Unnatural Amino Acid Photo-Crosslinking Sheds Light on Gating of the Mechanosensitive Ion Channel OSCA1.2
Abstract
1. Introduction
2. Results
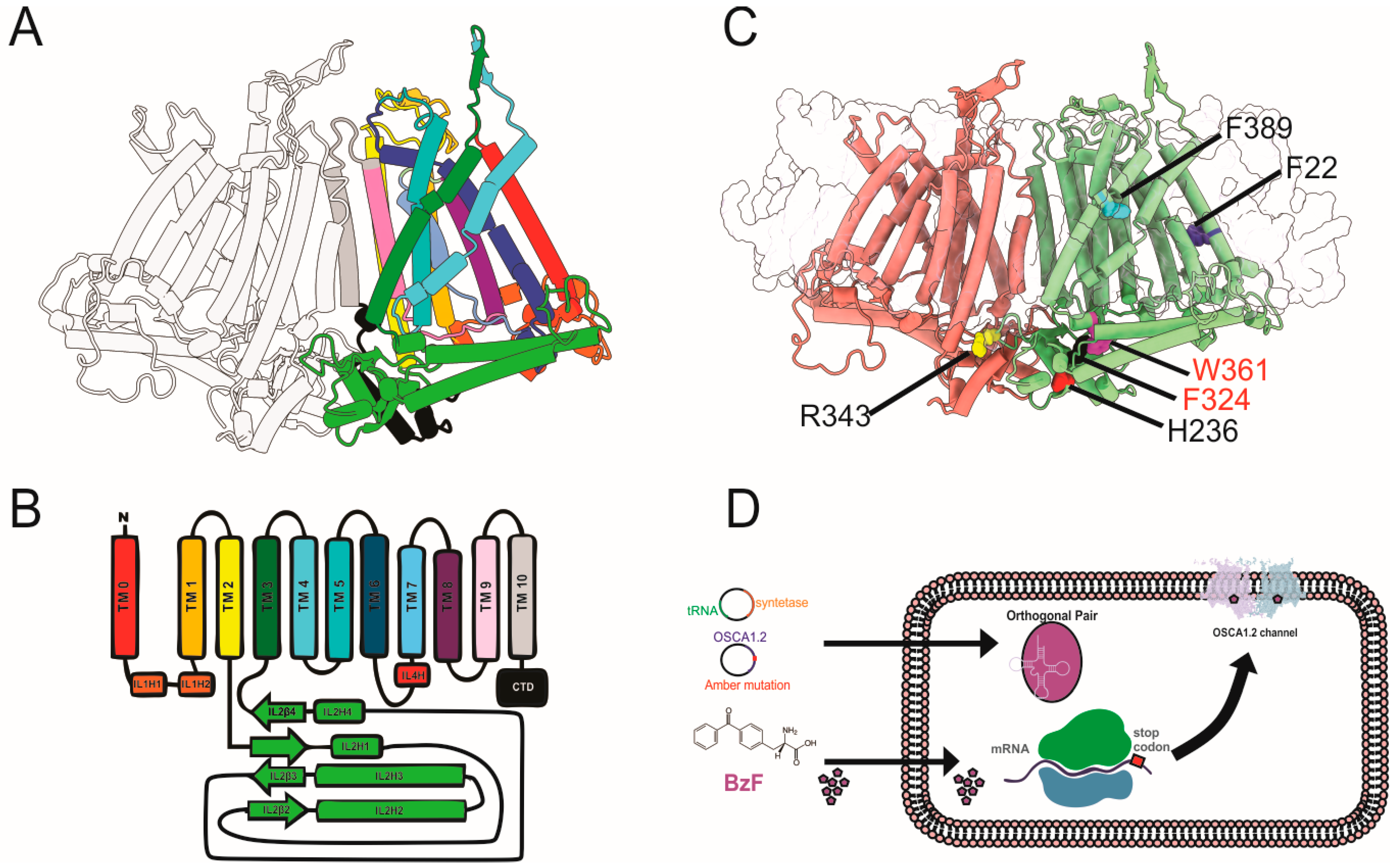
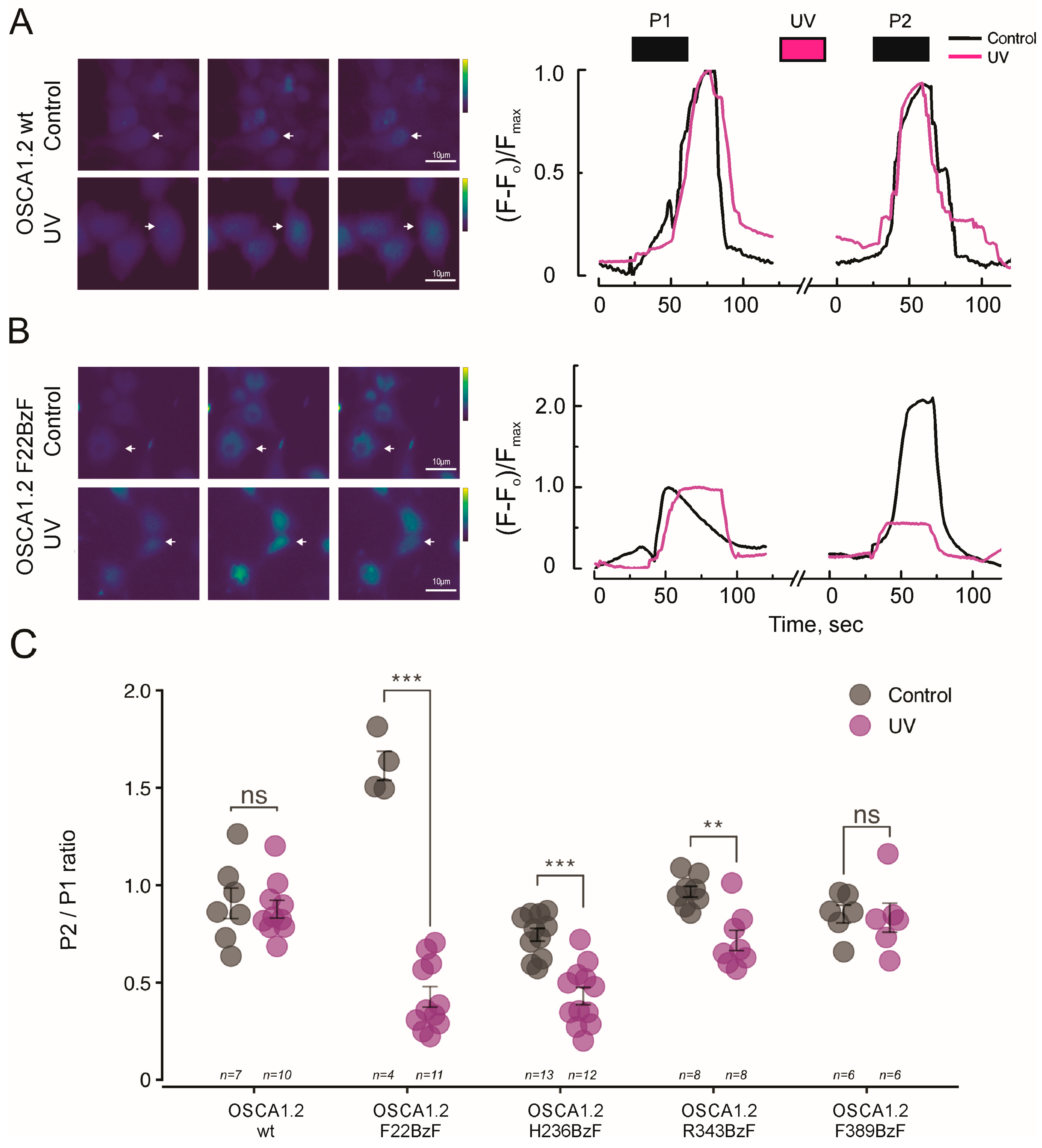
2.1. Conformational Dynamics at F22, H236, and R343 Is Required for OSCA1.2 Gating
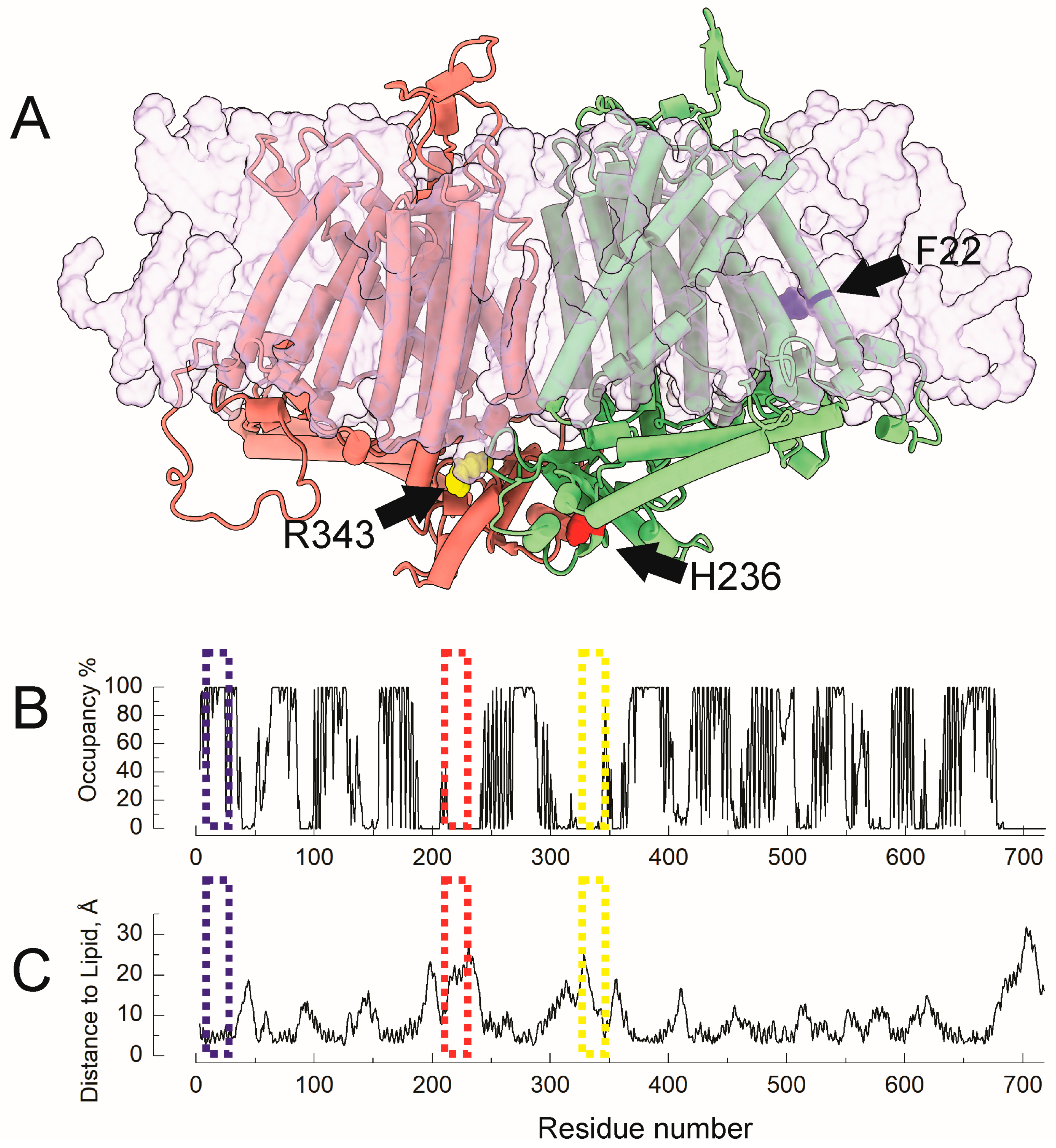
2.2. Molecular Dynamics Reveal Differential Lipid Accessibility for F22, R343, and H236
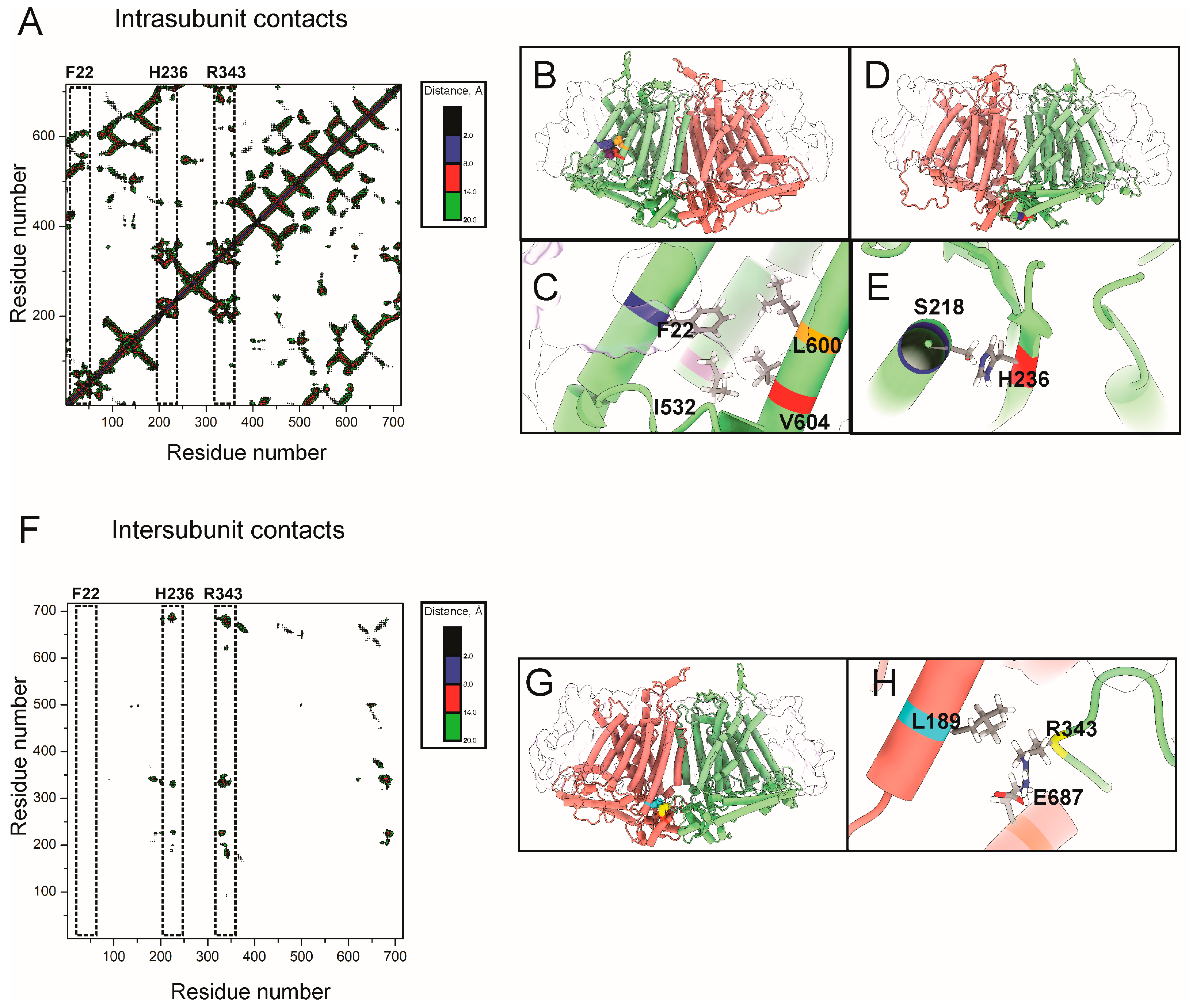
2.3. Contact and Correlation Analyses Suggest Dual Allosteric Pathways to Y519 in OSCA1
3. Discussion
4. Materials and Methods
4.1. Molecular Biology
4.2. Cell Culture, Transfection, and UAA Incorporation
4.3. Calcium Imaging
4.4. Molecular Simulations
4.5. Dynamic Cross Correlation Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnadóttir, J.; Chalfie, M. Eukaryotic mechanosensitive channels. Annu. Rev. Biophys. 2010, 39, 111–137. [Google Scholar] [CrossRef]
- Kefauver, J.M.; Ward, A.B.; Patapoutian, A. Discoveries in structure and physiology of mechanically activated ion channels. Nature 2020, 587, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Tian, W.; Kleist, T.; He, K.; Garcia, V.; Bai, F.; Hao, Y.; Luan, S.; Li, L. DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res. 2014, 24, 632–635. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yuan, F.; Wang, X.; Zhu, S.; Pei, Z.M. Evolution of osmosensing OSCA1 Ca2+ channel family coincident with plant transition from water to land. Plant Genome 2022, 15, e20198. [Google Scholar] [CrossRef] [PubMed]
