Microbial Lipopolysaccharide Regulates Host Development Through Insulin/IGF-1 Signaling
Abstract
1. Introduction
2. Results
2.1. Establishment of a High-Throughput Screening Platform for C. elegans Development
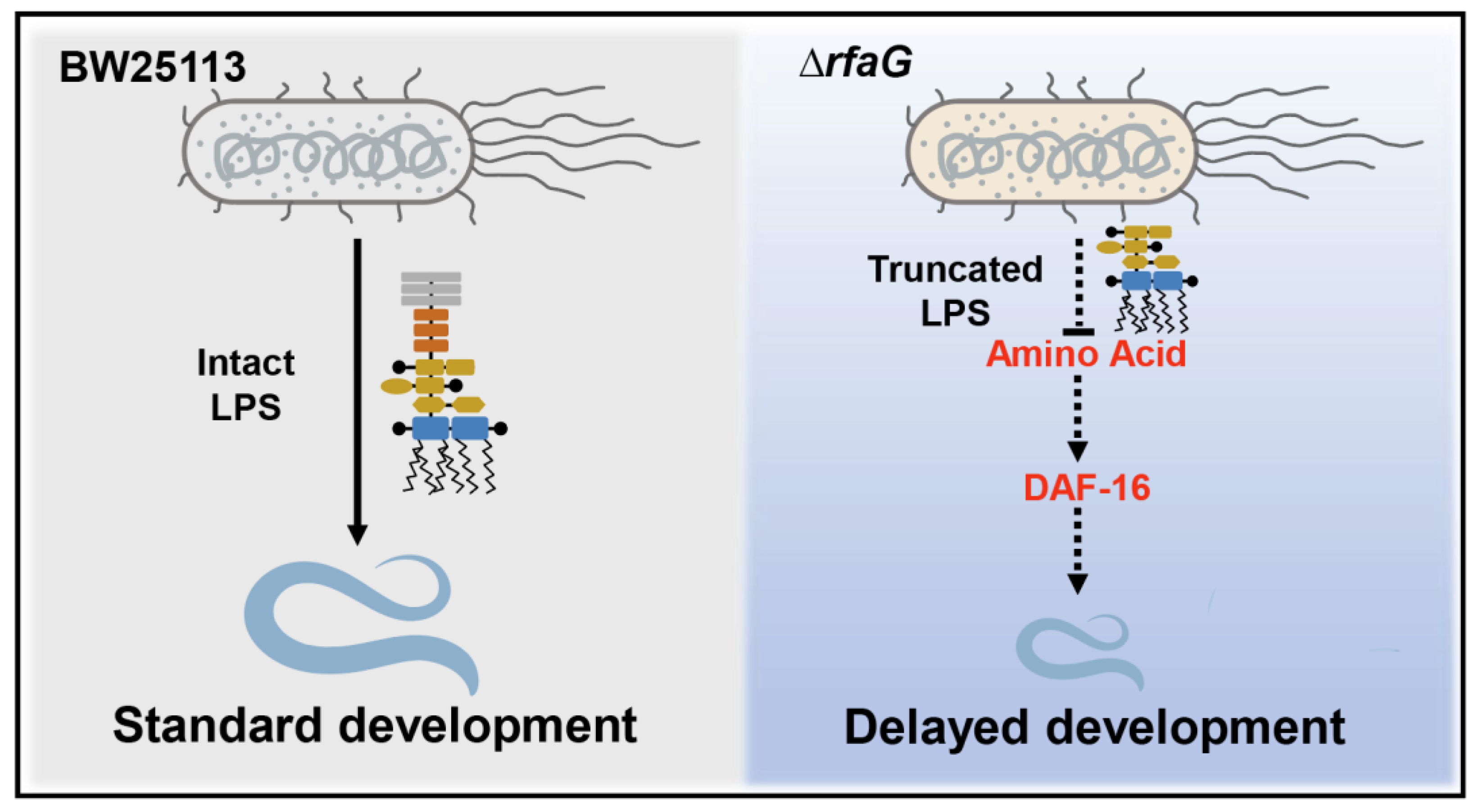
2.2. The LPS Biosynthesis Mutant ΔrfaG Impairs C. elegans Development
2.3. LPS Structure Mediates ΔrfaG-Induced Developmental Delay
2.4. The ΔrfaG Mutant Induced Developmental Defects via DAF-16
3. Discussion
4. Materials and Methods
4.1. C. elegans Strains and Maintenance
4.2. Bacterial Strains and Maintenance
4.3. Reagents
4.4. E. coli LPS Synthesis Mutants Screen in Liquid Culture System
4.5. Assessment of Bacterial Growth Rate
4.6. Assessment of Bacterial ROS Levels
4.7. Paraformaldehyde Treatment of Bacteria
4.8. Bacterial Mixing Assay
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beveridge Terry, J. Structures of Gram-Negative Cell Walls and Their Derived Membrane Vesicles. J. Bacteriol. 1999, 181, 4725–4733. [Google Scholar] [CrossRef] [PubMed]
- Caroff, M.; Karibian, D. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 2003, 338, 2431–2447. [Google Scholar] [CrossRef] [PubMed]
- Vatanen, T.; Kostic, A.D.; d’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hämäläinen, A.-M.; et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Wilksch, J.J.; Dunstan, R.A.; Mularski, A.; Wang, N.; Hocking, D.; Jebeli, L.; Cao, H.; Clements, A.; Jenney, A.W.J.; et al. LPS O Antigen Plays a Key Role in Klebsiella pneumoniae Capsule Retention. Microbiol. Spectr. 2022, 10, e0151721. [Google Scholar] [CrossRef]
