Dysregulated miRNAs Targeting Adiponectin Signaling in Colorectal Cancer
Abstract
1. Introduction
2. Results
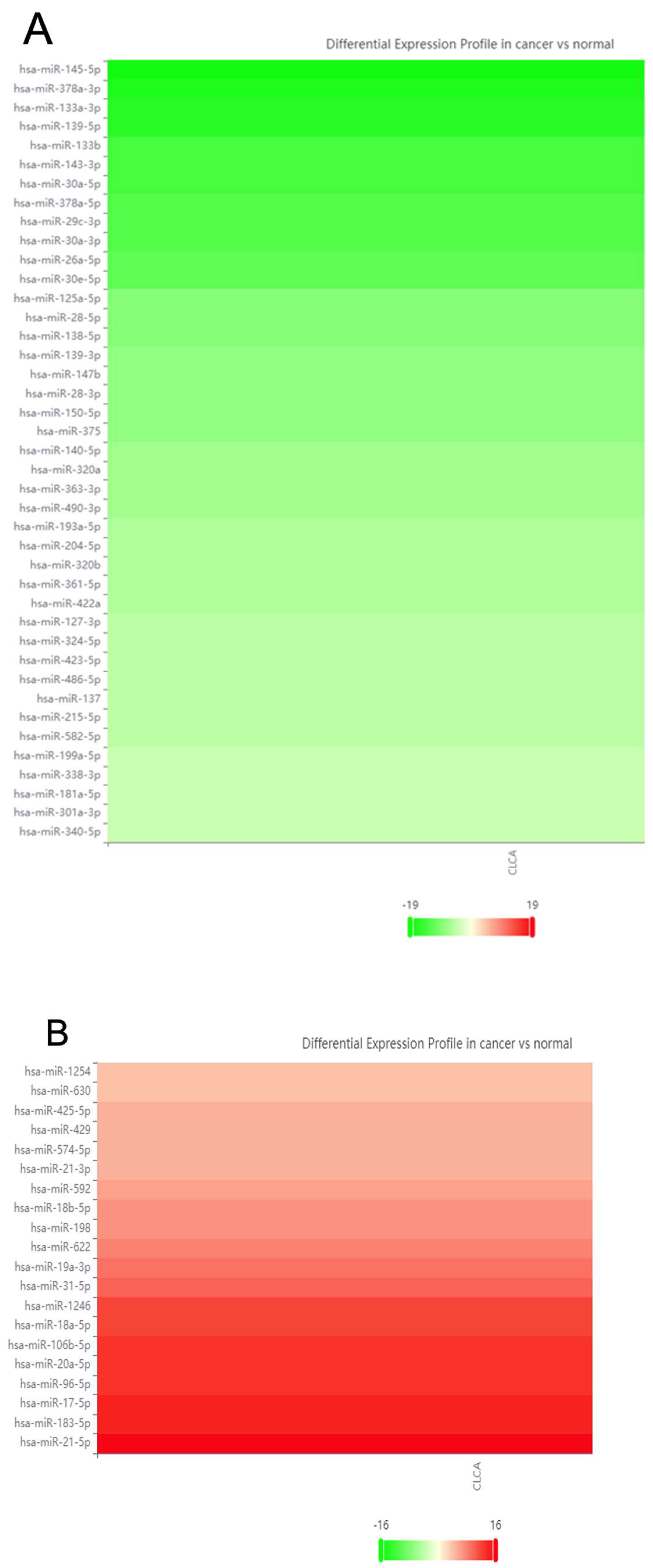
2.1. Database Cross-Referencing
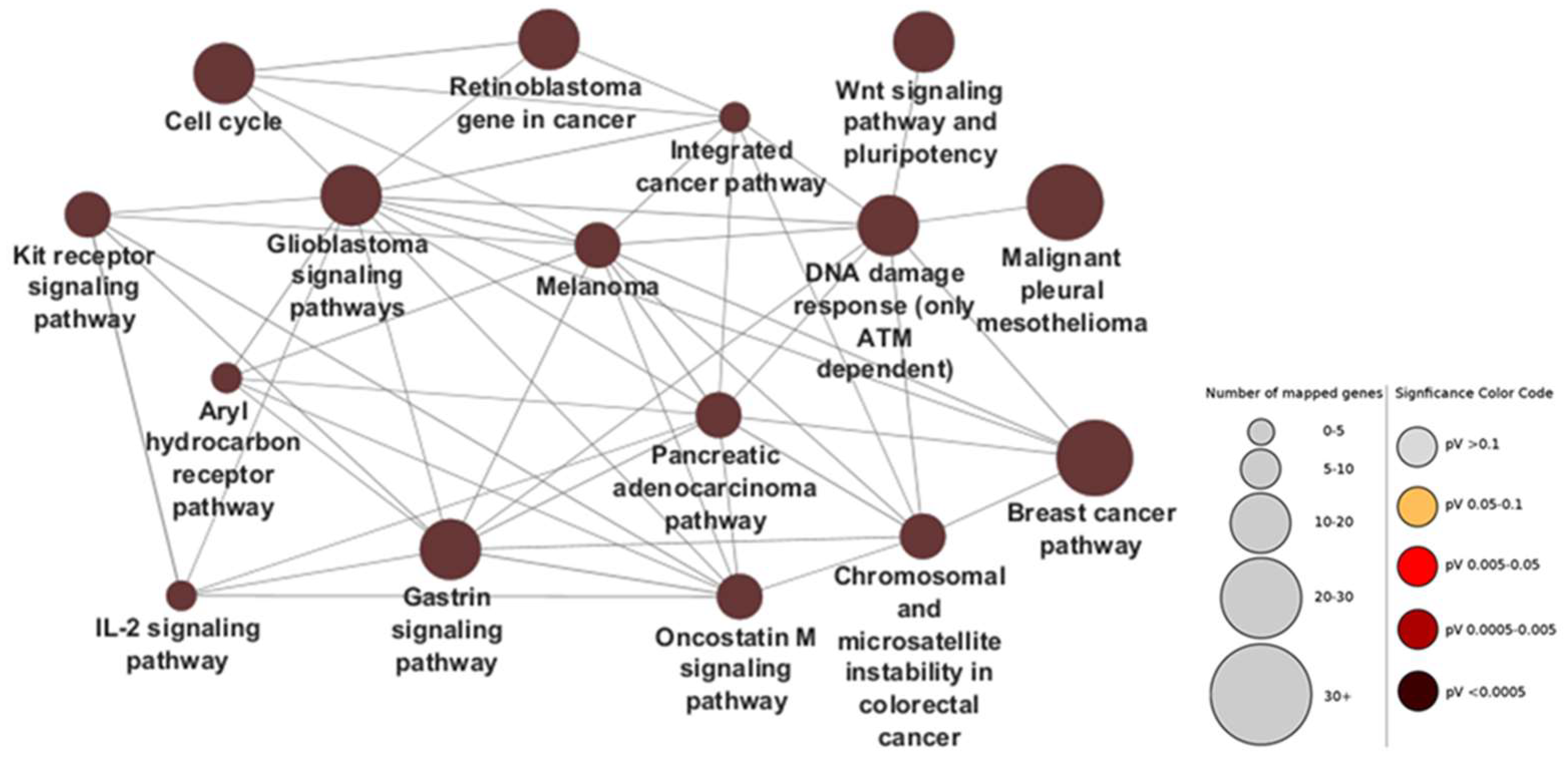
2.2. Ontology Networks
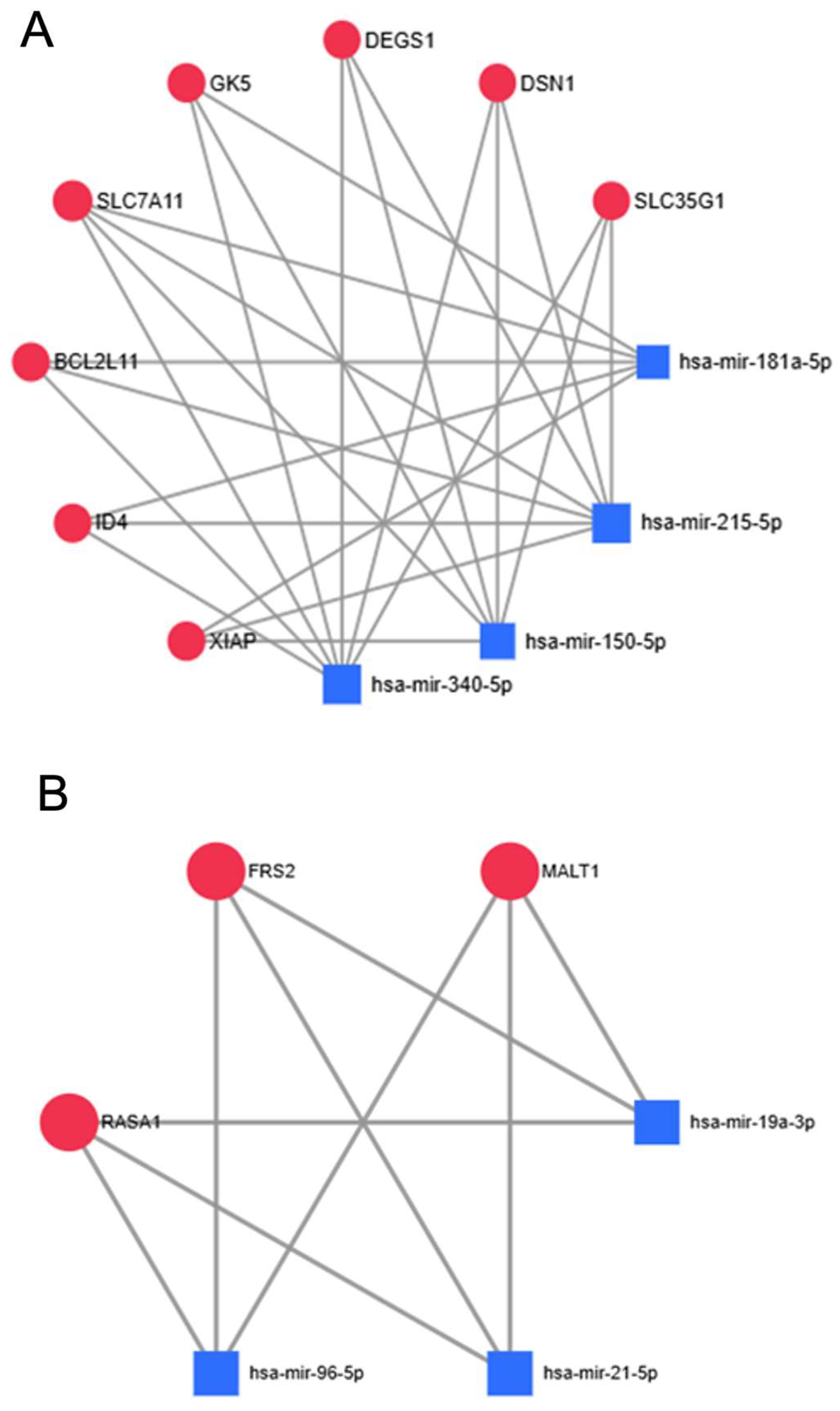
2.3. MirNET—Most Targeted Genes
2.4. Literature Review of Reproducibly Altered miRNAs in CRC
3. Discussion
4. Materials and Methods
4.1. Data Mining and Presentation
4.2. Data Source and Literature Search
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Okayama, H.; Harris, C.C. The Role of MicroRNAs in Colorectal Cancer. Cancer J. 2012, 18, 244–252. [Google Scholar] [CrossRef]
- Mehterov, N.; Sacconi, A.; Pulito, C.; Vladimirov, B.; Haralanov, G.; Pazardjikliev, D.; Nonchev, B.; Berindan-Neagoe, I.; Blandino, G.; Sarafian, V. A novel panel of clinically relevant miRNAs signature accurately differentiates oral cancer from normal mucosa. Front. Oncol. 2022, 12, 1072579. [Google Scholar] [CrossRef] [PubMed]
- Mulrane, L.; McGee, S.F.; Gallagher, W.M.; O’COnnor, D.P. miRNA Dysregulation in Breast Cancer. Cancer Res. 2013, 73, 6554–6562. [Google Scholar] [CrossRef] [PubMed]
- Zeng, K.; Zheng, W.; Mo, X.; Liu, F.; Li, M.; Liu, Z.; Zhang, W.; Hu, X. Dysregulated microRNAs involved in the progression of cervical neoplasm. Arch. Gynecol. Obstet. 2015, 292, 905–913. [Google Scholar] [CrossRef]
- Joshi, P.; Middleton, J.; Jeon, Y.-J.; Garofalo, M. MicroRNA in lung cancer. World J. Methodol. 2014, 4, 59–72. [Google Scholar] [CrossRef]
- Zhang, X.J.; Ye, H.; Zeng, C.W.; He, B.; Zhang, H.; Chen, Y.Q. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J. Hematol. Oncol. 2010, 3, 46. [Google Scholar] [CrossRef]
- Doghish, A.S.; Abulsoud, A.I.; Elshaer, S.S.; Abdelmaksoud, N.M.; Zaki, M.B.; El-Mahdy, H.A.; Ismail, A.; Fathi, D.; Elsakka, E.G. miRNAs as cornerstones in chronic lymphocytic leukemia pathogenesis and therapeutic resistance– An emphasis on the interaction of signaling pathways. Pathol.-Res. Pr. 2023, 243, 154363. [Google Scholar] [CrossRef]
- Mazeh, H.; Mizrahi, I.; Ilyayev, N.; Halle, D.; Brücher, B.L.; Bilchik, A.; Protic, M.; Daumer, M.; Stojadinovic, A.; Avital, I.; et al. The Diagnostic and Prognostic Role of microRNA in Colorectal Cancer—A Comprehensive review. J. Cancer 2013, 4, 281–295. [Google Scholar] [CrossRef]
- Ng, S.S.; Dong, Y.; Yu, J. MicroRNA dysregulation as a prognostic biomarker in colorectal cancer. Cancer Manag. Res. 2014, 6, 405–422. [Google Scholar] [CrossRef]
- Michael, M.Z.; O’Connor, S.M.; van Holst Pellekaan, N.G.; Young, G.P.; James, R.J. Reduced Accumulation of Specific mi-croRNAs in Colorectal Neoplasia. Mol. Cancer Res. 2003, 1, 882–891. [Google Scholar]
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Gunter, M.J.; Leitzmann, M.F. Obesity and colorectal cancer: Epidemiology, mechanisms and candidate genes. J. Nutr. Biochem. 2006, 17, 145–156. [Google Scholar] [CrossRef]
- Jochem, C.; Leitzmann, M. Obesity and Colorectal Cancer. In Obesity and Cancer; Pischon, T., Nimptsch, K., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 17–41. [Google Scholar] [CrossRef]
- Gulubova, M.; Vlaykova, T. Prognostic significance of mast cell number and microvascular density for the survival of patients with primary colorectal cancer. J. Gastroenterol. Hepatol. 2009, 24, 1265–1275. [Google Scholar] [CrossRef]
- Gonzalez-Gutierrez, L.; Motiño, O.; Barriuso, D.; de la Puente-Aldea, J.; Alvarez-Frutos, L.; Kroemer, G.; Palacios-Ramirez, R.; Senovilla, L. Obesity-Associated Colorectal Cancer. Int. J. Mol. Sci. 2024, 25, 8836. [Google Scholar] [CrossRef]
- Otani, K.; Kitayama, J.; Yasuda, K.; Nio, Y.; Iwabu, M.; Okudaira, S.; Aoki, J.; Yamauchi, T.; Kadowaki, T.; Nagawa, H. Adiponectin suppresses tumorigenesis in ApcMin/+ mice. Cancer Lett. 2010, 288, 177–182. [Google Scholar] [CrossRef]
- Wang, Z.V.; Scherer, P.E. Adiponectin, the past two decades. J. Mol. Cell Biol. 2016, 8, 93–100. [Google Scholar] [CrossRef]
- Kita, S.; Fukuda, S.; Maeda, N.; Shimomura, I. Japan Native adiponectin in serum binds to mammalian cells expressing T-cadherin, but not AdipoRs or calreticulin. eLife 2019, 8, e48675. [Google Scholar] [CrossRef] [PubMed]