- Murthy, S.E.; Dubin, A.E.; Whitwam, T.; Jojoa-Cruz, S.; Cahalan, S.M.; Mousavi, S.A.R.; Ward, A.B.; Patapoutian, A. OSCA/TMEM63 are an Evolutionarily Conserved Family of Mechanically Activated Ion Channels. eLife 2018, 7, e41844. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Wu, D.; Zhan, Y.; Li, F.; Zang, Y.Y.; Teng, X.Y.; Zhang, L.; Duan, G.F.; Wang, H.; Xu, R.; et al. Cation Channel TMEM63A Autonomously Facilitates Oligodendrocyte Differentiation at an Early Stage. Neurosci. Bull. 2025, 41, 615–632. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.L.; Li, J.Y.; Chen, X.; Liu, J.W.; Zhang, Q.; Liu, J.Y.; Wen, J.; Wang, N.; Lei, M.; Wei, J.P.; et al. Mechanosensitive channels TMEM63A and TMEM63B mediate lung inflation-induced surfactant secretion. J. Clin. Investig. 2024, 134, e174508. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Helman, G.; Murthy, S.E.; Ji, H.; Crawford, J.; Kubisiak, T.; Bent, S.J.; Xiao, J.; Taft, R.J.; Coombs, A.; et al. Heterozygous Variants in the Mechanosensitive Ion Channel TMEM63A Result in Transient Hypomyelination during Infancy. Am. J. Hum. Genet. 2019, 105, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Fukumura, S.; Hiraide, T.; Yamamoto, A.; Tsuchida, K.; Aoto, K.; Nakashima, M.; Saitsu, H. A novel de novo TMEM63A variant in a patient with severe hypomyelination and global developmental delay. Brain Dev. 2022, 44, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Chen, Y.; Ren, Y.; Yang, W.; Yu, W.; Zhou, Y.; Guo, J.; Hu, J.; Chen, X.; Gu, Y.; et al. Deep learning analyses of splicing variants identify the link of PCP4 with amyotrophic lateral sclerosis. Brain A J. Neurol. 2025, 148, 2331–2347. [Google Scholar] [CrossRef] [PubMed]
