1. Introduction
Trichomes, hair-like epidermal structures on plant aerial surfaces, serve critical ecological roles in mitigating herbivory, ultraviolet (UV) radiation, and water loss [
1,
2,
3]. In economically important crops such as cotton and tobacco, trichomes contribute to fiber production and the synthesis of secondary metabolites [
4]. In
Arabidopsis thaliana, trichome development has emerged as a model system for studying cell fate determination, differentiation, and the integration of hormonal and environmental signals. A core regulatory network centered on the MYB-bHLH-WD40 (MBW) complex—comprising
GLABRA1 (GL1),
GLABRA3 (GL3)/ENHANCER OF GLABRA3 (EGL3), and
TRANSPARENT TESTA GLABRA1 (TTG1)—activates trichome-promoting genes such as
GLABRA2 (GL2) [
5,
6,
7,
8]. Trichome patterning is administered by two competing models. The activation-inhibition mechanism involves the MBW complex inducing trichome initiation while stimulating inhibitors such as
TRIPTYCHON (TRY) and
CAPRICE (CPC), creating a self-organizing feedback loop to prevent adjacent cell differentiation [
9,
10,
11]. In contrast, the activator-depletion model proposes that trichome precursors sequester
TTG1, limiting its availability in neighboring cells [
10,
12].
Hormonal crosstalk intricately modulates trichome development. Gibberellins (GA) promote initiation by destabilizing DELLA repressors, which otherwise suppress GL1 and GIS family proteins [
13,
14]. Cytokinins (CK) antagonize GA signaling in a dose-dependent manner, reducing trichome density on mature leaves through mechanisms involving
ARABIDOPSIS RESPONSE REGULATOR (ARR) genes [
15,
16], while jasmonic acid (JA) synergizes with GA via JAZ protein degradation, releasing
GL3/EGL3 to activate trichome genes [
17]. This crosstalk ensures precise spatiotemporal control of trichome development, enabling adaptation to environmental and developmental cues. However, the integration of hormonal signals (e.g., GA, CK, and JA) with environmental factors remains incompletely resolved, particularly the roles of C2H2 zinc finger proteins like
GIS2 and
ZFP8 in mediating these interactions, highlighting the need for further research into these regulatory intersections.
C2H2 zinc finger proteins (ZFPs) function as key transcriptional regulators in trichome development by integrating hormonal signaling and environmental responses through sequence-specific DNA interactions. Plant C2H2 ZFPs are characterized by conserved cysteine (Cys) and histidine (His) residues that coordinate zinc ions, forming a “finger-like” DNA-binding domain [
18,
19]. This structural configuration enables sequence-specific interactions, often targeting AG/CT-rich motifs such as AGCTNAC, which are critical for regulating developmental and stress-responsive pathways [
20,
21,
22,
23,
24]. A hallmark of plant-specific C2H2 ZFPs, such as
GIS2 and
ZFP8, is the QALGGH pentapeptide motif within their zinc finger loops, which enhances DNA-binding specificity to plant cis-elements and distinguishes them from animal counterparts [
20,
25,
26]. Functionally, these proteins act as molecular hubs, integrating environmental and endogenous signals to regulate diverse processes, including development, hormone signaling, and abiotic stress responses [
27]. For example, the C2H2 ZFP
SUPERMAN maintains floral meristem boundaries by repressing
WUSCHEL expression [
28,
29], while
ZAT12 enhances oxidative stress tolerance by activating ROS-scavenging enzymes [
30]. Similarly,
STZ/ZAT10 integrates salt and drought responses through ABA-dependent pathways [
31]. A subset of C2H2 ZFPs, including
GIS,
GIS2, and
ZFP8, specifically regulate trichome development through hormonal signals.
GIS promotes trichome initiation on inflorescence stems through gibberellin (GA) signal upstream of
GLABRA1 (GL1) [
32], while
GIS2 and
ZFP8 regulate trichome development by integrating gibberellin (GA) and cytokinin (CK) pathways.
GIS2 and
ZFP8 exhibit distinct spatial roles:
GIS2 primarily influences floral organ trichomes, whereas
ZFP8 modulates trichomes on stems and leaves [
13,
33]. Despite their conserved QALGGH motif and overlapping regulatory roles, key questions remain unresolved. These include the direct transcriptional targets of
GIS2 and
ZFP8, their interactions with hormone pathways, and the evolutionary drivers of their functional divergence. Collectively, C2H2 ZFPs exemplify functional versatility, bridging growth regulation and stress adaptation. Their structural conservation, coupled with lineage-specific motifs like QALGGH, highlights their evolutionary specialization in plant gene regulatory networks. Future studies should prioritize elucidating their context-specific DNA-binding dynamics and signaling crosstalk in developmental and stress contexts, as addressed in this work.
Hormonal crosstalk between gibberellin (GA) and cytokinin (CK) signaling pathways represents an essential regulatory axis in trichome regulation, with the
GLABROUS INFLORESCENCE STEMS (GIS) family of C2H2 zinc finger proteins (ZFPs) acting as key molecular mediators. GA promotes trichome initiation by destabilizing DELLA repressor proteins, which suppress the expression of
GIS family genes (e.g.,
GIS,
GIS2, and
ZFP8) [
13,
32,
34]. Conversely, CK antagonizes GA signaling in a dose-dependent manner, reducing trichome density through mechanisms involving
ARABIDOPSIS RESPONSE REGULATOR (ARR) genes, though the molecular intermediates linking these pathways remain unidentified [
15,
16]. Recent studies highlight the hierarchical regulation within the
GIS family: GA and CK signals converge to regulate trichome density, with
GIS2 and
ZFP8 exhibiting distinct hormonal sensitivities [
13,
33,
35]. Functional redundancy and spatial specialization exist among
GIS paralogs;
GIS2 primarily regulates floral trichomes, whereas
ZFP8 governs stem and leaf trichomes, partially compensating for
GIS loss [
36].