- Obata, Y.; Kita, S.; Koyama, Y.; Fukuda, S.; Takeda, H.; Takahashi, M.; Fujishima, Y.; Nagao, H.; Masuda, S.; Tanaka, Y.; et al. Adiponectin/T-cadherin system enhances exosome biogenesis and decreases cellular ceramides by exosomal release. J. Clin. Investig. 2018, 3, e99680. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.X.; Holland, W.L. Adiponectin and its Hydrolase-Activated Receptors. J. Nat. Sci. 2017, 3, e396. [Google Scholar] [PubMed]
- Chaurasia, B.; Summers, S.A. Ceramides in Metabolism: Key Lipotoxic Players. Annu. Rev. Physiol. 2020, 83, 303–330. [Google Scholar] [CrossRef]
- Hammerschmidt, P.; Steculorum, S.M.; Bandet, C.L.; Del Río-Martín, A.; Steuernagel, L.; Kohlhaas, V.; Feldmann, M.; Varela, L.; Majcher, A.; Correia, M.Q.; et al. CerS6-dependent ceramide synthesis in hypothalamic neurons promotes ER/mitochondrial stress and impairs glucose homeostasis in obese mice. Nat. Commun. 2023, 14, 1–22. [Google Scholar] [CrossRef]
- Altura, B.M.; Shah, N.C.; Shah, G.J.; Zhang, A.; Li, W.; Zheng, T.; Perez-Albela, J.L.; Altura, B.T. Short-Term Mg Deficiency Upregulates Protein Kinase C Isoforms in Cardiovascular Tissues and Cells; Relation to NF-κB, Cytokines, Ceramide Sal-vage Sphingolipid Pathway and PKC-Zeta: Hypothesis and Review. Int. J. Clin. Exp. Med. 2014, 7, 1–21. [Google Scholar]
- Kim, J.; Yang, G.; Kim, Y.; Kim, J.; Ha, J. AMPK activators: Mechanisms of action and physiological activities. Exp. Mol. Med. 2016, 48, e224. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, W.; Song, Z.; Aji, G.; Liu, X.T.; Xia, P. Role of Sphingosine Kinase in Type 2 Diabetes Mellitus. Front. Endocrinol. 2021, 11, 627076. [Google Scholar] [CrossRef]
- Diniz, T.A.; Junior, E.A.d.L.; Teixeira, A.A.; Biondo, L.A.; da Rocha, L.A.F.; Valadão, I.C.; Silveira, L.S.; Cabral-Santos, C.; de Souza, C.O.; Neto, J.C.R. Aerobic training improves NAFLD markers and insulin resistance through AMPK-PPAR-α signaling in obese mice. Life Sci. 2021, 266, 118868. [Google Scholar] [CrossRef]
- Singh, V.; Ubaid, S. Role of Silent Information Regulator 1 (SIRT1) in Regulating Oxidative Stress and Inflammation. Inflammation 2020, 43, 1589–1598. [Google Scholar] [CrossRef]
- Ryu, J.; Galan, A.K.; Xin, X.; Dong, F.; Abdul-Ghani, M.A.; Zhou, L.; Wang, C.; Li, C.; Holmes, B.M.; Sloane, L.B.; et al. APPL1 Potentiates Insulin Sensitivity by Facilitating the Binding of IRS1/2 to the Insulin Receptor. Cell Rep. 2014, 7, 1227–1238. [Google Scholar] [CrossRef]
- Krapivinsky, G.; Krapivinsky, L.; Stotz, S.C.; Manasian, Y.; Clapham, D.E. POST, partner of stromal interaction molecule 1 (STIM1), targets STIM1 to multiple transporters. Proc. Natl. Acad. Sci. USA 2011, 108, 19234–19239. [Google Scholar] [CrossRef]
- Kline, S.L.; Cheeseman, I.M.; Hori, T.; Fukagawa, T.; Desai, A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 2006, 173, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, X.; Wang, P.; Chen, Q.; Xiong, L.; Tang, M.; Hong, C.; Lin, X.; Shi, K.; Liang, L.; et al. SRSF9 promotes colorectal cancer progression via stabilizing DSN1 mRNA in an m6A-related manner. J. Transl. Med. 2022, 20, 198. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Huang, S.; Ju, W.; Hou, Y.; Wang, Z.; Liu, Y.; Wu, L.; He, X. Elevated DSN1 expression is associated with poor survival in patients with hepatocellular carcinoma. Hum. Pathol. 2018, 81, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Wen, T.; Liu, D.; Wang, S.; Jiang, X.; Zhao, S.; Huang, G. DSN1 is a prognostic biomarker and correlated with clinical characterize in breast cancer. Int. Immunopharmacol. 2021, 101, 107605. [Google Scholar] [CrossRef]
- Pant, D.C.; Dorboz, I.; Schluter, A.; Fourcade, S.; Launay, N.; Joya, J.; Aguilera-Albesa, S.; Yoldi, M.E.; Casasnovas, C.; Willis, M.J.; et al. Loss of the sphingolipid desaturase DEGS1 causes hypomyelinating leukodystrophy. J. Clin. Investig. 2019, 129, 1240–1256. [Google Scholar] [CrossRef]
- Blackburn, N.B.; Michael, L.F.; Meikle, P.J.; Peralta, J.M.; Mosior, M.; McAhren, S.; Bui, H.H.; Bellinger, M.A.; Giles, C.; Kumar, S.; et al. Rare DEGS1 variant significantly alters de novo ceramide synthesis pathway. J. Lipid Res. 2019, 60, 1630–1639. [Google Scholar] [CrossRef]
- Zhang, D.; Tomisato, W.; Su, L.; Sun, L.; Choi, J.H.; Zhang, Z.; Wang, K.-W.; Zhan, X.; Choi, M.; Li, X.; et al. Skin-specific regulation of SREBP processing and lipid biosynthesis by glycerol kinase. Proc. Natl. Acad. Sci. USA 2017, 114, E5197–E5206. [Google Scholar] [CrossRef]