- Jojoa-Cruz, S.; Saotome, K.; Murthy, S.E.; Tsui, C.C.A.; Sansom, M.S.; Patapoutian, A.; Ward, A.B. Cryo-EM structure of the mechanically activated ion channel OSCA1.2. eLife 2018, 7, e41845. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, J.; Sun, L. Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2. Nat. Commun. 2018, 9, 5060. [Google Scholar] [CrossRef] [PubMed]
- Jojoa-Cruz, S.; Dubin, A.E.; Lee, W.H.; Ward, A.B. Structure-guided mutagenesis of OSCAs reveals differential activation to mechanical stimuli. eLife 2024, 12, RP93147. [Google Scholar] [CrossRef] [PubMed]
- Goettig, P.; Koch, N.G.; Budisa, N. Non-Canonical Amino Acids in Analyses of Protease Structure and Function. Int. J. Mol. Sci. 2023, 24, 14035. [Google Scholar] [CrossRef] [PubMed]
- Codding, S.J.; Trudeau, M.C. Photoinhibition of the hERG potassium channel PAS domain by ultraviolet light speeds channel closing. Biophys. J. 2024, 123, 2392–2405. [Google Scholar] [CrossRef] [PubMed]
- Coin, I. Application of non-canonical crosslinking amino acids to study protein–protein interactions in live cells. Curr. Opin. Chem. Biol. 2018, 46, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Horn, R. Slow photo-cross-linking kinetics of benzophenone-labeled voltage sensors of ion channels. Biochemistry 2001, 40, 10707–10716. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, M.H.; Poshtiban, A.; Klippenstein, V.; Ghisi, V.; Plested, A.J.R. Gating modules of the AMPA receptor pore domain revealed by unnatural amino acid mutagenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 13358–13367. [Google Scholar] [CrossRef] [PubMed]
