Impact of Copper Nanoparticles on Keratin 19 (KRT19) Gene Expression in Breast Cancer Subtypes: Integrating Experimental and Bioinformatics Approaches
Abstract
1. Introduction
1.1. Research Gap
1.2. Study Rationale and Objectives
2. Results
2.1. Characterization of Copper Nanoparticles
2.2. Cytotoxicity and Growth Inhibition by CuNPs
2.3. KRT19 Gene Expression Analysis
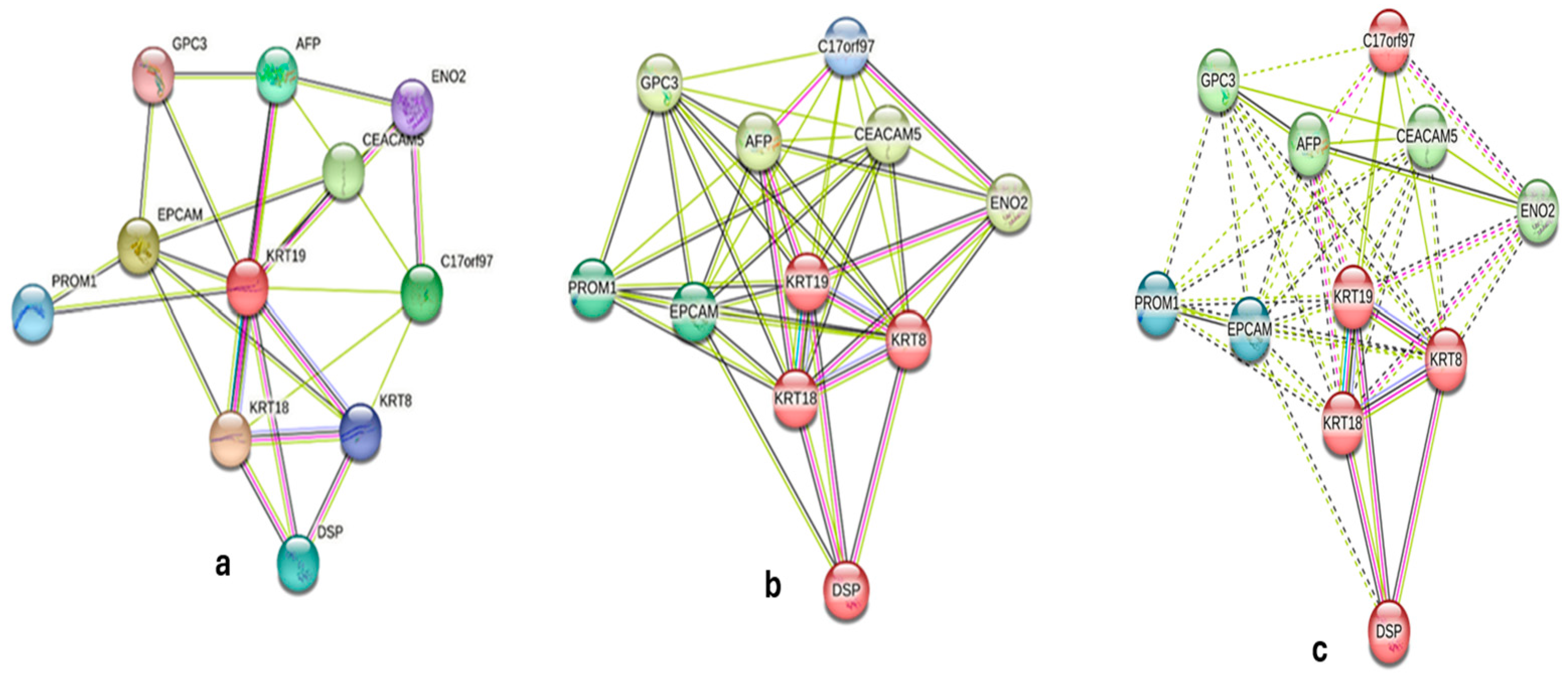
2.4. Bioinformatics Analysis
3. Discussion
Limitations and Future Directions
4. Materials and Methods
4.1. Cell Culture and CuNP Treatment
4.2. Copper Nanoparticle (CuNP) Synthesis
4.3. Cytotoxicity and Growth Inhibition Activity: IC50 Determination
4.4. RNA Extraction and cDNA Synthesis
4.5. Quantitative Real-Time PCR Analysis
4.6. Statistical and Bioinformatics Analysis
4.7. ΔΔCt Method for Relative Gene Expression
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jain, N.K.; Pathak, S.; Alam, M. Synthesis of Copper Nanoparticles by Pulsed Electrochemical Dissolution Process. Ind. Eng. Chem. Res. 2018, 58, 602–608. [Google Scholar] [CrossRef]
- Ahamed, M.; Akhtar, M.J.; Alhadlaq, H.A.; Alshamsan, A. Copper ferrite nanoparticle-induced cytotoxicity and oxidative stress in human breast cancer MCF-7 cells. Colloids Surf. B Biointerfaces 2016, 142, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Alsharif, S.; Bursch, K.; Parvathaneni, S.; Anastasakis, D.G.; Chahine, J.; Fallatah, A.; Nicolas, K.; Sharma, S.; Hafner, M.; et al. Keratin 19 regulates cell cycle pathway and sensitivity of breast cancer cells to CDK inhibitors. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Ervik, M.; Lam, F.; Laversanne, M.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today, version 1.1. International Agency for Research on Cancer: Lyon, France, 2024. Available online: https://gco.iarc.fr/today (accessed on 6 May 2025).