- McGarry, N.; Roe, D.; Smith, S.G.J. Synergy between Group 2 capsules and lipopolysaccharide underpins serum resistance in extra-intestinal pathogenic Escherichia coli. Microbiology 2024, 170. [Google Scholar] [CrossRef]
- Kumar, A.; Mallik, D.; Pal, S.; Mallick, S.; Sarkar, S.; Chanda, A.; Ghosh, A.S. Escherichia coli O8-antigen enhances biofilm formation under agitated conditions. FEMS Microbiol. Lett. 2015, 362, fnv112. [Google Scholar] [CrossRef]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.S.; Lee, H.; Lee, J.O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef]
- Alexander, C.; Zähringer, U. Chemical structure of lipid A—The primary immunomodulatory center of bacterial lipopolysaccharides. Trends Glycosci. Glycotechnol. 2002, 14, 69–86. [Google Scholar] [CrossRef]
- Ruhal, R.; Kataria, R. Biofilm patterns in gram-positive and gram-negative bacteria. Microbiol. Res. 2021, 251, 126829. [Google Scholar] [CrossRef] [PubMed]
- Mai-Prochnow, A.; Clauson, M.; Hong, J.; Murphy, A.B. Gram positive and Gram negative bacteria differ in their sensitivity to cold plasma. Sci. Rep. 2016, 6, 38610. [Google Scholar] [CrossRef]
- Varghese, M.; Balachandran, M. Antibacterial efficiency of carbon dots against Gram-positive and Gram-negative bacteria: A review. J. Environ. Chem. Eng. 2021, 9, 106821. [Google Scholar] [CrossRef]
- Yethon Jeremy, A.; Vinogradov, E.; Perry Malcolm, B.; Whitfield, C. Mutation of the Lipopolysaccharide Core Glycosyltransferase Encoded by waaG Destabilizes the Outer Membrane of Escherichia coli by Interfering with Core Phosphorylation. J. Bacteriol. 2000, 182, 5620–5623. [Google Scholar] [CrossRef]