- Chin, J.W.; Schultz, P.G. In vivo photocrosslinking with unnatural amino Acid mutagenesis. ChemBioChem A Eur. J. Chem. Biol. 2002, 3, 1135–1137. [Google Scholar] [CrossRef]
- Perozo, E.; Kloda, A.; Cortes, D.M.; Martinac, B. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat. Struct. Biol. 2002, 9, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.D.; Bavi, N.; Martinac, B. Biophysical Principles of Ion-Channel-Mediated Mechanosensory Transduction. Cell Rep. 2019, 29, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fakhoury, Z.; Sosso, G.C.; Habershon, S. Contact-Map-Driven Exploration of Heterogeneous Protein-Folding Paths. J. Chem. Theory Comput. 2024, 20, 8340–8353. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.A.A.I.; Dorbath, E.; Stock, G. Allosteric Communication Mediated by Protein Contact Clusters: A Dynamical Model. J. Chem. Theory Comput. 2024, 20, 10731–10739. [Google Scholar] [CrossRef] [PubMed]
- Ichiye, T.; Karplus, M. Collective motions in proteins: A covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins Struct. Funct. Bioinform. 1991, 11, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Arnold, G.E.; Ornstein, R.L. Molecular dynamics study of time-correlated protein domain motions and molecular flexibility: Cytochrome P450BM-3. Biophys. J. 1997, 73, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Carugo, O.; Djinović-Carugo, K. Structural biology: A golden era. PLoS Biol. 2023, 21, e3002187. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Bratesco, D.; McGee, F.A.; Colenso, C.K.; Zavala, K.; Granata, D.; Carnevale, V.; Opazo, J.C.; Brauchi, S.E. Sequence and structural conservation reveal fingerprint residues in TRP channels. eLife 2022, 11, e73645. [Google Scholar] [CrossRef] [PubMed]