- Yan, M.; Wu, Y.; Peng, W.; Fu, C.; Giunta, S.; Chang, M.; Zhang, G.; Dou, M.; Xia, S.; Li, H.; et al. Exposure to particulate matter 2.5 and cigarette smoke induces the synthesis of lipid droplets by glycerol kinase. Clin. Exp. Pharmacol. Physiol. 2021, 48, 498–507. [Google Scholar] [CrossRef]
- Parker, J.L.; Deme, J.C.; Kolokouris, D.; Kuteyi, G.; Biggin, P.C.; Lea, S.M.; Newstead, S. Molecular basis for redox control by the human cystine/glutamate antiporter system xc−. Nat. Commun. 2021, 12, 7147. [Google Scholar] [CrossRef]
- Lin, W.; Wang, C.; Liu, G.; Bi, C.; Wang, X.; Zhou, Q.; Jin, H. SLC7A11/xCT in Cancer: Biological Functions and Therapeutic Implications. Am. J. Cancer Res. 2020, 10, 3106–3126. [Google Scholar] [PubMed]
- Liu, M.-R.; Zhu, W.-T.; Pei, D.-S. System Xc−: A key regulatory target of ferroptosis in cancer. Investig. New Drugs 2021, 39, 1123–1131. [Google Scholar] [CrossRef]
- Tu, H.; Costa, M. XIAP’s Profile in Human Cancer. Biomolecules 2020, 10, 1493. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, H.; Noguchi, K.; Masumi, A.; Murakami, Y.; Uehara, Y. BimEL is an important determinant for induction of anoikis sensitivity by mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitors. Mol. Cancer Ther. 2004, 3, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Morton, D.J.; Carey, J.; Havrda, M.C.; Chaudhary, J. Inhibitor of differentiation 4 (ID4): From development to cancer. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2015, 1855, 92–103. [Google Scholar] [CrossRef]
- Chen, H.-J.; Yu, Y.; Sun, Y.-X.; Huang, C.-Z.; Li, J.-Y.; Liu, F.; Guo, G.-X.; Ye, Y.-B.; Ding, X. Id4 Suppresses the Growth and Invasion of Colorectal Cancer HCT116 Cells through CK18-Related Inhibition of AKT and EMT Signaling. J. Oncol. 2021, 2021, 6660486. [Google Scholar] [CrossRef]
- Carey, J.P.; Asirvatham, A.J.; Galm, O.; Ghogomu, T.A.; Chaudhary, J. Inhibitor of differentiation 4 (Id4) is a potential tumor suppressor in prostate cancer. BMC Cancer 2009, 9, 173. [Google Scholar] [CrossRef]
- Baker, L.A.; Holliday, H.; Swarbrick, A. ID4 controls luminal lineage commitment in normal mammary epithelium and inhibits BRCA1 function in basal-like breast cancer. Endocr.-Relat. Cancer 2016, 23, R381–R392. [Google Scholar] [CrossRef]
- O’NEill, T.J.; Tofaute, M.J.; Krappmann, D. Function and targeting of MALT1 paracaspase in cancer. Cancer Treat. Rev. 2023, 117, 102568. [Google Scholar] [CrossRef]
- Luo, L.Y.; Kim, E.; Cheung, H.W.; Weir, B.A.; Dunn, G.P.; Shen, R.R.; Hahn, W.C. The Tyrosine Kinase Adaptor Protein FRS2 Is Oncogenic and Amplified in High-Grade Serous Ovarian Cancer. Mol. Cancer Res. 2015, 13, 502–509. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Z.; Cao, L.; Fan, G.; Ji, R.; Zhang, L.; Daji, S.; Zhu, H.; Wang, Y.; Zhou, G. FRS2 regulated by miR-429 and miR-206 promotes angiogenesis in osteosarcoma. Gene 2023, 898, 148118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Wang, Q.; Su, B.; Xu, H.; Sun, Y.; Sun, P.; Li, R.; Peng, X.; Cai, J. Role of RASA1 in cancer: A review and update (Review). Oncol. Rep. 2020, 44, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhan, X.; Yan, D.; Wang, Z. Circulating MicroRNA-21 Is Involved in Lymph Node Metastasis in Cervical Cancer by Targeting RASA. Int. J. Gynecol. Cancer 2016, 26, 810–816. [Google Scholar] [CrossRef]
- Slaby, O.; Svoboda, M.; Fabian, P.; Smerdova, T.; Knoflickova, D.; Bednarikova, M.; Nenutil, R.; Vyzula, R. Altered Expression of miR-21, miR-31, miR-143 and miR-145 Is Related to Clinicopathologic Features of Colorectal Cancer. Oncology 2007, 72, 397–402. [Google Scholar] [CrossRef]
- Toiyama, Y.; Takahashi, M.; Hur, K.; Nagasaka, T.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-21 as a Diagnostic and Prognostic Biomarker in Colorectal Cancer. JNCI J. Natl. Cancer Inst. 2013, 105, 849–859. [Google Scholar] [CrossRef]
- Zhu, M.; Huang, Z.; Zhu, D.; Zhou, X.; Shan, X.; Qi, L.-W.; Wu, L.; Cheng, W.; Zhu, J.; Zhang, L.; et al. A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget 2017, 8, 17081–17091. [Google Scholar] [CrossRef]
- Sarver, A.L.; French, A.J.; Borralho, P.M.; Thayanithy, V.; Oberg, A.L.; Silverstein, K.A.; Morlan, B.W.; Riska, S.M.; Boardman, L.A.; Cunningham, J.M.; et al. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer 2009, 9, 401. [Google Scholar] [CrossRef]
- Chen, E.; Li, Q.; Wang, H.; Zhang, P.; Zhao, X.; Yang, F.; Yang, J. MiR-32 promotes tumorigenesis of colorectal cancer by targeting BMP5. Biomed. Pharmacother. 2018, 106, 1046–1051. [Google Scholar] [CrossRef]