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Keyvani, S.; Karimi, N.; Orafa, Z.; Bouzari, S.; Oloomi, M. Assessment of Cytokeratin-19 Gene Expression in Peripheral Blood of Breast Cancer Patients and Breast Cancer Cell Lines. Biomark Cancer. 2016, 8, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.J.; Padmanaban, V.; Silvestri, V.; Schipper, K.; Cohen, J.D.; Fairchild, A.N.; Gorin, M.A.; Verdone, J.E.; Pienta, K.J.; Bader, J.S.; et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci. USA 2016, 113, E854–E863. [Google Scholar] [CrossRef] [PubMed]
- Ju, J.-H.; Yang, W.; Lee, K.-M.; Oh, S.; Nam, K.; Shim, S.; Shin, S.Y.; Gye, M.C.; Chu, I.-S.; Shin, I. Regulation of Cell Proliferation and Migration by Keratin19-Induced Nuclear Import of Early Growth Response-1 in Breast Cancer Cells. Clin. Cancer Res. 2013, 19, 4335–4346. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.K.; Kim, K.; Yang, G.M.; Choi, H.Y.; Cho, S.G. Cytokeratin 19 (KRT19) has a role in the reprogramming of cancer stem cell-like cells to less aggressive and more drug-sensitive cells. Int. J. Mol. Sci. 2018, 19, 1423. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.K.; Choi, H.Y.; Kim, B.W.; Dayem, A.A.; Yang, G.-M.; Kim, K.S.; Yin, Y.F.; Cho, S.-G. KRT19 directly interacts with β-catenin/RAC1 complex to regulate NUMB-dependent NOTCH signaling pathway and breast cancer properties. Oncogene 2017, 36, 332–349. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Meng, H.; Xing, G.; Chen, C.; Zhao, Y.; Jia, G.; Wang, T.; Yuan, H.; Ye, C.; Zhao, F.; et al. Acute toxicological effects of copper nanoparticles in vivo. Toxicol. Lett. 2006, 163, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Dong, Q.; Zhang, Y.; Hu, Q.; Zhang, Z.; Mao, W.; Qi, W.; Yang, Y.; Yang, Z. The clinical significance of the expression of PROM1/CD133, NOTCH2 and CDK4 in triple-negative breast cancer. Res. Square 2023. [Google Scholar] [CrossRef]