- Perozo, E.; Cortes, D.M.; Sompornpisut, P.; Kloda, A.; Martinac, B. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature 2002, 418, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Perozo, E.; Rees, D.C. Structure and mechanism in prokaryotic mechanosensitive channels. Curr. Opin. Struct. Biol. 2003, 13, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Le, S.C.; Liang, P.; Lowry, A.J.; Yang, H. Gating and Regulatory Mechanisms of TMEM16 Ion Channels and Scramblases. Front. Physiol. 2021, 12, 787773. [Google Scholar] [CrossRef] [PubMed]
- Brohawn, S.G.; Campbell, E.B.; MacKinnon, R. Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel. Nature 2014, 516, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Brohawn, S.G.; Su, Z.; MacKinnon, R. Mechanosensitivity is mediated directly by the lipid membrane in TRAAK and TREK1 K+ channels. Proc. Natl. Acad. Sci. USA 2014, 111, 3614–3619. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lin, C.; Chen, X.; Li, S.; Li, X.; Xiao, B. Structure deformation and curvature sensing of PIEZO1 in lipid membranes. Nature 2022, 604, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Shan, Y.; Cox, C.D.; Pei, D. A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity. Nat. Commun. 2023, 14, 3943. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, D.; Kang, Y.; Wu, J.