- Barua, S.; Yamashino, T.; Hasegawa, T.; Yokoyama, K.; Torii, K.; Ohta, M. Involvement of surface polysaccharides in the organic acid resistance of Shiga Toxin-producing Escherichia coli O157:H7. Mol. Microbiol. 2002, 43, 629–640. [Google Scholar] [CrossRef]
- Sivamaruthi, B.S.; Prasanth, M.I.; Balamurugan, K. Alterations in Caenorhabditis elegans and Cronobacter sakazakii lipopolysaccharide during interaction. Arch. Microbiol. 2015, 197, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Kamaladevi, A.; Balamurugan, K. Lipopolysaccharide of Klebsiella pneumoniae attenuates immunity of Caenorhabditis elegans and evades by altering its supramolecular structure. RSC Adv. 2016, 6, 30070–30080. [Google Scholar] [CrossRef]
- Ma, J.; Xu, X.; Wang, R.; Yan, H.; Yao, H.; Zhang, H.; Jiang, S.; Xu, A. Lipopolysaccharide exposure induces oxidative damage in Caenorhabditis elegans: Protective effects of carnosine. BMC Pharmacol. Toxicol. 2020, 21, 85. [Google Scholar] [CrossRef]
- Zhang, X.; Li, W.; Feng, K.; Xiao, J.; Du, J.; Cao, Y.; Chen, Y. Immunomodulatory effect of pentagalloyl glucose in LPS-stimulated RAW264.7 macrophages and PAO1-induced Caenorhabditis elegans. Exp. Gerontol. 2021, 150, 111388. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, Y.; Xu, J.; Feng, J.; He, X. Propacin, a coumarinolignoid isolated from durian, inhibits the lipopolysaccharide-induced inflammatory response in macrophages through the MAPK and NF-κB pathways. Food Funct. 2020, 11, 596–605. [Google Scholar] [CrossRef]