- Kawai, T.; Yasuchika, K.; Ishii, T.; Katayama, H.; Yoshitoshi, E.Y.; Ogiso, S.; Kita, S.; Yasuda, K.; Fukumitsu, K.; Mizumoto, M.; et al. Keratin 19, a cancer stem cell marker in human hepatocellular carcinoma. Clin. Cancer Res. 2015, 21, 3081–3091. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Deng, Y.; Li, G. Keratin 19 (Krt19) is a novel marker gene for epicardial cells. Front. Genet. 2024, 15, 1385867. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]

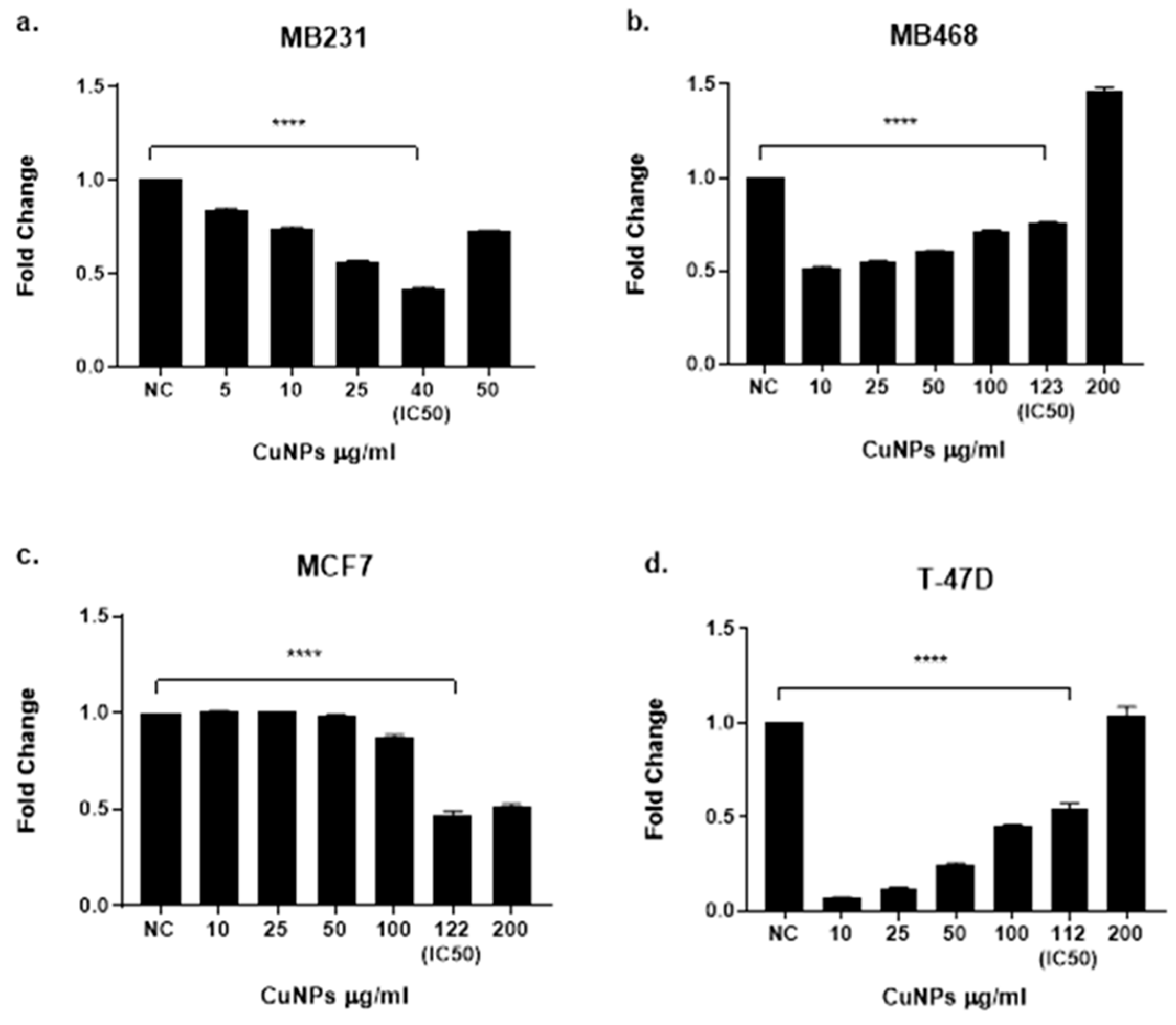


| Cell Line | Hormone Receptors | Subtype | IC50 (µg/mL) | IC50 (mM) | Viability at 1800 µg/mL (%) | Sensitivity Rank | R2 |
|---|---|---|---|---|---|---|---|
| MDA-MB-231 | ER−, PR−, HER2− | TNBC | 40 | 0.629 | <10 | Highest (1) | 0.9958 |
| MDA-MB-468 | ER−, PR−, HER2− | TNBC Basal | 123 | 1.932 | ~30 | Low (4) | 0.9919 |
| MCF7 | ER+, PR+, HER2− | Luminal | 122 | 1.920 | ~28 | Low (3) | 0.9882 |
| T47D | ER+, PR+, HER2− | Luminal | 112 | 1.764 | ~15 | Moderate (2) | 0.9929 |
| Gene | Confidence Score | Role in Breast Cancer |
|---|---|---|
| KRT8 | 0.95 | Cytoskeletal organization |
| KRT18 | 0.92 | Cytoskeletal organization |
| EPCAM | 0.888 | Cell adhesion, epithelial marker |
| CEACAM5 | 0.85 | Tumor progression |
| C17orf97 | 0.8 | Cytoskeletal organization |
| AFP | 0.78 | Tumor marker |
| DSP | 0.77 | Cell adhesion |
| PROM1 | 0.76 | Cancer stem cell marker |
| ENO2 | 0.75 | Tumor progression |
| GPC3 | 0.74 | Tumor progression |
| Gene | Co-Expression Strength | Role in Breast Cancer |
|---|---|---|
| KRT18 | Strong (dark red) | Cytoskeletal organization |
| KRT8 | Strong (dark red) | Cytoskeletal organization |
| EPCAM | Moderate (medium pink) | Cell adhesion, epithelial marker |
| LGALS3 | Moderate (medium pink) | Metastasis, cell migration |
| Characteristic | MDA-MB-231 | MDA-MB-468 | T47D | MCF7 |
|---|---|---|---|---|
| ATCC Catalog | HTB-26 | HTB-132 | HTB-133 | HTB-22 |
| Molecular Subtype | Triple-negative | Triple-negative (Basal A) | Luminal | Luminal |
| Hormone Receptors | ||||
| ER | Negative | Negative | Positive | Positive |
| PR | Negative | Negative | Positive | Positive |
| HER2 | Negative | Negative | Negative | Negative |
| Key Markers | ||||