-X.; Yao, F.; Pan, C.; Yan, Z.; Song, C.; Chen, L. Structure of the mechanosensitive OSCA channels. Nat. Struct. Mol. Biol. 2018, 25, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhou, Z.; Jin, R.; Dai, F.; Ge, Y.; Ju, X.; Ma, X.; He, S.; Yuan, L.; Wang, Y.; et al. Mechanical activation opens a lipid-lined pore in OSCA ion channels. Nature 2024, 628, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Jojoa-Cruz, S.; Burendei, B.; Lee, W.H.; Ward, A.B. Structure of mechanically activated ion channel OSCA2.3 reveals mobile elements in the transmembrane domain. Structure 2024, 32, 157–167.e5. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Zhang, M.; Chen, M.; Guo, X.; Li, Y.; Zhang, M.; Pei, D. Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels. Nat. Commun. 2024, 15, 7504. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Rawson, S.; Shen, Z.; Tamilselvan, E.; Smith, H.E.; Halford, J.; Shen, C.; Murthy, S.E.; Ulbrich, M.H.; Sotomayor, M.; et al. TMEM63 proteins function as monomeric high-threshold mechanosensitive ion channels. Neuron 2023, 111, 3195–3210.e7. [Google Scholar] [CrossRef] [PubMed]
- Edelstein, A.D.; Tsuchida, M.A.; Amodaj, N.; Pinkard, H.; Vale, R.D.; Stuurman, N. Advanced methods of microscope control using μManager software. J. Biol. Methods 2014, 1, e10. [Google Scholar] [CrossRef] [PubMed]
- Miura, K. Bleach correction ImageJ plugin for compensating the photobleaching of time-lapse sequences. F1000Research 2020, 9, 1494. [Google Scholar] [CrossRef] [PubMed]
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D., Jr.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [PubMed]
