De Novo Hybrid Assembly of the Tripterygium wilfordii Mitochondrial Genome Provides the Chromosomal Mitochondrial DNA Structure and RNA Editing Events
Abstract
1. Introduction
2. Results
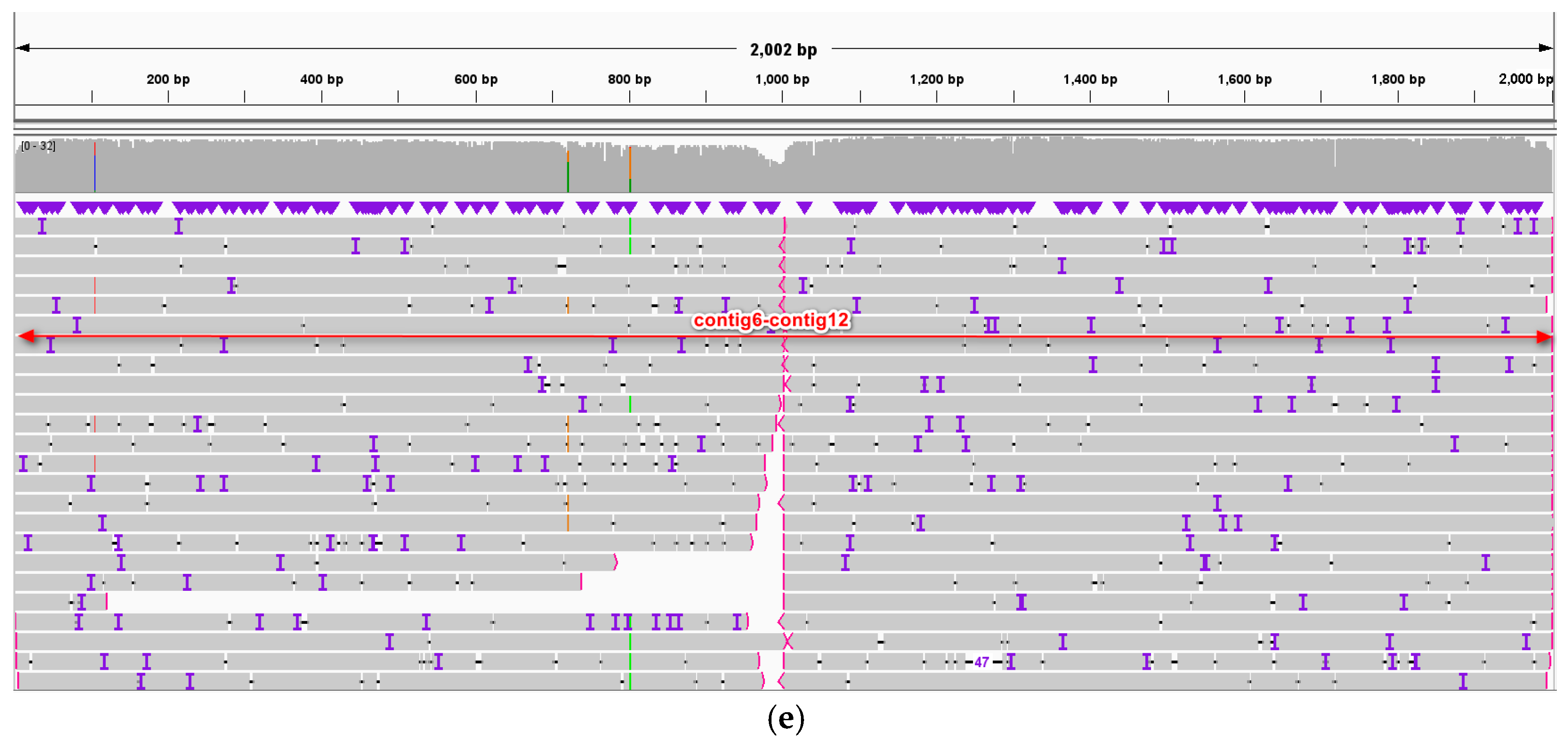
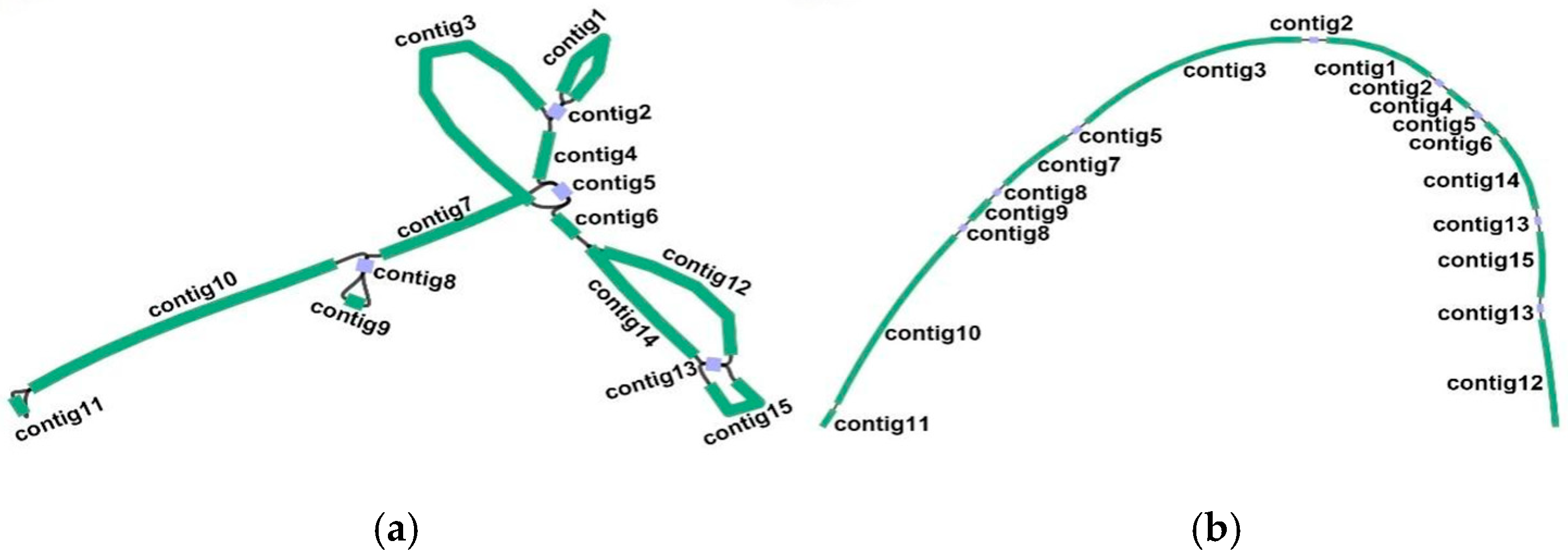
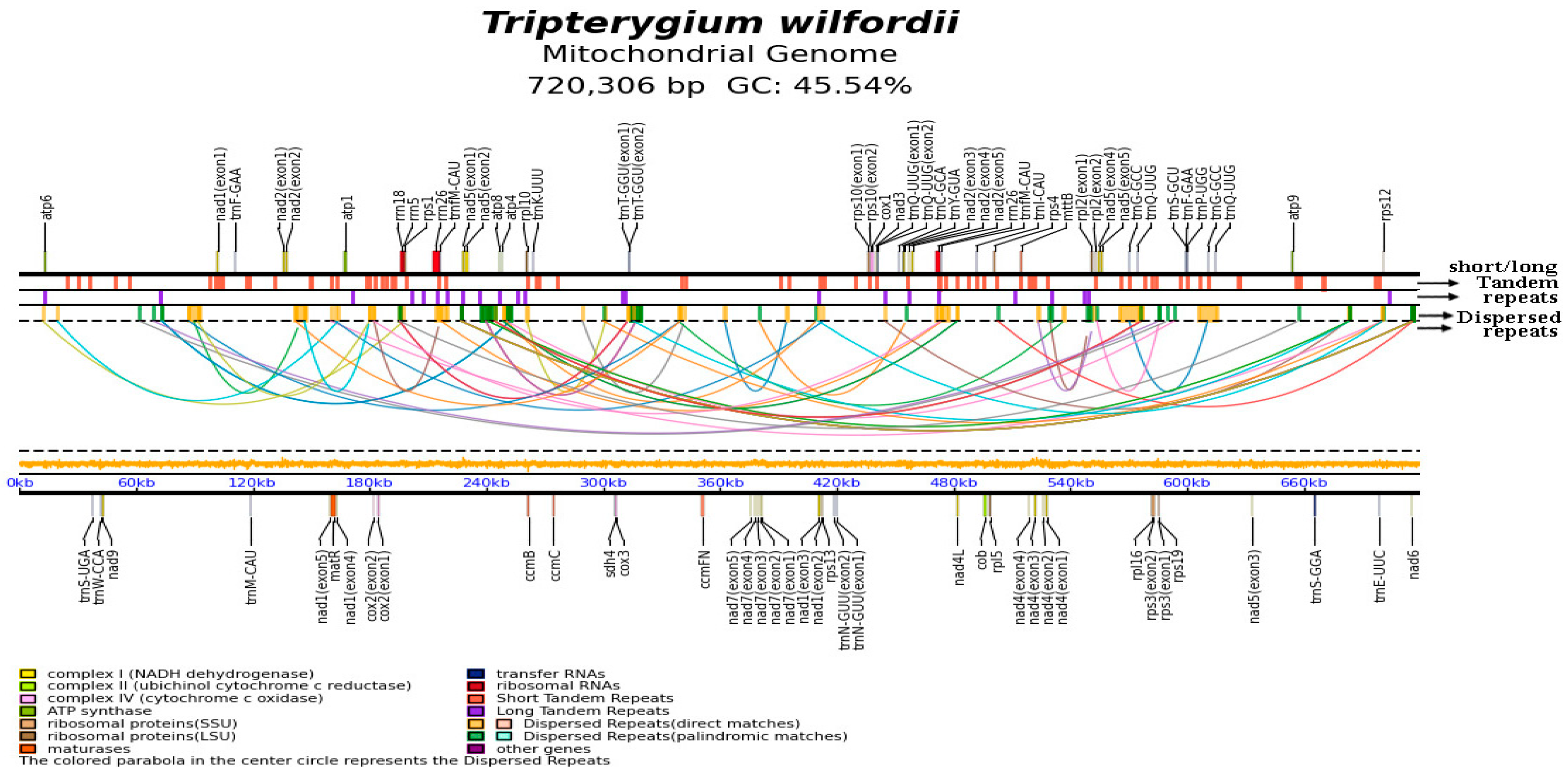