- Aballay, A.; Drenkard, E.; Hilbun, L.R.; Ausubel, F.M. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr. Biol. 2003, 13, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-P.; Ma, X.; Liu, J.-L.; Liu, A.-L. Exploring the potential use of Caenorhabditis elegans as an animal model for evaluating chemical-induced intestinal dysfunction. Toxicol. Appl. Pharmacol. 2024, 493, 117140. [Google Scholar] [CrossRef]
- Kumar, A.; Baruah, A.; Tomioka, M.; Iino, Y.; Kalita, M.C.; Khan, M. Caenorhabditis elegans: A model to understand host-microbe interactions. Cell. Mol. Life Sci. 2019, 77, 1229–1249. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.A.; Curran, S.P. Gene-diet interactions and aging in C. elegans. Exp. Gerontol. 2016, 86, 106–112. [Google Scholar] [CrossRef]
- Park, Y.-J.; Yeon, J.; Cho, J.; Kim, D.-Y.; Bai, X.; Oh, Y.; Kim, J.; Nam, H.; Hwang, H.; Heo, W.; et al. PIEZO acts in an intestinal valve to regulate swallowing in C. elegans. Nat. Commun. 2024, 15, 10072. [Google Scholar] [CrossRef]
- Berg, M.; Stenuit, B.; Ho, J.; Wang, A.; Parke, C.; Knight, M.; Alvarez-Cohen, L.; Shapira, M. Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. ISME J. 2016, 10, 1998–2009. [Google Scholar] [CrossRef]
- Berg, M.; Zhou, X.Y.; Shapira, M. Host-specific functional significance of Caenorhabditis gut commensals. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Asan, A.; Raiders, S.A.; Priess, J.R. Morphogenesis of the C. elegans Intestine Involves Axon Guidance Genes. PLoS Genet. 2016, 12, e1005950. [Google Scholar] [CrossRef] [PubMed]
- Stroustrup, N.; Ulmschneider, B.E.; Nash, Z.M.; López-Moyado, I.F.; Apfeld, J.; Fontana, W. The Caenorhabditis elegans Lifespan Machine. Nat. Methods 2013, 10, 665–670. [Google Scholar] [CrossRef]