Despite these advances, critical knowledge gaps persist. The regulatory cascades downstream of
GIS2 and
ZFP8, including their putative targets such as
PR-4 and
SPL15 (already characterized for
GIS), remain uncharacterized. Furthermore, the mechanisms by which
GIS2 and
ZFP8 differentially integrate GA and CK signals are unclear, as are the evolutionary drivers underlying their promoter-binding specificity and sub-functionalization. Although
ZFP5 has been identified as a GA-responsive regulator linking
GIS and
ZFP8 [
37,
38,
39], it is unknown whether
GIS2 and
ZFP8 directly mediate hormone signaling or require additional intermediaries. Additionally, the extent of functional redundancy and divergence among
GIS family members in fine-tuning trichome development in response to environmental cues remains unresolved. Addressing these questions will clarify how hormonal and developmental signals orchestrate trichome patterning through the
GIS network, offering broader insights into plant developmental plasticity.
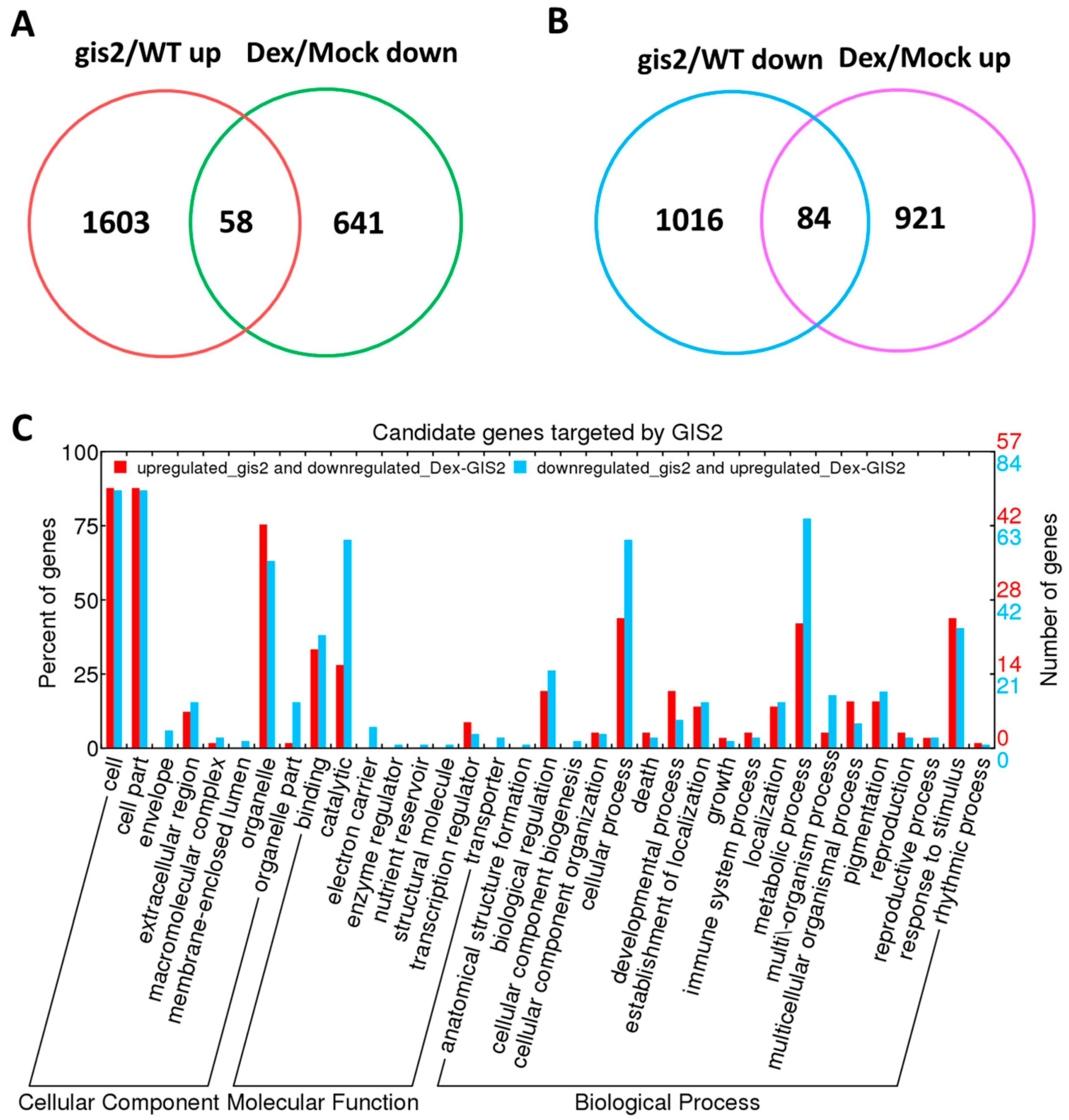
Trichome development, a key determinant of plant and environment interactions, is dynamically regulated by hormonal and environmental cues through specialized transcription factors. C2H2 zinc finger proteins play a crucial role in coordinating growth and stress adaptation; however, the mechanistic basis by which homologs GIS2 and ZFP8 integrate gibberellin (GA) and cytokinin (CK) crosstalk to modulate trichome patterning remains unclear. This study addresses this gap by pursuing three objectives: identifying transcriptional targets of GIS2 and ZFP8, using chromatin immunoprecipitation (ChIP) and DEX-CHX methods integrated with transcriptomic profiling, and examining their roles in sterol metabolism (SQE5), senescence (SEN1), and stress adaptation (PR-4, WRKY25); investigating how GA and CK signaling influence their activity, through hormone responsiveness assays, in the context of promoter-binding specificity and functional divergence; and comparing potential redundancy and specialization between GIS2 and ZFP8 by analyzing single and double mutants (gis2 zfp8) and RNAi lines, focusing on shared targets like SPL15 that influence trichome patterning during developmental phase transitions. By synthesizing ChIP-qPCR, microarray analysis, and mutant characterization, this work examines how GIS2 and ZFP8 may have evolved divergent regulatory networks while retaining overlapping functions in trichome development. The significance of this research lies in its potential to expand the trichome regulatory framework, offering mechanistic insights into how C2H2 zinc finger proteins balance redundancy and specialization to coordinate development with environmental responses. Notably, GIS2 regulation of SQE5 and SEN1 suggests possible links between trichome morphogenesis and drought or ABA signaling, while ZFP8 regulation of PR-4 (pathogen defense) may connect trichome traits to biotic resilience. Elucidating GIS2 and ZFP8 networks could provide a conceptual foundation for future strategies such as CRISPR-based approaches to engineer crops with optimized trichome features and enhanced adaptability to environmental challenges.