- Wu, J.; Ji, X.; Zhu, L.; Jiang, Q.; Wen, Z.; Xu, S.; Shao, W.; Cai, J.; Du, Q.; Zhu, Y.; et al. Up-regulation of microRNA-1290 impairs cytokinesis and affects the reprogramming of colon cancer cells. Cancer Lett. 2013, 329, 155–163. [Google Scholar] [CrossRef]
- Imaoka, H.; Toiyama, Y.; Fujikawa, H.; Hiro, J.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Mori, T.; Kato, T.; et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann. Oncol. 2016, 27, 1879–1886. [Google Scholar] [CrossRef]
- Gao, S.; Chen, L.; Lu, W.; Zhang, L.; Wang, L.; Zhu, H. miR-888 functions as an oncogene and predicts poor prognosis in colorectal cancer. Oncol. Lett. 2018, 15, 9101–9109. [Google Scholar] [CrossRef]
- Qu, Y.L.; Wang, H.F.; Sun, Z.Q.; Tang, Y.; Han, X.N.; Yu, X.B.; Liu, K. Up-regulated miR-155-5p promotes cell proliferation, invasion and metastasis in colorectal carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 6988. [Google Scholar] [PubMed] [PubMed Central]
- Song, C.; Wang, X.; Zhao, X.; Ai, J.; Qi, Y.; Chen, A. MicroRNA-325-3p contributes to colorectal carcinoma by targeting cytokeratin 18. Oncol. Lett. 2021, 21, 248. [Google Scholar] [CrossRef]
- Liu, W.; Qian, K.; Wei, X.; Deng, H.; Zhao, B.; Chen, Q.; Zhang, J.; Liu, H. miR-27a promotes proliferation, migration, and invasion of colorectal cancer by targeting FAM172A and acts as a diagnostic and prognostic biomarker. Oncol. Rep. 2017, 37, 3554–3564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-J.; Li, Y.; Zhou, H.; Xiao, H.-X.; Zhou, T. miR-20a is an independent prognostic factor in colorectal cancer and is involved in cell metastasis. Mol. Med. Rep. 2014, 10, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Chen, X.; Liu, Z.; Tian, X.; Huo, Z. miR-106a Reduces 5-Fluorouracil (5-FU) Sensitivity of Colorectal Cancer by Targeting Dual-Specificity Phosphatases 2 (DUSP2). Med. Sci. Monit. 2018, 24, 4944–4951. [Google Scholar] [CrossRef] [PubMed]
- Fasihi, A.; Soltani, B.M.; Ranjbaran, Z.S.; Bahonar, S.; Norouzi, R.; Nasiri, S. Hsa-miR-942 fingerprint in colorectal cancer through Wnt signaling pathway. Gene 2019, 712, 143958. [Google Scholar] [CrossRef]
- Zhou, W.; Li, X.; Liu, F.; Xiao, Z.; He, M.; Shen, S.; Liu, S. MiR-135a promotes growth and invasion of colorectal cancer via metastasis suppressor 1 in vitro. Acta Biochim. Biophys. Sin. 2012, 44, 838–846. [Google Scholar] [CrossRef]
- Li, J.; Liang, H.; Bai, M.; Ning, T.; Wang, C.; Fan, Q.; Wang, Y.; Fu, Z.; Wang, N.; Liu, R.; et al. Correction: miR-135b Promotes Cancer Progression by Targeting Transforming Growth Factor Beta Receptor II (TGFBR2) in Colorectal Cancer. PLoS ONE 2015, 10, e0145589. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Zhang, Q.; Lin, R. miR-182 contributes to cell proliferation, invasion and tumor growth in colorectal cancer by targeting DAB2IP. Int. J. Biochem. Cell Biol. 2019, 111, 27–36. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Lin, Y.-M.; Chung, H.-C.; Lang, Y.-D.; Lin, C.-J.; Huang, J.; Wang, W.-C.; Lin, F.-M.; Chen, Z.; Huang, H.-D.; et al. miR-103/107 Promote Metastasis of Colorectal Cancer by Targeting the Metastasis Suppressors DAPK and KLF4. Cancer Res. 2012, 72, 3631–3641. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, M.; Shan, X.; Zhou, X.; Wang, T.; Zhang, J.; Tao, J.; Cheng, W.; Chen, G.; Li, J.; et al. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene 2019, 687, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Zou, J.; Dong, Z.; Zeng, Q.; Wu, D.; Liu, L. Up-regulated miR-17 promotes cell proliferation, tumour growth and cell cycle progression by targeting the RND3 tumour suppressor gene in colorectal carcinoma. Biochem. J. 2012, 442, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shi, K.; Wang, Y.; Song, M.; Zhou, W.; Tu, H.; Lin, Z. Clinical value of integrated-signature miRNAs in colorectal cancer: miRNA expression profiling analysis and experimental validation. Oncotarget 2015, 6, 37544–37556. [Google Scholar] [CrossRef] [PubMed]
- Pellatt, D.F.; Stevens, J.R.; Wolff, R.K.; Mullany, L.E.; Herrick, J.S.; Samowitz, W.; Slattery, M.L. Expression Profiles of miRNA Subsets Distinguish Human Colorectal Carcinoma and Normal Colonic Mucosa. Clin. Transl. Gastroenterol. 2016, 7, e152. [Google Scholar] [CrossRef]