- Guzman, D.M.; Chakka, K.; Shi, T.; Marron, A.; Fiorito, A.E.; Rahman, N.S.; Ro, S.; Sucich, D.G.; Pierce, J.T. Transgenerational effects of alcohol on behavioral sensitivity to alcohol in Caenorhabditis elegans. PLoS ONE 2022, 17, e0271849. [Google Scholar] [CrossRef] [PubMed]
- Noma, K.; Jin, Y. Optogenetic mutagenesis in Caenorhabditis elegans. Nat. Commun. 2015, 6, 8868. [Google Scholar] [CrossRef]
- Davies, A.G.; McIntire, S.L.; Using, C. elegans to screen for targets of ethanol and behavior-altering drugs. Biol. Proced. Online 2004, 6, 113–119. [Google Scholar] [CrossRef][Green Version]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef]
- Parker, C.T.; Pradel, E.; Schnaitman, C.A. Identification and sequences of the lipopolysaccharide core biosynthetic genes rfaQ, rfaP, and rfaG of Escherichia coli K-12. J. Bacteriol. 1992, 174, 930–934. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kadam, S.K.; Rehemtulla, A.; Sanderson, K.E. Cloning of rfaG, B, I, and J genes for glycosyltransferase enzymes for synthesis of the lipopolysaccharide core of Salmonella typhimurium. J. Bacteriol. 1985, 161, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.T.; Kloser, A.W.; Schnaitman, C.A.; Stein, M.A.; Gottesman, S.; Gibson, B.W. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 1992, 174, 2525–2538. [Google Scholar] [CrossRef] [PubMed]
- Seregina, T.A.; Petrushanko, I.Y.; Shakulov, R.S.; Zaripov, P.I.; Makarov, A.A.; Mitkevich, V.A.; Mironov, A.S. The Inactivation of LPS Biosynthesis Genes in E. coli Cells Leads to Oxidative Stress. Cells 2022, 11, 2667. [Google Scholar] [CrossRef]