- Purisima, E.O.; Nilar, S.H. A simple yet accurate boundary element method for continuum dielectric calculations. J. Comput. Chem. 1995, 16, 681–689. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. A Publ. Protein Soc. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.J.; Rodrigues, A.P.C.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S.D. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duran-Morales, S.; Reyes-Lizana, R.; Fernández, G.; Loncon-Pavez, M.; Duarte, Y.; Marquez-Miranda, V.; Diaz-Franulic, I. Unnatural Amino Acid Photo-Crosslinking Sheds Light on Gating of the Mechanosensitive Ion Channel OSCA1.2. Int. J. Mol. Sci. 2025, 26, 7121. https://doi.org/10.3390/ijms26157121
Duran-Morales S, Reyes-Lizana R, Fernández G, Loncon-Pavez M, Duarte Y, Marquez-Miranda V, Diaz-Franulic I. Unnatural Amino Acid Photo-Crosslinking Sheds Light on Gating of the Mechanosensitive Ion Channel OSCA1.2. International Journal of Molecular Sciences. 2025; 26(15):7121. https://doi.org/10.3390/ijms26157121
Chicago/Turabian StyleDuran-Morales, Scarleth, Rachel Reyes-Lizana, German Fernández, Macarena Loncon-Pavez, Yorley Duarte, Valeria Marquez-Miranda, and Ignacio Diaz-Franulic. 2025. "Unnatural Amino Acid Photo-Crosslinking Sheds Light on Gating of the Mechanosensitive Ion Channel OSCA1.2" International Journal of Molecular Sciences 26, no. 15: 7121. https://doi.org/10.3390/ijms26157121
APA StyleDuran-Morales, S., Reyes-Lizana, R., Fernández, G., Loncon-Pavez, M., Duarte, Y., Marquez-Miranda, V., & Diaz-Franulic, I. (2025). Unnatural Amino Acid Photo-Crosslinking Sheds Light on Gating of the Mechanosensitive Ion Channel OSCA1.2. International Journal of Molecular Sciences, 26(15), 7121. https://doi.org/10.3390/ijms26157121