- Huang, L.; Wang, X.; Wen, C.; Yang, X.; Song, M.; Chen, J.; Wang, C.; Zhang, B.; Wang, L.; Iwamoto, A.; et al. Hsa-miR-19a is associated with lymph metastasis and mediates the TNF-α induced epithelial-to-mesenchymal transition in colorectal cancer. Sci. Rep. 2015, 5, 13350. [Google Scholar] [CrossRef]
- Kang, E.; Jung, S.C.; Nam, S.K.; Park, Y.; Seo, S.H.; Park, K.U.; Oh, H.-K.; Kim, D.-W.; Kang, S.-B.; Lee, H.S. Tissue miR-200c-3p and circulating miR-1290 as potential prognostic biomarkers for colorectal cancer. Sci. Rep. 2022, 12, 2295. [Google Scholar] [CrossRef]
- Pan, Z.; Xie, R.; Song, W.; Gao, C. MicroRNA-592 promotes cell proliferation, migration and invasion in colorectal cancer by directly targeting SPARC. Mol. Med. Rep. 2021, 23, 1. [Google Scholar] [CrossRef]
- Rapti, S.-M.; Kontos, C.K.; Papadopoulos, I.N.; Scorilas, A. High miR-96 levels in colorectal adenocarcinoma predict poor prognosis, particularly in patients without distant metastasis at the time of initial diagnosis. Tumor Biol. 2016, 37, 11815–11824. [Google Scholar] [CrossRef]
- Ji, D.; Chen, Z.; Li, M.; Zhan, T.; Yao, Y.; Zhang, Z.; Xi, J.; Yan, L.; Gu, J. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol. Cancer 2014, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Deng, Y.-N.; Wang, X.; Xia, Z.; He, Y.; Zhang, P.; Syed, S.E.; Li, Q.; Liang, S. miR-26a enhances colorectal cancer cell growth by targeting RREB1 deacetylation to activate AKT-mediated glycolysis. Cancer Lett. 2021, 521, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, R.; Kawada, K.; Sakai, Y. The Molecular Basis and Therapeutic Potential of Let-7 MicroRNAs against Colorectal Cancer. Can. J. Gastroenterol. Hepatol. 2018, 2018, 5769591. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.-B.; Qiu, H.; Chen, J.-M.; Shi, W.; Chen, Y.-S. MicroRNA-202-5p functions as a tumor suppressor in colorectal carcinoma by directly targeting SMARCC1. Gene 2018, 676, 329–335. [Google Scholar] [CrossRef]
- Guo, L.; Fu, J.; Sun, S.; Zhu, M.; Zhang, L.; Niu, H.; Chen, Z.; Zhang, Y.; Guo, L.; Wang, S. MicroRNA-143-3p inhibits colorectal cancer metastases by targeting ITGA6 and ASAP3. Cancer Sci. 2018, 110, 805–816. [Google Scholar] [CrossRef]
- Tazawa, H.; Tsuchiya, N.; Izumiya, M.; Nakagama, H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. USA 2007, 104, 15472–15477. [Google Scholar] [CrossRef]
- Zhang, G.; Xia, S.; Tian, H.; Liu, Z.; Zhou, T. Clinical significance of miR-22 expression in patients with colorectal cancer. Med. Oncol. 2012, 29, 3108–3112. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, J.; Li, C.; Kong, J.; Wang, J.; Wu, Y.; Xu, E.; Lai, M. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett. 2015, 356, 781–790. [Google Scholar] [CrossRef]
- Cong, J.; Gong, J.; Yang, C.; Xia, Z.; Zhang, H. miR-22 Suppresses Tumor Invasion and Metastasis in Colorectal Cancer by Targeting NLRP3. Cancer Manag. Res. 2020, 12, 5419–5429. [Google Scholar] [CrossRef]
- Sachdeva, M.; Mo, Y.-Y. miR-145-mediated suppression of cell growth, invasion and metastasis. Am. J. Transl. Res. 2010, 2, 170–180. [Google Scholar]
- Wang, W.; Ji, G.; Xiao, X.; Chen, X.; Qin, W.-W.; Yang, F.; Li, Y.-F.; Fan, L.-N.; Xi, W.-J.; Huo, Y.; et al. Epigenetically regulated miR-145 suppresses colon cancer invasion and metastasis by targeting LASP1. Oncotarget 2016, 7, 68674–68687. [Google Scholar] [CrossRef]
- Liang, Z.; Li, X.; Liu, S.; Li, C.; Wang, X.; Xing, J. MiR-141–3p inhibits cell proliferation, migration and invasion by targeting TRAF5 in colorectal cancer. Biochem. Biophys. Res. Commun. 2019, 514, 699–705. [Google Scholar] [CrossRef]
- Tong, S.J.; Zhang, X.Y.; Guo, H.F.; Yang, J.; Qi, Y.P.; Lu, S. Study on effects of miR-141-3p in proliferation, migration, invasion and apoptosis of colon cancer cells by inhibiting Bcl2. Clin. Transl. Oncol. 2021, 23, 2526–2535. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Yu, X.; Chen, Y.; Cai, W. MiR-1-3p Suppresses Colorectal Cancer Cell Proliferation and Metastasis by Inhibiting YWHAZ-Mediated Epithelial–Mesenchymal Transition. Front. Oncol. 2021, 11, 634596. [Google Scholar] [CrossRef] [PubMed]
- Vychytilova-Faltejskova, P.; Merhautova, J.; Machackova, T.; Gutierrez-Garcia, I.; Garcia-Solano, J.; Radova, L.; Brchnelova, D.; Slaba, K.; Svoboda, M.; Halamkova, J.; et al. MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogenesis 2017, 6, 399. [Google Scholar] [CrossRef]