- Taylor, P.L.; Blakely, K.M.; de Leon, G.P.; Walker, J.R.; McArthur, F.; Evdokimova, E.; Zhang, K.; Valvano, M.A.; Wright, G.D.; Junop, M.S. Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants. J. Biol. Chem. 2008, 283, 2835–2845. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef]
- Zhou, J.; Xin-Yu, L.; Yu-Jia, L.; Ji, F.; Yong, W.; Han-Ming, S.; Lu, G.-D. Full-coverage regulations of autophagy by ROS: From induction to maturation. Autophagy 2022, 18, 1240–1255. [Google Scholar] [CrossRef]
- Cui, Q.; Wang, J.-Q.; Assaraf, Y.G.; Ren, L.; Gupta, P.; Wei, L.; Ashby, C.R.; Yang, D.-H.; Chen, Z.-S. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist. Updates 2018, 41, 1–25. [Google Scholar] [CrossRef]
- Shpilka, T.; Haynes, C.M. The mitochondrial UPR: Mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 2018, 19, 109–120. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Q.; Tian, G.; Zhou, X.; Chen, P.; Chen, B.; Shan, Z.; Qi, B. Neuronal PRDX-2-Mediated ROS Signaling Regulates Food Digestion via peripheral UPRmt Activation. Nat. Commun. 2024, 15, 10582. [Google Scholar] [CrossRef]
- Beydoun, S.; Choi, H.S.; Dela-Cruz, G.; Kruempel, J.; Huang, S.; Bazopoulou, D.; Miller, H.A.; Schaller, M.L.; Evans, C.R.; Leiser, S.F. An alternative food source for metabolism and longevity studies in Caenorhabditis elegans. Commun. Biol. 2021, 4, 258. [Google Scholar] [CrossRef]
- Feng, M.; Gao, B.; Ruiz, D.; Garcia, L.R.; Sun, Q. Bacterial vitamin B6 is required for post-embryonic development in C. elegans. Commun. Biol. 2024, 7, 367. [Google Scholar] [CrossRef]
