negatively modulates transforming growth factor-beta1 (TGF-beta1) responsiveness in epithelial cells. J. Biol. Chem. 2006, 281, 11506–11514. [Google Scholar] [CrossRef] [PubMed]
- Ferreras, L.; Moles, A.; Situmorang, G.R.; El Masri, R.; Wilson, I.L.; Cooke, K.; Thompson, E.; Kusche-Gullberg, M.; Vivès, R.R.; Sheerin, N.S.; et al. Heparan sulfate in chronic kidney diseases: Exploring the role of 3-O-sulfation. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 839–848. [Google Scholar] [CrossRef]
- Shu, D.Y.; Wojciechowski, M.C.; Lovicu, F.J. Bone Morphogenetic Protein-7 Suppresses TGFβ2-Induced Epithelial-Mesenchymal Transition in the Lens: Implications for Cataract Prevention. Investig. Ophthalmol. Vis. Sci. 2017, 58, 781–796. [Google Scholar] [CrossRef]
- Mody, A.A.; Wordinger, R.J.; Clark, A.F. Role of ID Proteins in BMP4 Inhibition of Profibrotic Effects of TGF-β2 in Human TM Cells. Investig. Ophthalmol. Vis. Sci. 2017, 58, 849–859. [Google Scholar] [CrossRef]
- Balamurugan, K.; Sterneck, E. The many faces of C/EBPδ and their relevance for inflammation and cancer. Int. J. Biol. Sci. 2013, 9, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, J.; Deng, Y.; Yang, Q.; Wei, Q.; Ye, D.; Rong, X.; Guo, J. CCAAT/Enhancer-Binding Proteins in Fibrosis: Complex Roles Beyond Conventional Understanding. Research 2022, 2022, 9891689. [Google Scholar] [CrossRef]
- Chang, S.S.; Cheng, C.C.; Chen, Y.R.; Chen, F.W.; Cheng, Y.M.; Wang, J.M. Epithelial CEBPD activates fibronectin and enhances macrophage adhesion in renal ischemia-reperfusion injury. Cell Death Discov. 2024, 10, 328. [Google Scholar] [CrossRef]
- Li, T.; Lin, S.; Zhu, Y.; Ye, D.; Rong, X.; Wang, L. Basic biology and roles of CEBPD in cardiovascular disease. Cell Death Discov. 2025, 11, 102. [Google Scholar] [CrossRef]
- Hori, N.; Okada, K.; Takakura, Y.; Takano, H.; Yamaguchi, N.; Yamaguchi, N. Vestigial-like family member 3 (VGLL3), a cofactor for TEAD transcription factors, promotes cancer cell proliferation by activating the Hippo pathway. J. Biol. Chem. 2020, 295, 8798–8807. [Google Scholar] [CrossRef]
- Horii, Y.; Matsuda, S.; Toyota, C.; Morinaga, T.; Nakaya, T.; Tsuchiya, S.; Ohmuraya, M.; Hironaka, T.; Yoshiki, R.; Kasai, K.; et al. VGLL3 is a mechanosensitive protein that promotes cardiac fibrosis through liquid-liquid phase separation. Nat. Commun. 2023, 14, 550. [Google Scholar] [CrossRef]
- Yao, Y.; Zuo, X.; Shao, F.; Yu, K.; Liang, Q. Jaceosidin attenuates the progression of hepatic fibrosis by inhibiting the VGLL3/HMGB1/TLR4 signaling pathway. Phytomedicine 2024, 128, 155502. [Google Scholar] [CrossRef]
- Falaleeva, M.; Welden, J.R.; Duncan, M.J.; Stamm, S. C/D-box snoRNAs form methylating and non-methylating ribonucleoprotein complexes: Old dogs show new tricks. Bioessays 2017, 39, 1600264. [Google Scholar] [CrossRef]
- Kiss, A.M.; Jády, B.E.; Bertrand, E.; Kiss, T. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell Biol. 2004, 24, 5797–5807. [Google Scholar] [CrossRef]