- Chen, X.; Xu, X.; Pan, B.; Zeng, K.; Xu, M.; Liu, X.; He, B.; Pan, Y.; Sun, H.; Wang, S. miR-150-5p suppresses tumor progression by targeting VEGFA in colorectal cancer. Aging 2018, 10, 3421–3437. [Google Scholar] [CrossRef]
- Yang, L.; Men, W.-L.; Yan, K.-M.; Tie, J.; Nie, Y.-Z.; Xiao, H.-J. MiR-340-5p is a potential prognostic indicator of colorectal cancer and modulates ANXA3. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4837–4845. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, H.; Song, Y.; Gu, Q.; Zhang, L.; Xie, Q.; Xu, J.; Zhang, M. MiR-325 Promotes Oxaliplatin-Induced Cytotoxicity Against Colorectal Cancer Through the HSPA12B/PI3K/AKT/Bcl-2 Pathway. Dig. Dis. Sci. 2020, 66, 2651–2660. [Google Scholar] [CrossRef]
- Li, Y.; Kuscu, C.; Banach, A.; Zhang, Q.; Pulkoski-Gross, A.; Kim, D.; Liu, J.; Roth, E.; Li, E.; Shroyer, K.R.; et al. miR-181a-5p Inhibits Cancer Cell Migration and Angiogenesis via Downregulation of Matrix Metalloproteinase-14. Cancer Res. 2015, 75, 2674–2685. [Google Scholar] [CrossRef]
- Li, C.; Zhao, L.; Chen, Y.; He, T.; Chen, X.; Mao, J.; Li, C.; Lyu, J.; Meng, Q.H. MicroRNA-21 promotes proliferation, migration, and invasion of colorectal cancer, and tumor growth associated with down-regulation of sec23a expression. BMC Cancer 2016, 16, 605. [Google Scholar] [CrossRef]
- Wu, W.; Yang, J.; Feng, X.; Wang, H.; Ye, S.; Yang, P.; Tan, W.; Wei, G.; Zhou, Y. MicroRNA-32 (miR-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Mol. Cancer 2013, 12, 30. [Google Scholar] [CrossRef]
- Cellura, D.; Pickard, K.; Quaratino, S.; Parker, H.; Strefford, J.; Thomas, G.; Mitter, R.; Mirnezami, A.; Peake, N. miR-19–Mediated Inhibition of Transglutaminase-2 Leads to Enhanced Invasion and Metastasis in Colorectal Cancer. Mol. Cancer Res. 2015, 13, 1095–1105. [Google Scholar] [CrossRef]
- Yu, F.-B.; Sheng, J.; Yu, J.-M.; Liu, J.-H.; Qin, X.-X.; Mou, B. MiR-19a-3p regulates the Forkhead box F2-mediated Wnt/β-catenin signaling pathway and affects the biological functions of colorectal cancer cells. World J. Gastroenterol. 2020, 26, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tu, R.; Li, K.; Ye, P.; Cui, X. Tumor Suppressor PTPRJ Is a Target of miR-155 in Colorectal Cancer. J. Cell. Biochem. 2017, 118, 3391–3400. [Google Scholar] [CrossRef] [PubMed]
- Meerson, A.; Yehuda, H. Leptin and insulin up-regulate miR-4443 to suppress NCOA1 and TRAF4, and decrease the invasiveness of human colon cancer cells. BMC Cancer 2016, 16, 882. [Google Scholar] [CrossRef] [PubMed]
- Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13421–13426. [Google Scholar] [CrossRef]
- Liu, M.-W.; Su, M.-X.; Zhang, W.; Wang, Y.H.; Qin, L.-F.; Liu, X.; Tian, M.-L.; Qian, C.-Y. Effect of Melilotus suaveolens extract on pulmonary microvascular permeability by downregulating vascular endothelial growth factor expression in rats with sepsis. Mol. Med. Rep. 2015, 11, 3308–3316. [Google Scholar] [CrossRef]
- Xing, Y.; Jing, H.; Zhang, Y.; Suo, J.; Qian, M. MicroRNA-141-3p affected proliferation, chemosensitivity, migration and invasion of colorectal cancer cells by targeting EGFR. Int. J. Biochem. Cell Biol. 2020, 118, 105643. [Google Scholar] [CrossRef]
- Jiang, R.; Chen, X.; Ge, S.; Wang, Q.; Liu, Y.; Chen, H.; Xu, J.; Wu, J. MiR-21-5p Induces Pyroptosis in Colorectal Cancer via TGFBI. Front. Oncol. 2021, 10, 610545. [Google Scholar] [CrossRef]
- Jiang, X.; Finucane, H.K.; Schumacher, F.R.; Schmit, S.L.; Tyrer, J.P.; Han, Y.; Michailidou, K.; Lesseur, C.; Kuchenbaecker, K.B.; Dennis, J.; et al. Shared heritability and functional enrichment across six solid cancers. Nat. Commun. 2019, 10, 431. [Google Scholar] [CrossRef]
- He, Z.; Dang, J.; Song, A.; Cui, X.; Ma, Z.; Zhang, Y. The involvement of miR-150/β-catenin axis in colorectal cancer progression. Biomed. Pharmacother. 2020, 121, 109495. [Google Scholar] [CrossRef]
- Li, C.; Du, X.; Xia, S.; Chen, L. MicroRNA-150 inhibits the proliferation and metastasis potential of colorectal cancer cells by targeting iASPP. Oncol. Rep. 2018, 40, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhao, X.; Zhou, Y.; Hu, Y. miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol. Rep. 2012, 28, 1346–1352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-L.; Xie, F.-J.; Tang, C.-H.; Xu, W.-R.; Ding, X.-S.; Liang, J. miR-340 suppresses tumor growth and enhances chemosensitivity of colorectal cancer by targeting RLIP76. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2875–2886. [Google Scholar] [PubMed]