3. Discussion
Plant tissue and organ morphogenesis is dependent on an exact coordination of cell cycle progression and differentiation, which is mediated by conserved regulators such as cyclin-dependent kinases (CDKs) and cyclins [
46]. Metabolic signals, including sugar availability, help to further refine these mechanisms through combining growth with developmental barriers to ensure appropriate cellular patterns [
47,
48]. Trichome development in
Arabidopsis thaliana provides a model of this coordination in which epidermal cells move from mitotic division to endoreduplication—a cell cycle variant that increases nuclear DNA content without cytokinesis. This change starts trichome differentiation; trichome size and branching complexity are determined by endoreduplication cycle count, hence defining important features for stress resilience [
49]. Recent studies show that, along with nuclear expansion with trichome morphogenesis, histone acetyltransferases like GCN5 and ubiquitin-mediated proteolysis of cell cycle inhibitors dynamically control this process [
50,
51]. Phytohormones, particularly gibberellins (GAs) and jasmonates (JAs), influence trichome density by connecting developmental cues with stimulation from the environment. GA signaling induces trichome initiation by degrading DELLA repressors via the GID1 receptor, releasing inhibition on trichome-promoting factors [
52] (
Figure 8A). JA, on the other hand, balances defense priority with growth by altering trichome elongation through MYC transcription factors [
53].
The interaction between these pathways provides adaptive plasticity, allowing plants to enhance trichome-mediated defenses in response to biotic or abiotic stress while preserving developmental homeostasis [
54]. The C2H2 zinc finger transcription factors GIS, GIS2, and ZFP8, which direct trichome patterning across many plant organs, are fundamental members of this regulatory network. Acting upstream of important regulators like GLABRA1 (GL1), GIS, and GIS2 mostly control trichome initiation on inflorescence stems and branches, integrating GA and cytokinin signals [
41]. However, ZFP8 controls trichome development on leaves by mediating cytokinin responses downstream of GL3 and TRY [
38]. Our findings extend this paradigm by demonstrating that GIS2 and ZFP8 exhibit bifunctional regulatory roles, directly activating stress-adaptive genes (e.g.,
SQE5;
Figure 7A,B,
Table 1) while repressing senescence-associated markers (e.g.,
SEN1;
Figure 7C,D,
Table 2). Spatial specificity is evident: GIS2 predominantly functions in inflorescence stems (
Figure 1A,B and
Figure 6C,D), where GA stabilizes its protein (
Figure 8A) and upregulates
SQE5 (
Figure 8B), whereas ZFP8 governs leaf trichomes through cytokinin/ABA pathways (
Figure 5C and
Figure 9A,B). This divergence mirrors evolutionary sub-functionalization, as seen in cotton homologs, where ZFP8 homologs regulate fiber development [
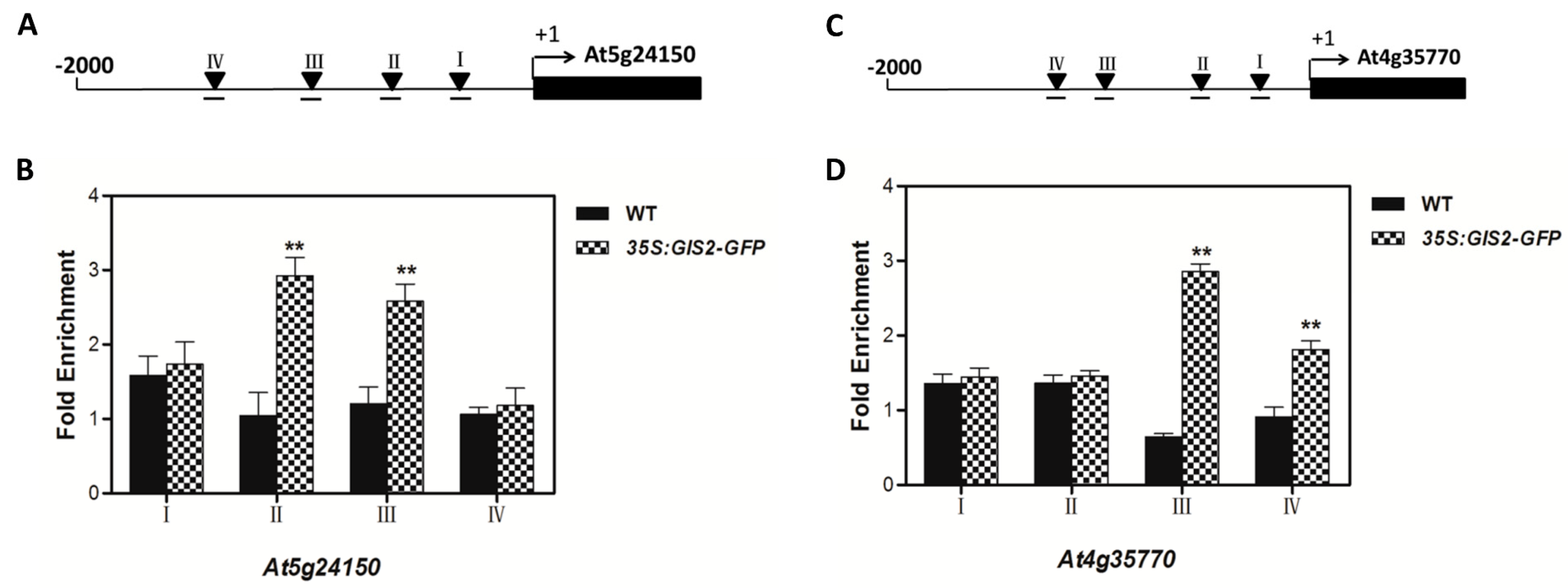
55]. The integration of hormonal cues (e.g., GA-induced GIS2 protein accumulation) and environmental signals (e.g., nutrient stress) suggests that these transcription factors balance developmental programs with stress adaptation. While ChIP-qPCR supports GIS2 binding to
At5g24150 and
At4g35770 promoters, future work will apply EMSA and yeast one-hybrid assays as used in our previous studies to further validate these interactions [
41,
56].