- Chauhan, W.; Sudharshan, S.J.; Kafle, S.; Zennadi, R. SnoRNAs: Exploring Their Implication in Human Diseases. Int. J. Mol. Sci. 2024, 25, 7202. [Google Scholar] [CrossRef] [PubMed]
- Galardi, S.; Fatica, A.; Bachi, A.; Scaloni, A.; Presutti, C.; Bozzoni, I. Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol. Cell Biol. 2002, 22, 6663–6668. [Google Scholar] [CrossRef] [PubMed]
- Moss, T.; Stefanovsky, V.Y. At the center of eukaryotic life. Cell 2002, 109, 545–548. [Google Scholar] [CrossRef] [PubMed]
- Catez, F.; Dalla Venezia, N.; Marcel, V.; Zorbas, C.; Lafontaine, D.L.J.; Diaz, J.J. Ribosome biogenesis: An emerging druggable pathway for cancer therapeutics. Biochem. Pharmacol. 2019, 159, 74–81. [Google Scholar] [CrossRef]
- Prakash, V.; Carson, B.B.; Feenstra, J.M.; Dass, R.A.; Sekyrova, P.; Hoshino, A.; Petersen, J.; Guo, Y.; Parks, M.M.; Kurylo, C.M.; et al. Ribosome biogenesis during cell cycle arrest fuels EMT in development and disease. Nat. Commun. 2019, 10, 2110. [Google Scholar] [CrossRef] [PubMed]
- Herrera, J.; Henke, C.A.; Bitterman, P.B. Extracellular matrix as a driver of progressive fibrosis. J. Clin. Investig. 2018, 128, 45–53. [Google Scholar] [CrossRef]
- Zahr, H.C.; Jaalouk, D.E. Exploring the Crosstalk Between LMNA and Splicing Machinery Gene Mutations in Dilated Cardiomyopathy. Front. Genet. 2018, 9, 231. [Google Scholar] [CrossRef] [PubMed]
- Swift, J.; Ivanovska, I.L.; Buxboim, A.; Harada, T.; Dingal, P.C.; Pinter, J.; Pajerowski, J.D.; Spinler, K.R.; Shin, J.W.; Tewari, M.; et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 2013, 341, 1240104. [Google Scholar] [CrossRef] [PubMed]
- Tschumperlin, D.J.; Ligresti, G.; Hilscher, M.B.; Shah, V.H. Mechanosensing and fibrosis. J. Clin. Investig. 2018, 128, 74–84. [Google Scholar] [CrossRef]
- Ba, J.; Zheng, C.; Lai, Y.; He, X.; Pan, Y.; Zhao, Y.; Xie, H.; Wu, B.; Deng, X.; Wang, N. High matrix stiffness promotes senescence of type II alveolar epithelial cells by lysosomal degradation of lamin A/C in pulmonary fibrosis. Respir. Res. 2025, 26, 128. [Google Scholar] [CrossRef]
- Shichino, S.; Ueha, S.; Hashimoto, S.; Otsuji, M.; Abe, J.; Tsukui, T.; Deshimaru, S.; Nakajima, T.; Kosugi-Kanaya, M.; Shand, F.H.; et al. Transcriptome network analysis identifies protective role of the LXR/SREBP-1c axis in murine pulmonary fibrosis. JCI Insight 2019, 4, e122163. [Google Scholar] [CrossRef]
- Liang, L.M.; Xiong, L.; Cheng, P.P.; Chen, S.J.; Feng, X.; Zhou, Y.Y.; Niu, Q.; Wang, M.; Chen, Q.; Song, L.J.; et al. Splicing factor SRSF6 mediates pleural fibrosis. JCI Insight 2021, 6, e146197. [Google Scholar] [CrossRef]
- Lu, M.Y.; Hsieh, P.L.; Chao, S.C.; Fang, C.Y.; Ohiro, Y.; Liao, Y.W.; Yu, C.C.; Chang, M.T. Targeting MetaLnc9/miR-143/FSCN1 axis inhibits oxidative stress and myofibroblast transdifferentiation in oral submucous fibrosis. J. Dent. Sci. 2024, 19, 1416–1425. [Google Scholar] [CrossRef]