- Fernandes de Abreu, D.A.; Caballero, A.; Fardel, P.; Stroustrup, N.; Chen, Z.; Lee, K.; Keyes, W.D.; Nash, Z.M.; López-Moyado, I.F.; Vaggi, F.; et al. An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology. PLoS Genet. 2014, 10, e1004225. [Google Scholar] [CrossRef] [PubMed]
- Zečić, A.; Braeckman, B.P. DAF-16/FoxO in Caenorhabditis elegans and Its Role in Metabolic Remodeling. Cells 2020, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.C.; Faber, E.; Bats, S.H.; Murillo, T.; Speidel, Y.; Coombs, N.; Josenhans, C. Helicobacter pylori modulates host cell responses by CagT4SS-dependent translocation of an intermediate metabolite of LPS inner core heptose biosynthesis. PLoS Pathog. 2017, 13, e1006514. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
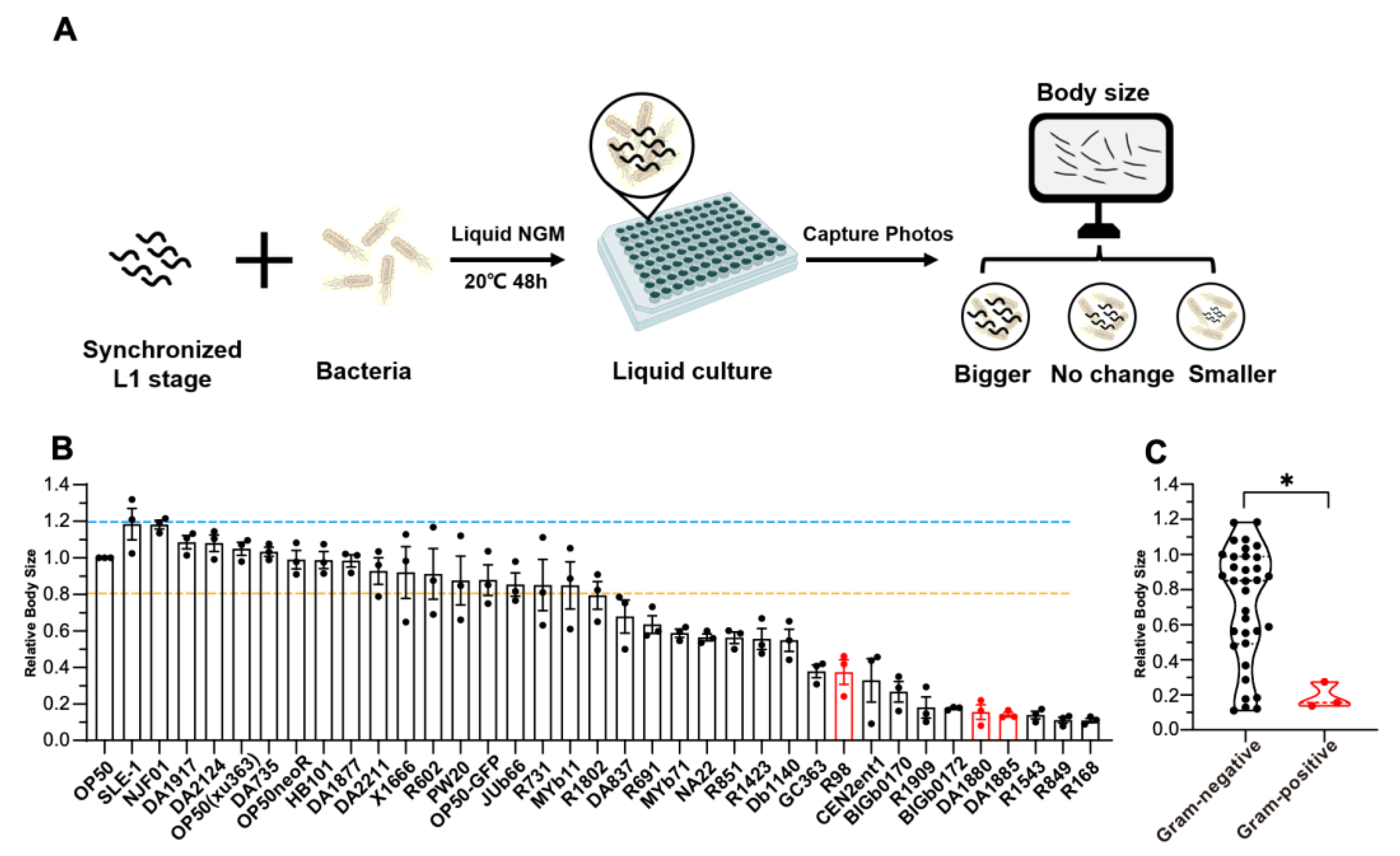
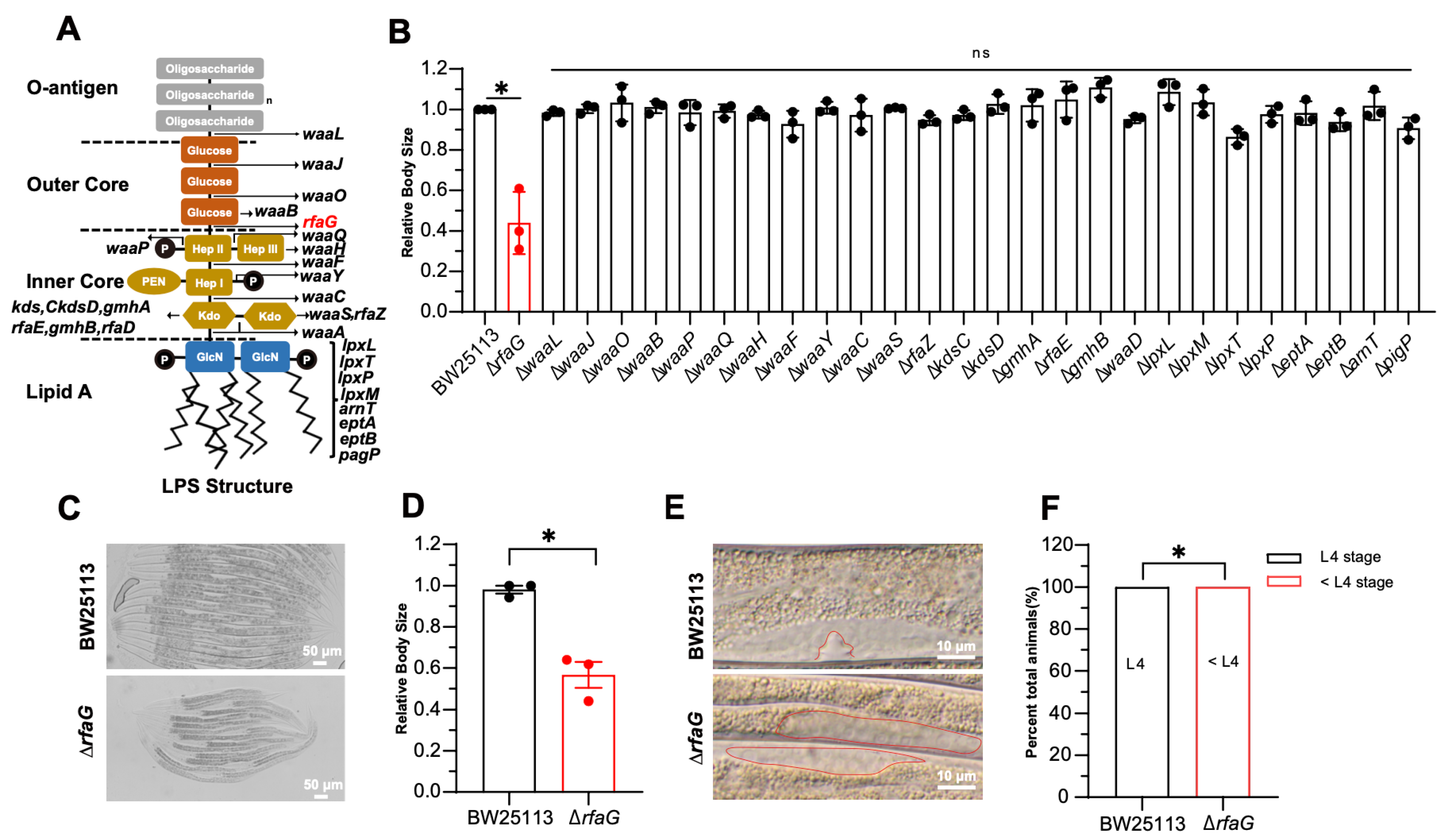
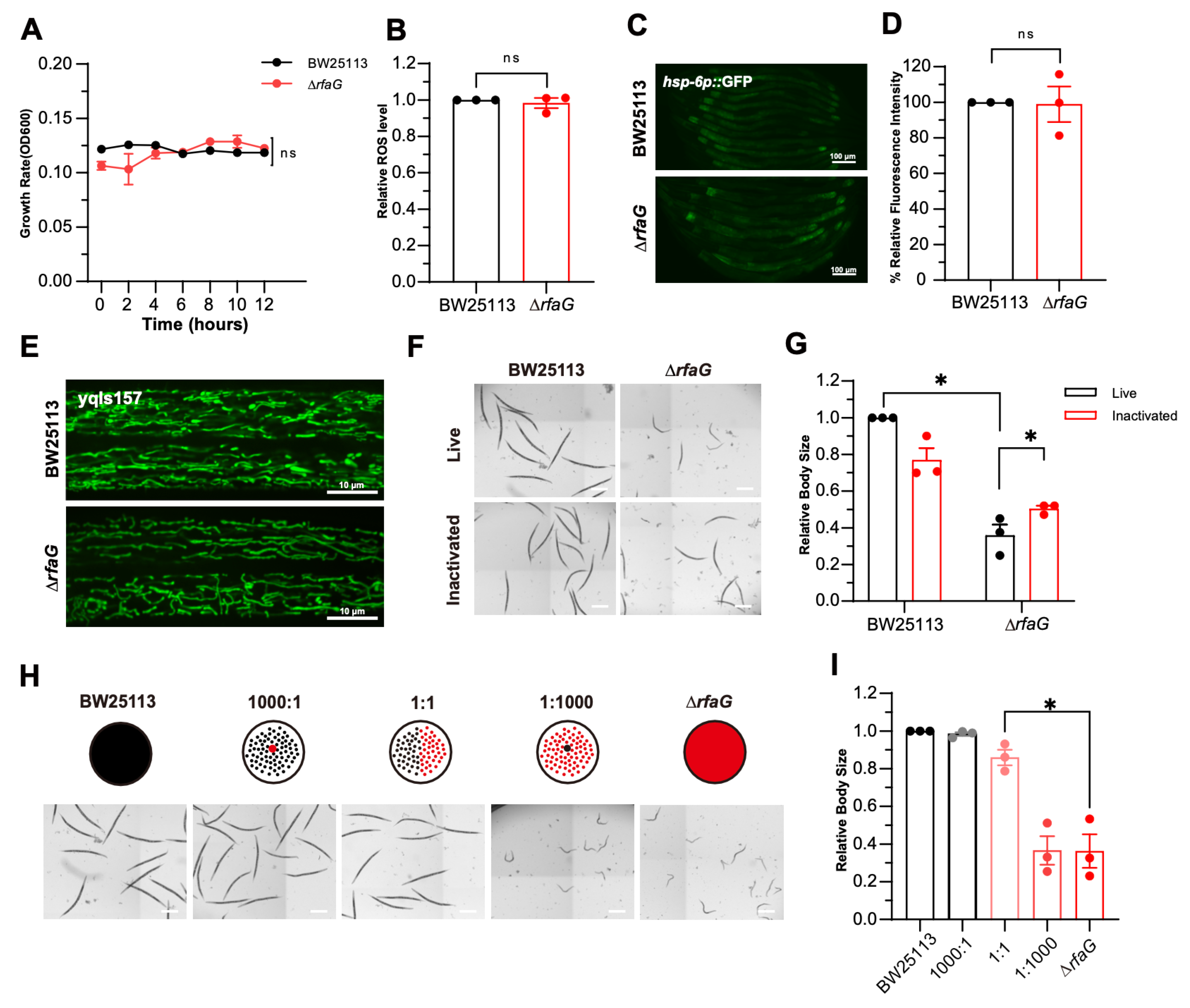
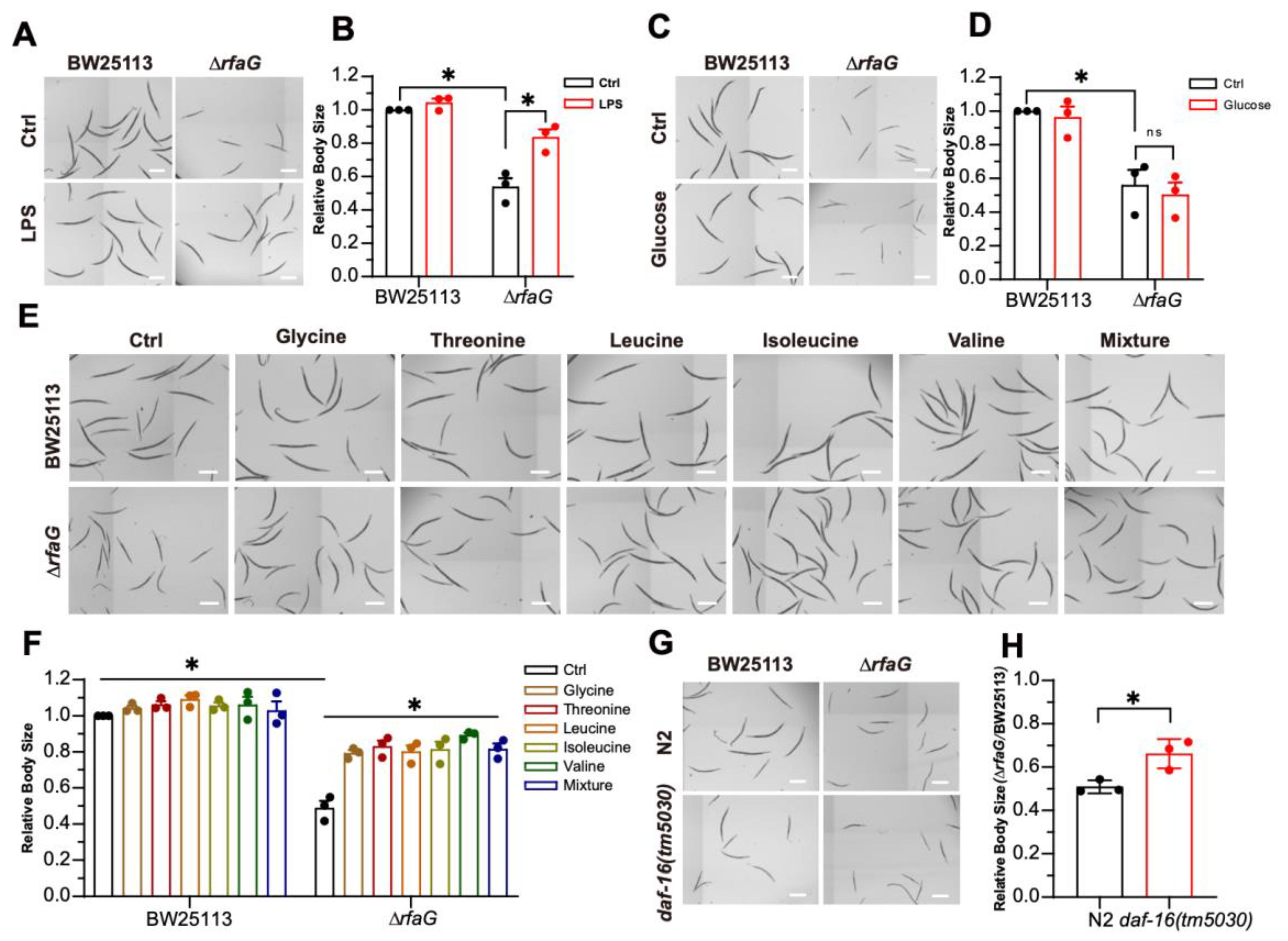
| Strain | Gram | Genus |
|---|---|---|
| OP50 | negative | Escherichia |
| SLE-1 | negative | Escherichia |
| NJF01 | negative | Escherichia |
| DA1917 | negative | Escherichia |
| DA2124 | negative | Escherichia |
| OP50(xu363) | negative | Escherichia |
| DA735 | negative | Escherichia |
| OP50-NeoR | negative | Escherichia |
| HB101 | negative | Escherichia |
| DA1877 | negative | Comamonas |
| DA2211 | negative | Escherichia |
| X1666 | negative | Escherichia |
| R602 | negative | Acidovorax |
| PW20 | negative | Escherichia |
| OP50-GFP | negative | Escherichia |
| JUb66 | negative | Lelliottia |
| R731 | negative | Sphingomonas |