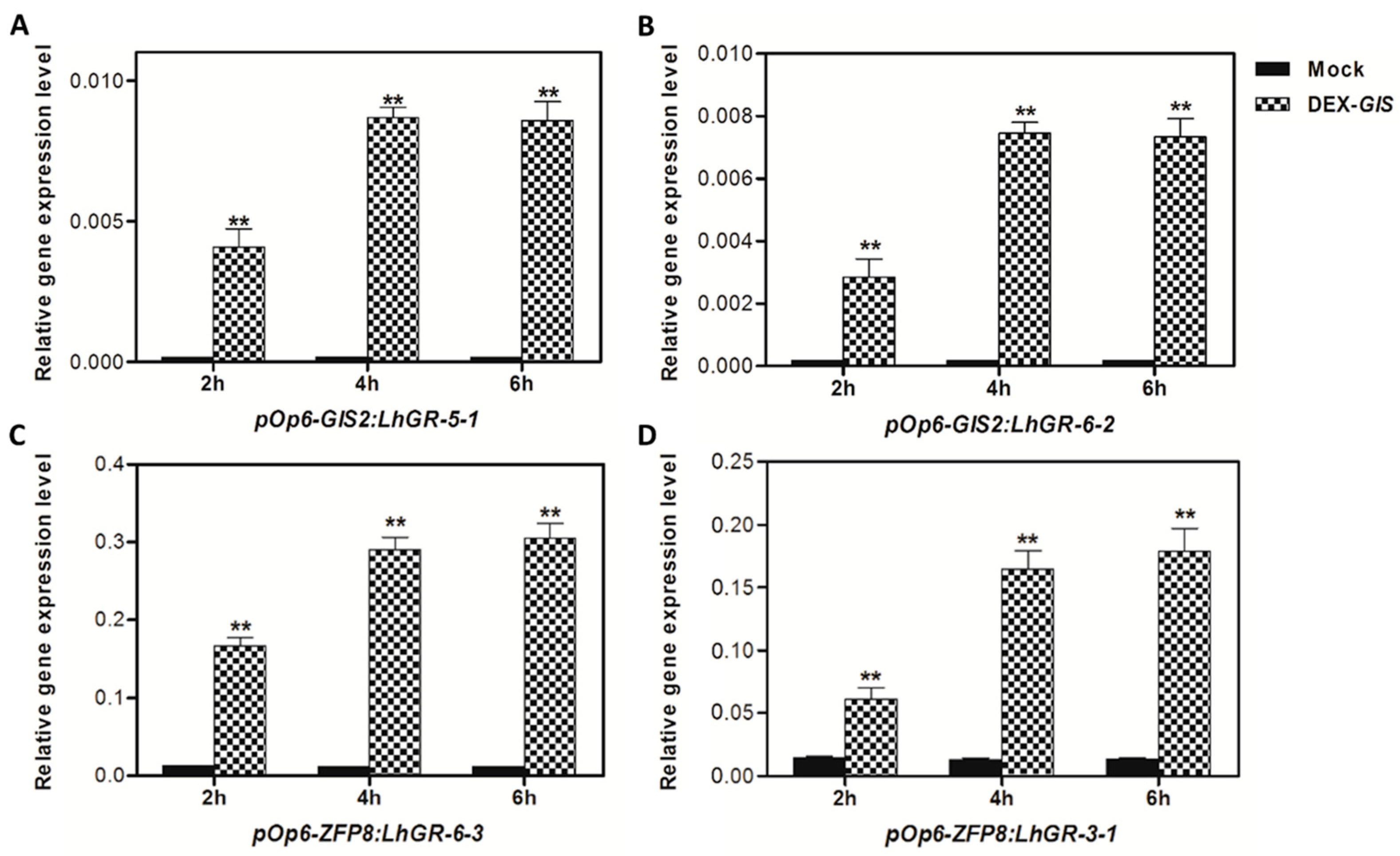
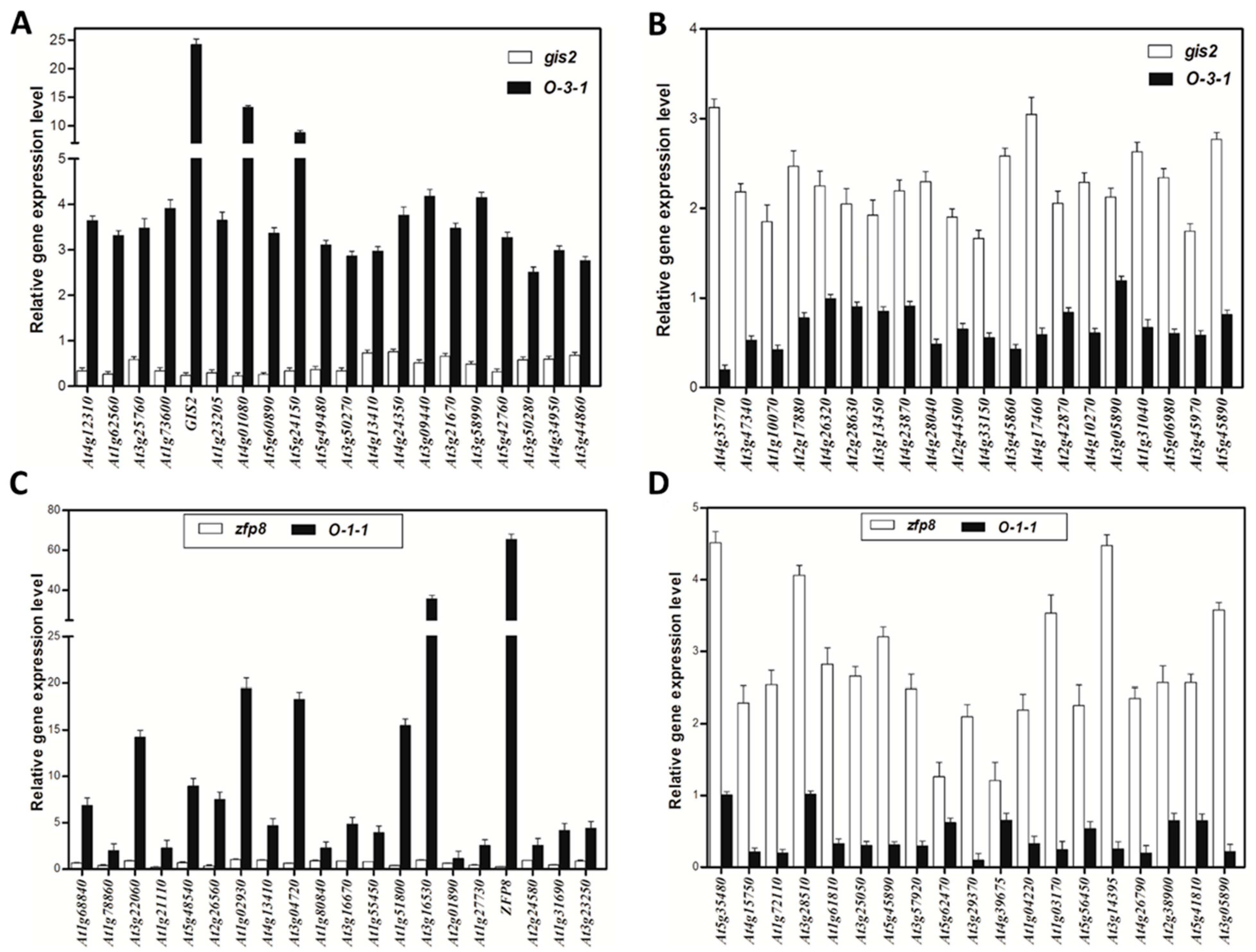
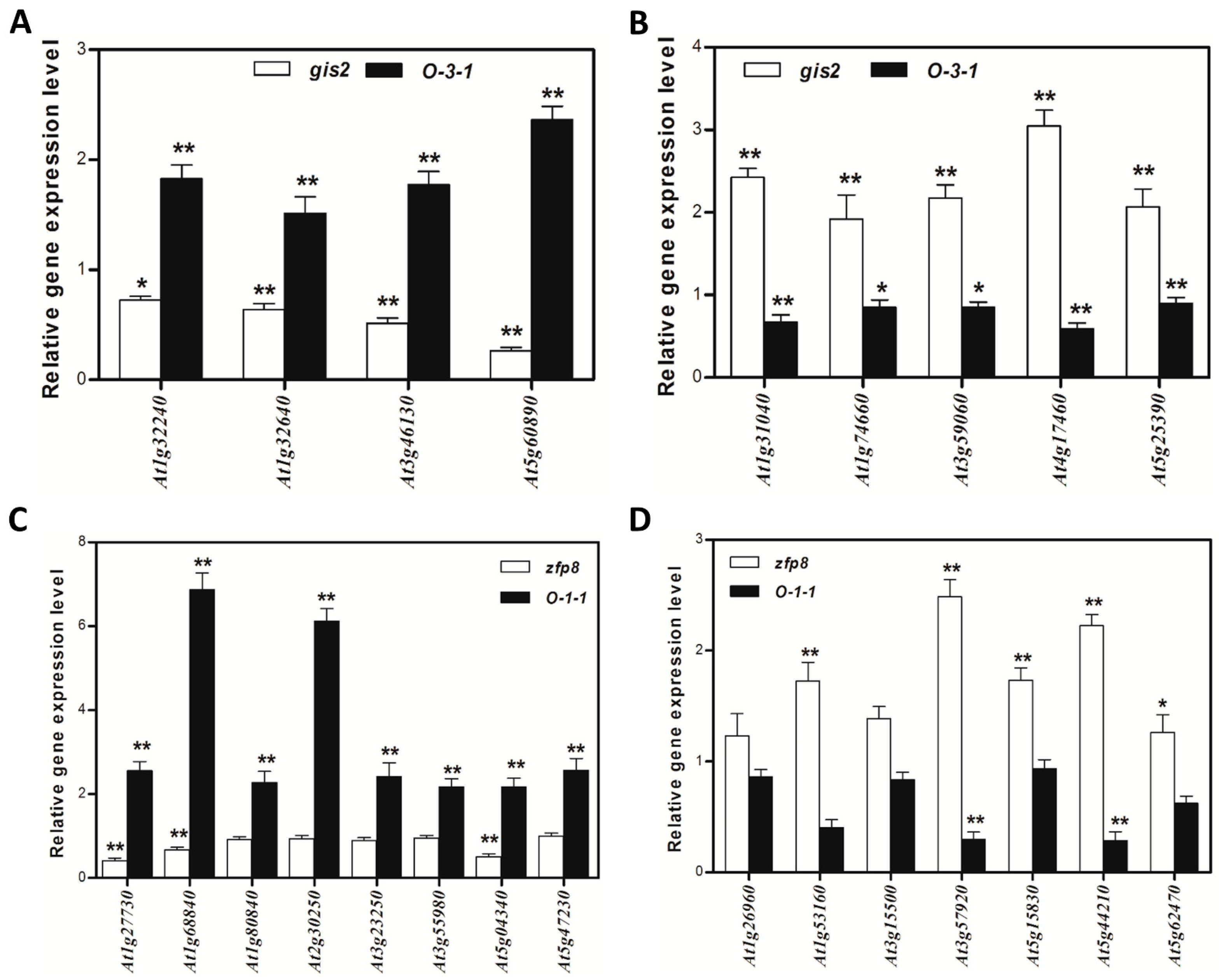
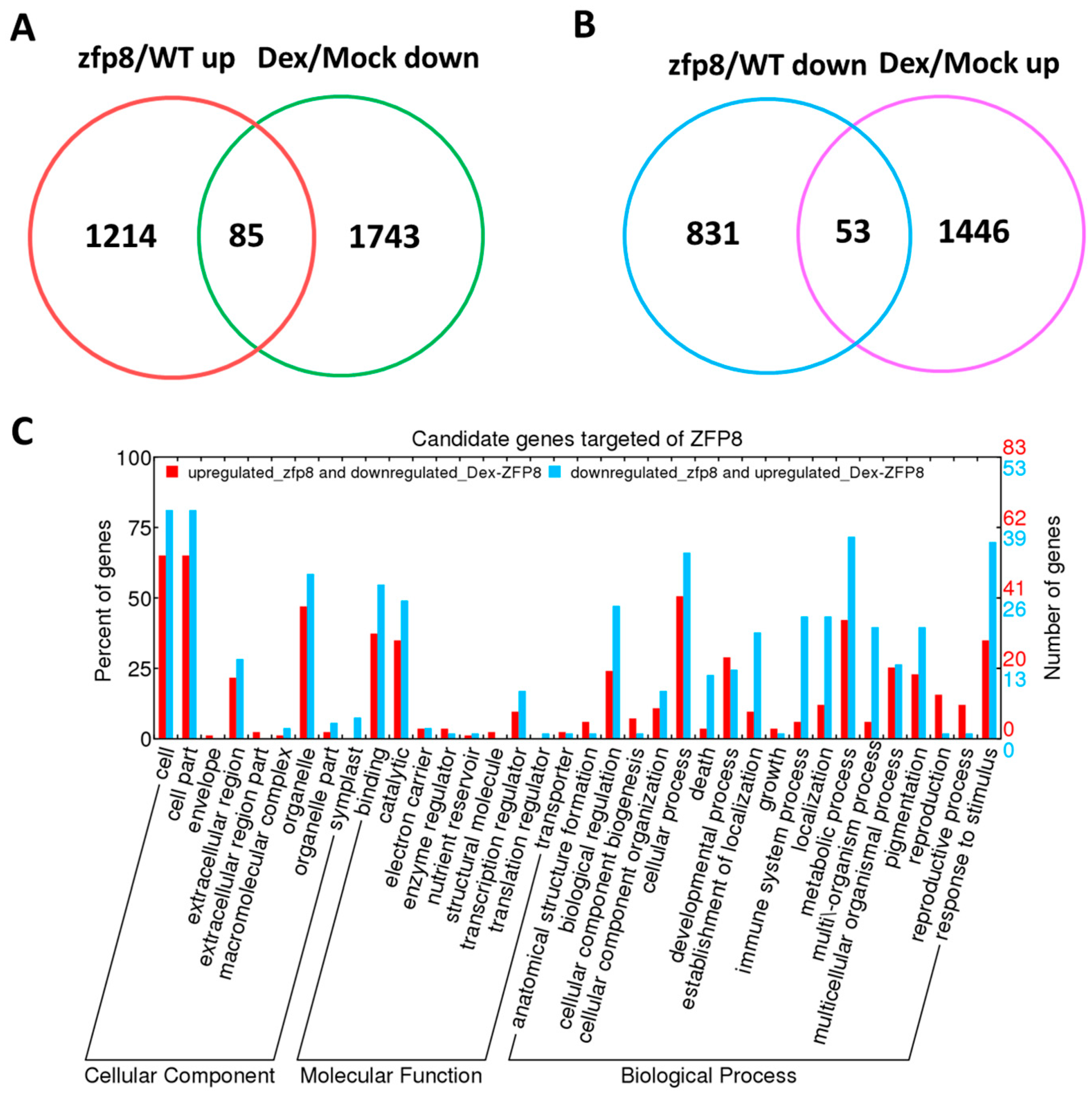
Transcriptomic analyses and dexamethasone-inducible overexpression systems demonstrate that GIS2 and ZFP8 control trichome development by overlapping but spatially different mechanisms. Microarray profiling of
gis2 and
zfp8 mutants revealed 142 and 138 target genes, respectively (
Figure 3 and
Figure 5), enhanced in catalytic activity, metabolic control, and stress adaptation—processes fundamental for trichome-mediated abiotic