- Arivazhagan, R.; Lee, J.; Bayarsaikhan, D.; Kwak, P.; Son, M.; Byun, K.; Salekdeh, G.H.; Lee, B. MicroRNA-340 inhibits the proliferation and promotes the apoptosis of colon cancer cells by modulating REV3L. Oncotarget 2017, 9, 5155–5168. [Google Scholar] [CrossRef]
- Takeyama, H.; Yamamoto, H.; Yamashita, S.; Wu, X.; Takahashi, H.; Nishimura, J.; Haraguchi, N.; Miyake, Y.; Suzuki, R.; Murata, K.; et al. Decreased miR-340 Expression in Bone Marrow Is Associated with Liver Metastasis of Colorectal Cancer. Mol. Cancer Ther. 2014, 13, 976–985. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, X.; Li, Q.; Jin, Y.; Liu, X.; Wang, F.; Mao, Y.; Hua, D. Integrated analysis identifies low microRNA-215 expression as associated with a poor prognosis of patients with colorectal cancer through the IKβ-α signaling pathway. Transl. Cancer Res. 2020, 9, 5233–5244. [Google Scholar] [CrossRef]
- Machackova, T.; Vychytilova-Faltejskova, P.; Souckova, K.; Trachtova, K.; Brchnelova, D.; Svoboda, M.; Kiss, I.; Prochazka, V.; Kala, Z.; Slaby, O. MiR-215-5p Reduces Liver Metastasis in an Experimental Model of Colorectal Cancer through Regulation of ECM-Receptor Interactions and Focal Adhesion. Cancers 2020, 12, 3518. [Google Scholar] [CrossRef]
- Tang, X.; Shi, X.; Wang, N.; Peng, W.; Cheng, Z. MicroRNA-215-3p Suppresses the Growth, Migration, and Invasion of Colorectal Cancer by Targeting FOXM1. Technol. Cancer Res. Treat. 2019, 18, 1533033819874776. [Google Scholar] [CrossRef]
- Han, P.; Li, J.-W.; Zhang, B.-M.; Lv, J.-C.; Li, Y.-M.; Gu, X.-Y.; Yu, Z.-W.; Jia, Y.-H.; Bai, X.-F.; Li, L.; et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol. Cancer 2017, 16, 9. [Google Scholar] [CrossRef]
- Zhao, S.; Mi, Y.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Xu, Y.; Cai, S.; Li, X.; Li, D. Highly-metastatic colorectal cancer cell released miR-181a-5p-rich extracellular vesicles promote liver metastasis by activating hepatic stellate cells and remodelling the tumour microenvironment. J. Extracell. Vesicles 2022, 11, e12186. [Google Scholar] [CrossRef]
- Yue, C.; Chen, J.; Li, Z.; Li, L.; Chen, J.; Guo, Y. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J. Exp. Clin. Cancer Res. 2020, 39, 240. [Google Scholar] [CrossRef]
- Ge, T.; Xiang, P.; Mao, H.; Tang, S.; Zhou, J.; Zhang, Y. Inhibition of miR-96 enhances the sensitivity of colorectal cancer cells to oxaliplatin by targeting TPM1. Exp. Ther. Med. 2020, 20, 2134–2140. [Google Scholar] [CrossRef]
- He, P.Y.; Yip, W.K.; Jabar, M.F.; Mohtarrudin, N.; Dusa, N.M.; Seow, H.F. Effect of the miR-96-5p inhibitor and mimic on the migration and invasion of the SW480-7 colorectal cancer cell line. Oncol. Lett. 2019, 18, 1949–1960. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, R.; Yang, F.; Cheng, R.; Chen, X.; Cui, S.; Gu, Y.; Sun, W.; You, C.; Liu, Z.; et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol. Cancer 2017, 16, 53. [Google Scholar] [CrossRef]
- Li, H.; Huang, B. miR-19a targeting CLCA4 to regulate the proliferation, migration, and invasion of colorectal cancer cells. Eur. J. Histochem. 2022, 66, 3381. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Ai, R.; Mu, S.; Niu, X.; Guo, Z.; Liu, L. MiR-19a suppresses ferroptosis of colorectal cancer cells by targeting IREB2. Bioengineered 2022, 13, 12021–12029. [Google Scholar] [CrossRef]
- Li, J.; Chen, H.; Sun, G.; Zhang, X.; Ye, H.; Wang, P. Role of miR-21 in the diagnosis of colorectal cancer: Meta-analysis and bioinformatics. Pathol.-Res. Pr. 2023, 248, 154670. [Google Scholar] [CrossRef]
- Cao, L.-Q.; Yang, X.-W.; Chen, Y.-B.; Zhang, D.-W.; Jiang, X.-F.; Xue, P. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol. Cancer 2019, 18, 148. [Google Scholar] [CrossRef]
- Chen, D.; Guo, Y.; Chen, Y.; Guo, Q.; Chen, J.; Li, Y.; Zheng, Q.; Jiang, M.; Xi, M.; Cheng, L. LncRNA growth arrest-specific transcript 5 targets miR-21 gene and regulates bladder cancer cell proliferation and apoptosis through PTEN. Cancer Med. 2020, 9, 2846–2858. [Google Scholar] [CrossRef]
- Ding, S.; Zheng, Y.; Xu, Y.; Zhao, X.; Zhong, C. MiR-21/PTEN Signaling Modulates the Chemo-Sensitivity to 5-Fluorouracil in Human Lung Adenocarcinoma A549 Cells. Int. J. Clin. Exp. Pathol. 2019, 12, 2339–2352. [Google Scholar]