| MYb11 | negative | Pseudomonas |
| R1802 | negative | Acidovorax |
| DA837 | negative | Escherichia |
| R691 | negative | Chryseobacterium |
| MYb71 | negative | Ochrobactrum |
| NA22 | negative | Escherichia |
| R851 | negative | Sphingomonas |
| R1423 | negative | Chryseobacterium |
| Db1140 | negative | Serratia |
| GC363 | negative | Escherichia |
| R98 | positive | Agrococcus |
| CEN2ent1 | negative | Enterobacter |
| BIGb0170 | negative | Sphingobacterium |
| R1909 | negative | Chryseobacterium |
| BIGb0172 | negative | Comamonas |
| DA1880 | positive | Bacillus |
| DA1885 | positive | Bacillus |
| R1543 | negative | Pedobacter |
| R849 | negative | Azospirillum |
| R168 | negative | Azospirillum |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teng, L.; Zhang, J. Microbial Lipopolysaccharide Regulates Host Development Through Insulin/IGF-1 Signaling. Int. J. Mol. Sci. 2025, 26, 7399. https://doi.org/10.3390/ijms26157399
Teng L, Zhang J. Microbial Lipopolysaccharide Regulates Host Development Through Insulin/IGF-1 Signaling. International Journal of Molecular Sciences. 2025; 26(15):7399. https://doi.org/10.3390/ijms26157399
Chicago/Turabian StyleTeng, Lijuan, and Jingyan Zhang. 2025. "Microbial Lipopolysaccharide Regulates Host Development Through Insulin/IGF-1 Signaling" International Journal of Molecular Sciences 26, no. 15: 7399. https://doi.org/10.3390/ijms26157399
APA StyleTeng, L., & Zhang, J. (2025). Microbial Lipopolysaccharide Regulates Host Development Through Insulin/IGF-1 Signaling. International Journal of Molecular Sciences, 26(15), 7399. https://doi.org/10.3390/ijms26157399