| EGFR | Positive | Strongly Positive | Low | Low |
| Vimentin | Positive | Less mesenchymal | - | Low |
| CD44 | Positive | Lower expression | - | - |
| Ki67 | Low | Moderate | Moderate | Moderate |
| CK5/6 | - | Positive | Low | Low |
| EpCAM | - | Positive | Positive | Strongly Positive |
| KRT19 | Low | Moderate | Positive | Strongly Positive |
| Special Features | ||||
| Phenotype | CD44+/CD24−/low | Grape-like morphology | Epithelial | Drug resistant (ABCG2+) |
| Other Features | Mesenchymal, Invasive | Lacks E-cadherin, Wild-type p53 | Differentiated epithelial | Differentiated epithelial |
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Amplicon Size (bp) | Tm (°C) |
|---|---|---|---|---|
| KRT19 | GCGAGCTAGAGGTGAAGATC | AGTGCTCCCAGACGCAAG | 198 | 60 |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC | 226 | 60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taha, S.; Sultan, A.; Aljishi, M.; Greish, K. Impact of Copper Nanoparticles on Keratin 19 (KRT19) Gene Expression in Breast Cancer Subtypes: Integrating Experimental and Bioinformatics Approaches. Int. J. Mol. Sci. 2025, 26, 7269. https://doi.org/10.3390/ijms26157269
Taha S, Sultan A, Aljishi M, Greish K. Impact of Copper Nanoparticles on Keratin 19 (KRT19) Gene Expression in Breast Cancer Subtypes: Integrating Experimental and Bioinformatics Approaches. International Journal of Molecular Sciences. 2025; 26(15):7269. https://doi.org/10.3390/ijms26157269
Chicago/Turabian StyleTaha, Safa, Ameera Sultan, Muna Aljishi, and Khaled Greish. 2025. "Impact of Copper Nanoparticles on Keratin 19 (KRT19) Gene Expression in Breast Cancer Subtypes: Integrating Experimental and Bioinformatics Approaches" International Journal of Molecular Sciences 26, no. 15: 7269. https://doi.org/10.3390/ijms26157269
APA StyleTaha, S., Sultan, A., Aljishi, M., & Greish, K. (2025). Impact of Copper Nanoparticles on Keratin 19 (KRT19) Gene Expression in Breast Cancer Subtypes: Integrating Experimental and Bioinformatics Approaches. International Journal of Molecular Sciences, 26(15), 7269. https://doi.org/10.3390/ijms26157269