- He, Q.; Ye, A.; Ye, W.; Liao, X.; Qin, G.; Xu, Y.; Yin, Y.; Luo, H.; Yi, M.; Xian, L.; et al. Cancer-secreted exosomal miR-21-5p induces angiogenesis and vascular permeability by targeting KRIT1. Cell Death Dis. 2021, 12, 576. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J.; et al. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Res. 2019, 79, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, S.; Huang, R.; Zhou, Y.; Zou, Y.; Yang, W.; Zhang, J. Adiponectin inhibits LPS-induced nucleus pulposus cell pyroptosis through the miR-135a-5p/TXNIP signaling pathway. Aging 2023, 15, 13680–13692. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cheng, Y.; Xiaoran, Y.; Chen, F.; Jie, W.; Yahui, H.; Yue, W.; Dong, L.; Yumei, L.; Cheng, F.; et al. Globular adiponectin induces esophageal adenocarcinoma cell pyroptosis via the miR-378a-3p/UHRF1 axis. Environ. Toxicol. 2024, 40, 429–444. [Google Scholar] [CrossRef]
- Li, Y.; Cai, X.; Guan, Y.; Wang, L.; Wang, S.; Li, Y.; Fu, Y.; Gao, X.; Su, G.; Karmazyn, M. Adiponectin Upregulates MiR-133a in Cardiac Hypertrophy through AMPK Activation and Reduced ERK1/2 Phosphorylation. PLoS ONE 2016, 11, e0148482. [Google Scholar] [CrossRef]
- Duan, H.; Zhang, X.; Song, R.; Liu, T.; Zhang, Y.; Yu, A. Upregulation of miR-133a by adiponectin inhibits pyroptosis pathway and rescues acute aortic dissection. Acta Biochim. Biophys. Sin. 2020, 52, 988–997. [Google Scholar] [CrossRef]
- Xie, B.; Ding, Q.; Han, H.; Wu, D. miRCancer: A microRNA–cancer association database constructed by text mining on literature. Bioinformatics 2013, 29, 638–644. [Google Scholar] [CrossRef]
- Xu, F.; Wang, Y.; Ling, Y.; Zhou, C.; Wang, H.; Teschendorff, A.E.; Zhao, Y.; Zhao, H.; He, Y.; Zhang, G.; et al. dbDEMC 3.0: Functional Exploration of Differentially Expressed miRNAs in Cancers of Human and Model Organisms. Genom. Proteom. Bioinform. 2022, 20, 446–454. [Google Scholar] [CrossRef]
- Chan, S.K.; Griffith, O.L.; Tai, I.T.; Jones, S.J. Meta-analysis of Colorectal Cancer Gene Expression Profiling Studies Identifies Consistently Reported Candidate Biomarkers. Cancer Epidemiol. Biomark. Prev. 2008, 17, 543–552. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Lin, Y.-C.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA–target interactions. Nucleic Acids Res. 2021, 50, D222–D230. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Agrawal, A.; Balcı, H.; Hanspers, K.; Coort, S.L.; Martens, M.; Slenter, D.N.; Ehrhart, F.; Digles, D.; Waagmeester, A.; Wassink, I.; et al. WikiPathways 2024: Next generation pathway database. Nucleic Acids Res. 2023, 52, D679–D689. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef] [PubMed]
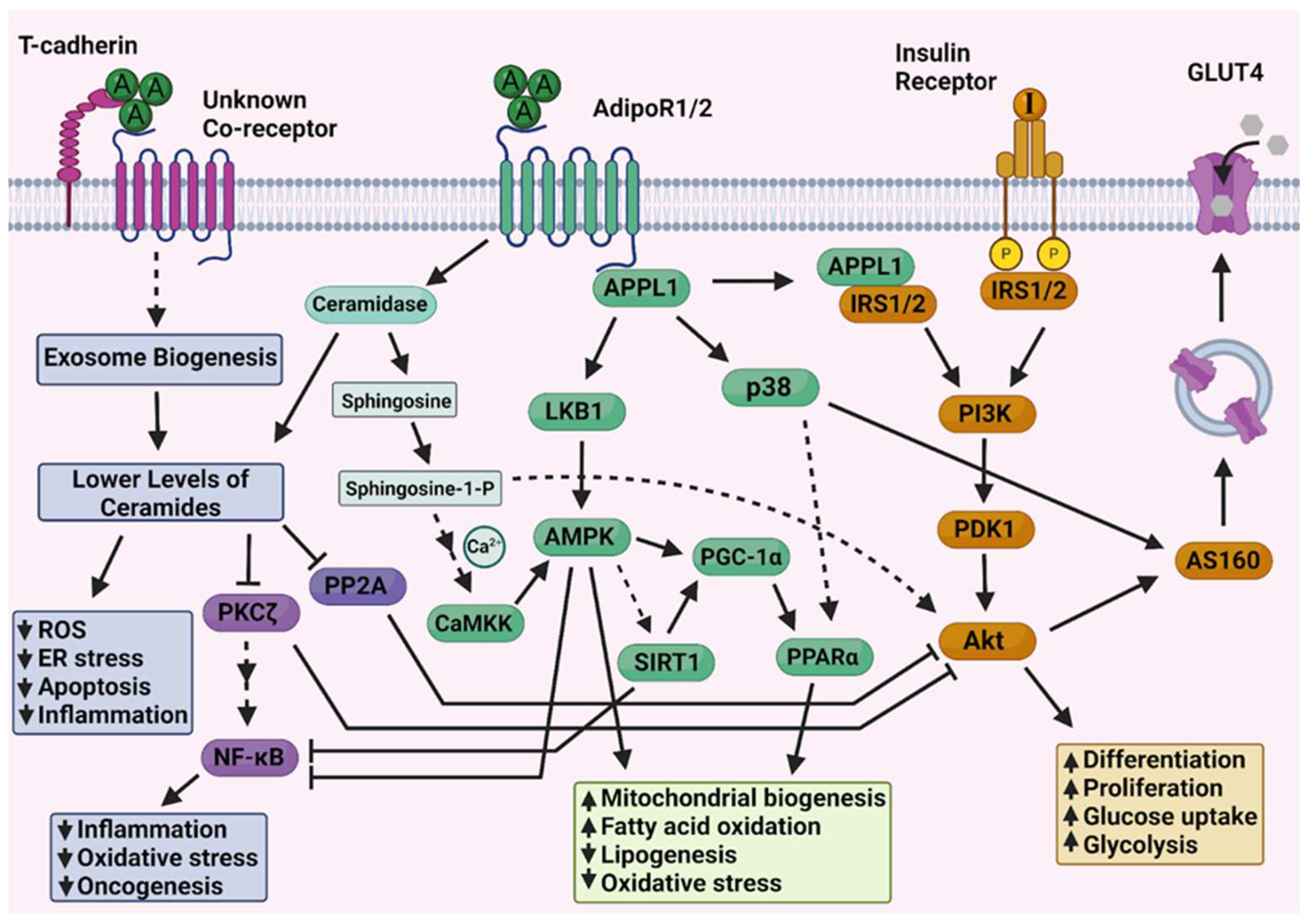

| miRNA | Target Genes from Pathway | Profile | VC Score |
|---|---|---|---|
| hsa-miR-215-5p | APPL1 | downregulated | −5 |
| hsa-miR-340-5p | APPL1/AKT1 | downregulated | −4 |
| hsa-miR-181a-5p | CDH13 | downregulated | −4 |
| hsa-miR-150-5p | PRKAB1/ADIPOR2 | downregulated | −8 |
| hsa-miR-96-5p | APPL1/IRS1/AKT1 | upregulated | 13 |
| hsa-miR-19a-3p | PPARA/PRKAA1/AKT1/ADIPOR2 | upregulated | 9 |
| hsa-miR-21-5p | PPARA/PRKAB2/APPL1 | upregulated | 16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbolov, M.; Slavova, S.; Nedeva, N.; Ivanov, K.; Kolev, N.; Komosinska-Vassev, K.; Ivanova, D.; Vankova, D.; Kiselova-Kaneva, Y. Dysregulated miRNAs Targeting Adiponectin Signaling in Colorectal Cancer. Int. J. Mol. Sci. 2025, 26, 7196. https://doi.org/10.3390/ijms26157196
Barbolov M, Slavova S, Nedeva N, Ivanov K, Kolev N, Komosinska-Vassev K, Ivanova D, Vankova D, Kiselova-Kaneva Y. Dysregulated miRNAs Targeting Adiponectin Signaling in Colorectal Cancer. International Journal of Molecular Sciences. 2025; 26(15):7196. https://doi.org/10.3390/ijms26157196
Chicago/Turabian StyleBarbolov, Momchil, Svetla Slavova, Neda Nedeva, Krasimir Ivanov, Nikola Kolev, Katarzyna Komosinska-Vassev, Diana Ivanova, Deyana Vankova, and Yoana Kiselova-Kaneva. 2025. "Dysregulated miRNAs Targeting Adiponectin Signaling in Colorectal Cancer" International Journal of Molecular Sciences 26, no. 15: 7196. https://doi.org/10.3390/ijms26157196
APA StyleBarbolov, M., Slavova, S., Nedeva, N., Ivanov, K., Kolev, N., Komosinska-Vassev, K., Ivanova, D., Vankova, D., & Kiselova-Kaneva, Y. (2025). Dysregulated miRNAs Targeting Adiponectin Signaling in Colorectal Cancer. International Journal of Molecular Sciences, 26(15), 7196. https://doi.org/10.3390/ijms26157196