- Naito, Y.; Sakamoto, N.; Oue, N.; Yashiro, M.; Sentani, K.; Yanagihara, K.; Hirakawa, K.; Yasui, W. MicroRNA-143 regulates collagen type III expression in stromal fibroblasts of scirrhous type gastric cancer. Cancer Sci. 2014, 105, 228–235. [Google Scholar] [CrossRef]
- Li, C.; Li, J.; Xue, K.; Zhang, J.; Wang, C.; Zhang, Q.; Chen, X.; Gao, C.; Yu, X.; Sun, L. MicroRNA-143-3p promotes human cardiac fibrosis via targeting sprouty3 after myocardial infarction. J. Mol. Cell Cardiol. 2019, 129, 281–292. [Google Scholar] [CrossRef]
- Shen, Y.; Ning, J.; Zhao, L.; Liu, W.; Wang, T.; Yu, J.; Wang, Y. Matrix remodeling associated 7 proteins promote cutaneous wound healing through vimentin in coordinating fibroblast functions. Inflamm. Regen. 2023, 43, 5. [Google Scholar] [CrossRef]
- Ahadome, S.D.; Abraham, D.J.; Rayapureddi, S.; Saw, V.P.; Saban, D.R.; Calder, V.L.; Norman, J.T.; Ponticos, M.; Daniels, J.T.; Dart, J.K. Aldehyde dehydrogenase inhibition blocks mucosal fibrosis in human and mouse ocular scarring. JCI Insight 2016, 1, e87001. [Google Scholar] [CrossRef]
- Lukaszuk, T.; Krawczuk, J. Importance of feature selection stability in the classifier evaluation on high-dimensional genetic data. PeerJ 2024, 12, e18405. [Google Scholar] [CrossRef]
- Kalousis, A.; Prados, J.; Hilario, M. Stability of feature selection algorithms: A study on high-dimensional spaces. Knowl. Inf. Syst. 2007, 12, 95–116. [Google Scholar] [CrossRef]
- Dong, B.; Liu, X.; Yu, S. Utilizing machine learning algorithms to identify biomarkers associated with diabetic nephropathy: A review. Medicine 2024, 103, e37235. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lalman, C.; Stabler, K.R.; Yang, Y.; Walker, J.L. Supervised Machine-Based Learning and Computational Analysis to Reveal Unique Molecular Signatures Associated with Wound Healing and Fibrotic Outcomes to Lens Injury. Int. J. Mol. Sci. 2025, 26, 7422. https://doi.org/10.3390/ijms26157422
Lalman C, Stabler KR, Yang Y, Walker JL. Supervised Machine-Based Learning and Computational Analysis to Reveal Unique Molecular Signatures Associated with Wound Healing and Fibrotic Outcomes to Lens Injury. International Journal of Molecular Sciences. 2025; 26(15):7422. https://doi.org/10.3390/ijms26157422
Chicago/Turabian StyleLalman, Catherine, Kylie R. Stabler, Yimin Yang, and Janice L. Walker. 2025. "Supervised Machine-Based Learning and Computational Analysis to Reveal Unique Molecular Signatures Associated with Wound Healing and Fibrotic Outcomes to Lens Injury" International Journal of Molecular Sciences 26, no. 15: 7422. https://doi.org/10.3390/ijms26157422
APA StyleLalman, C., Stabler, K. R., Yang, Y., & Walker, J. L. (2025). Supervised Machine-Based Learning and Computational Analysis to Reveal Unique Molecular Signatures Associated with Wound Healing and Fibrotic Outcomes to Lens Injury. International Journal of Molecular Sciences, 26(15), 7422. https://doi.org/10.3390/ijms26157422