resilience. Notably, GIS2 and ZFP8 exhibit bifunctional regulatory roles, activating subsets of genes (e.g.,
SQE5 and
PR-4) while repressing others (e.g.,
SEN1 and
SPL15) to balance trichome differentiation with broader physiological demands (
Figure 2,
Figure 3,
Figure 4,
Figure 5,
Figure 6 and
Figure 7,
Table 1,
Table 2,
Table 3 and
Table 4). For instance, GIS2 directly activates
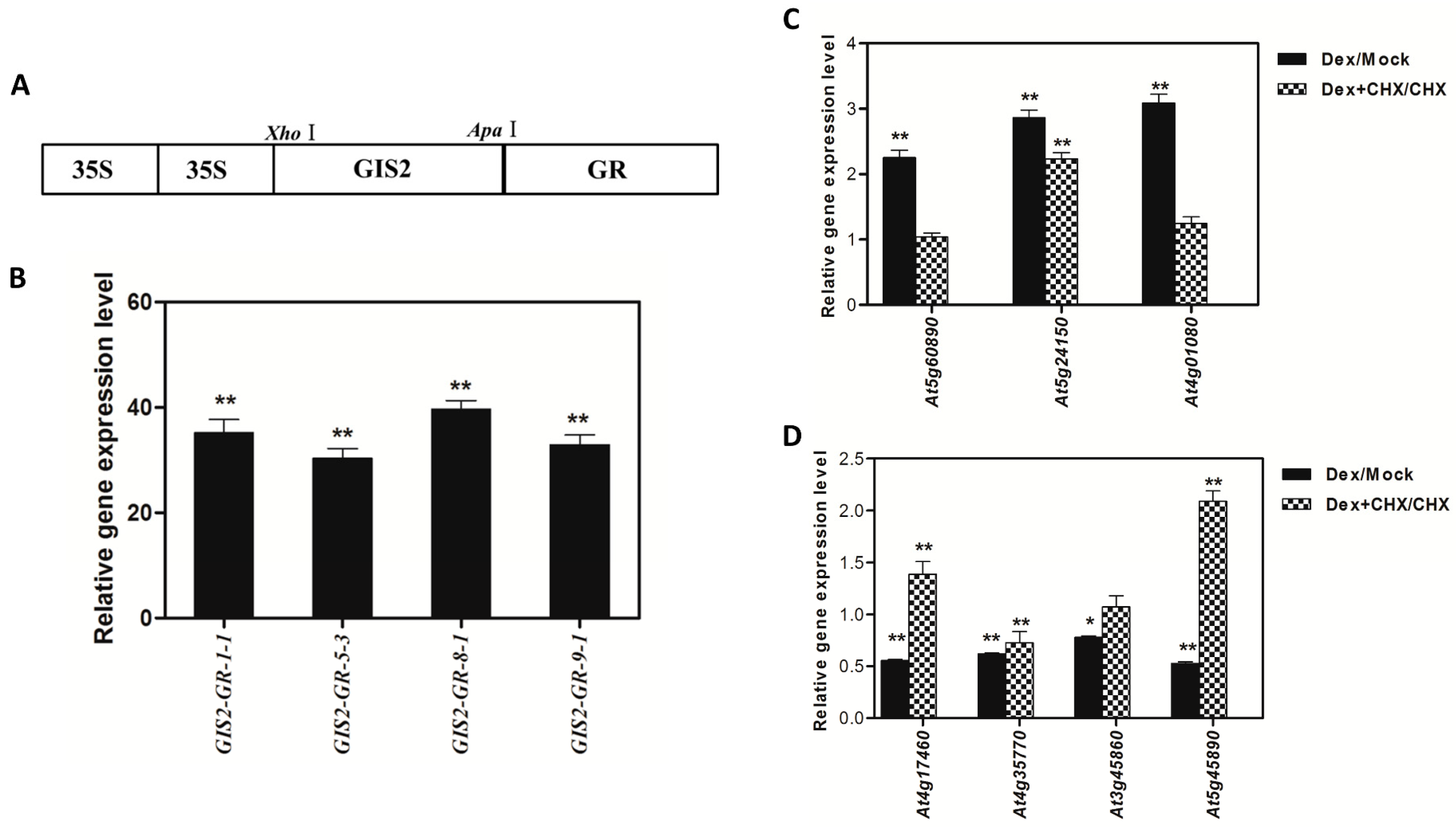
At5g24150 (
SQE5), a sterol biosynthesis enzyme critical for drought tolerance (
Figure 7A,B), while repressing
At4g35770 (
SEN1), a senescence marker regulated by ABA and phosphate starvation (
Figure 7C,D). This dual functionality positions GIS2 as a molecular integrator linking trichome morphogenesis to stress adaptation—a mechanism conserved in homologs like GIS3, which enhances trichome initiation across tissues [
39,
41].
ChIP assays confirmed the direct binding of GIS2 to conserved C2H2 motifs in the promoters of
SQE5 and
SEN1 (
Figure 7), establishing its role as a transcriptional hub. Similarly, ZFP8 modulates stress-responsive targets such as
PR-4 (pathogenesis-related protein) and
SPL15 (squamosa promoter-binding protein), the latter co-regulated by GIS (
Figure 2D and
Figure 9B;
Table 8). This regulatory overlap suggests functional convergence, particularly in coordinating trichome development with phase transitions. For example,
SPL15 is a mediator of shoot meristem phase transitions [
57], which is repressed by both GIS2 and ZFP8 (
Figure 4D), implying synchronized timing of trichome patterning and reproductive growth. Recent studies in cotton homologs further underscore the conserved role of ZFP8 in fiber development and photosynthetic efficiency [
55], suggesting evolutionary selection for stress-responsive trichome regulation.
The spatial and functional divergence between GIS2 and ZFP8 raises questions about redundancy and crosstalk. Does
SPL15 act as a central hub, synchronizing trichome development and flowering? Do interactions with TOE1/TOE2 transcription factors [
39] enable compensatory regulation under stress? Addressing these questions requires comparative analyses of DNA-binding landscapes across tissues and conditions. CRISPR-based editing or single-cell transcriptomics [
58] could resolve spatiotemporal dynamics of these networks in crops like cotton, where trichome density correlates with drought tolerance. Furthermore, elucidating the ecological roles of
SQE5 (sterol-mediated drought resilience) and
PR-4 (antifungal defense) could inform strategies to engineer crops with optimized trichome traits. By bridging mechanistic insights from
Arabidopsis (
Figure 1,
Figure 2,
Figure 3,
Figure 4,
Figure 5,
Figure 6,
Figure 7,
Figure 8 and
Figure 9) with translational applications, this work advances both basic research and agricultural innovation in a changing climate.
The regulatory overlap between ZFP8 and GIS2 is evidenced by their common targets, such as
At3g04720 (
PR-4) and
At3g57920 (
SPL15), the latter serving as a crucial integrator of developmental signals at the shoot apical meristem [
57]. While ZFP8 primarily directs trichome formation on stem leaves [
38], its targets are enriched in ABA signaling and drought defense pathways (
Figure 9A), supporting trichome roles in stress mitigation [
41]. Conversely, GIS2 links trichome development to sterol metabolism via
SQE5 (
Figure 7A,B) and senescence regulation via
SEN1 (
Figure 7C,D), suggesting broader roles in growth-phase transitions. This functional divergence is suggested by tissue-enriched expression and regulatory profiles: GIS2 showed elevated expression and downstream activation in inflorescence stems (
Figure 1A,B and
Figure 6C,D), while ZFP8-associated gene ontology terms and target enrichment were more prominent in leaf-related stress pathways (
Figure 5C and
Figure 9A,B). However, further studies using tissue-specific reporters or functional assays are needed to confirm precise spatial regulation. ZFP8 regulation of SPL15, a key mediator of phase transitions and flowering (
Figure 4D;
Table 8), suggests it may coordinate trichome development with reproductive timing [
57]. While GIS2 did not appear to directly regulate SPL15, its overlap with ZFP8 in targeting other developmental genes points to potential functional convergence. GA signaling stabilizes GIS2 protein (
Figure 8A), which coincides with increased expression of its downstream target SQE5 (
Figure 8B), a sterol biosynthesis gene associated with drought adaptation [
41]. This suggests that GA may indirectly enhance GIS2-mediated transcriptional regulation. SEN1 repression by GIS2 appears independent of GA signaling (
Figure 8B). Given previous reports linking SEN1 to ABA and phosphate starvation responses [
43,
44], its regulation may involve alternative hormonal or nutrient-responsive pathways, highlighting the potential for context-dependent control. Likewise, ZFP8’s interaction with stress-responsive TFs such as
AT1G68840 (
RAV2) and
AT2G30250 (
WRKY25) (
Figure 4C,D;
Table 7) expands its role beyond trichomes to abiotic adaptation. Recent research shows that these TFs interact with TOE1/TOE2 transcription factors [
39] to integrate environmental cues with GA and cytokinin signals, optimizing trichome patterning. This aligns with broader evidence that C2H2 TFs act as nodal points in hormonal crosstalk, balancing developmental precision with stress resilience [
59].
The spatial and functional divergence between GIS2 and ZFP8 raises questions about redundancy or crosstalk. Does
SPL15 function as a central hub, synchronizing trichome development and flowering? Do TOE1/TOE2 interactions facilitate compensatory regulation under stress? Addressing these questions necessitates comparative analyses of their binding landscapes across tissues and conditions. Furthermore, exploring the ecological relevance of key target genes such as SQE5, previously associated with drought tolerance [
42], and PR-4, known for its role in pathogen defense [
59], may provide insights to guide crop engineering strategies. While our study supports their transcriptional regulation by GIS2 and ZFP8, physiological validation under stress conditions remains a goal for future work. These mechanisms can be better understood with the help of new tools made possible by recent developments in single-cell transcriptomics and CRISPR-based spatial profiling [
58], which combine basic discoveries with agricultural innovations.
Our study establishes
GIS2 and
ZFP8 as central regulators integrating developmental and environmental cues to modulate trichome patterning. Their bifunctional roles as transcriptional activators and repressors—evidenced by targets such as
SQE5 (sterol biosynthesis and drought adaptation) and
SEN1 (senescence and ABA/phosphate signaling)—position them at the intersection of trichome morphogenesis and stress adaptation. Spatial specificity (inflorescence stems vs. leaves) and target divergence enable functional specialization, with GIS2 coordinating GA-dependent
SQE5 activation and ABA-mediated
SEN1 repression, while ZFP8 regulates stress-responsive pathways like
PR-4 and
SPL15 (
Figure 2,
Figure 7,
Figure 8 and
Figure 9). These findings extend the known roles of GIS family proteins by linking trichome development to drought resilience via
SQE5 and pathogen defense via
PR-4. Future studies leveraging CRISPR-based editing or single-cell transcriptomics could dissect the spatiotemporal dynamics of these networks in crops like cotton or maize, where trichome density correlates with stress tolerance. By resolving how GIS2 and ZFP8 balance redundancy and specialization, this work provides a framework to engineer crops with optimized epidermal traits, addressing yield stability in climate-variable environments.