2.1. Graph-Based Mitogenome and General Genomic Features
2.2. Tandem Repeat Analysis
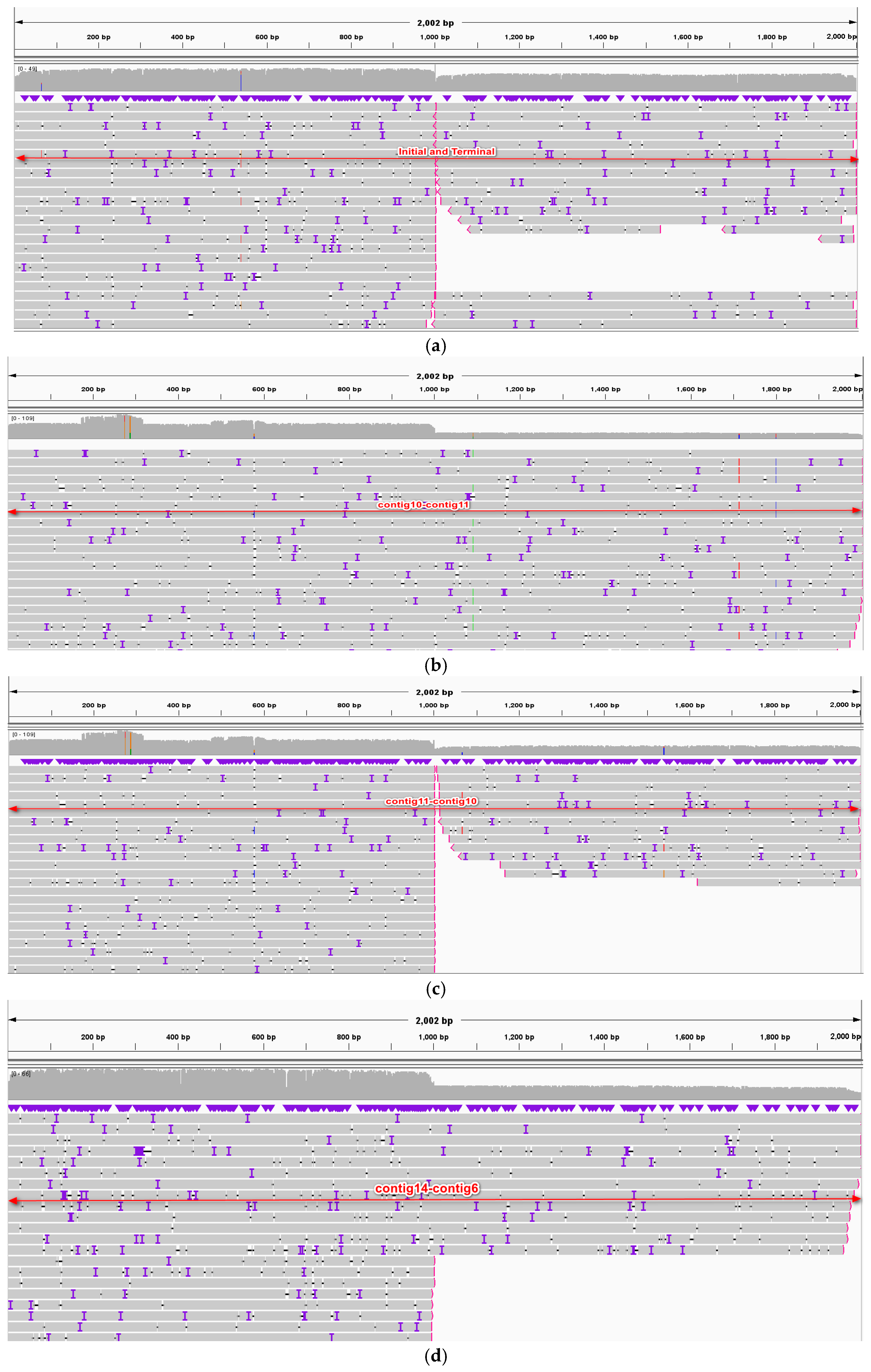
2.3. Recombinations Mediated by Repetitive Sequences
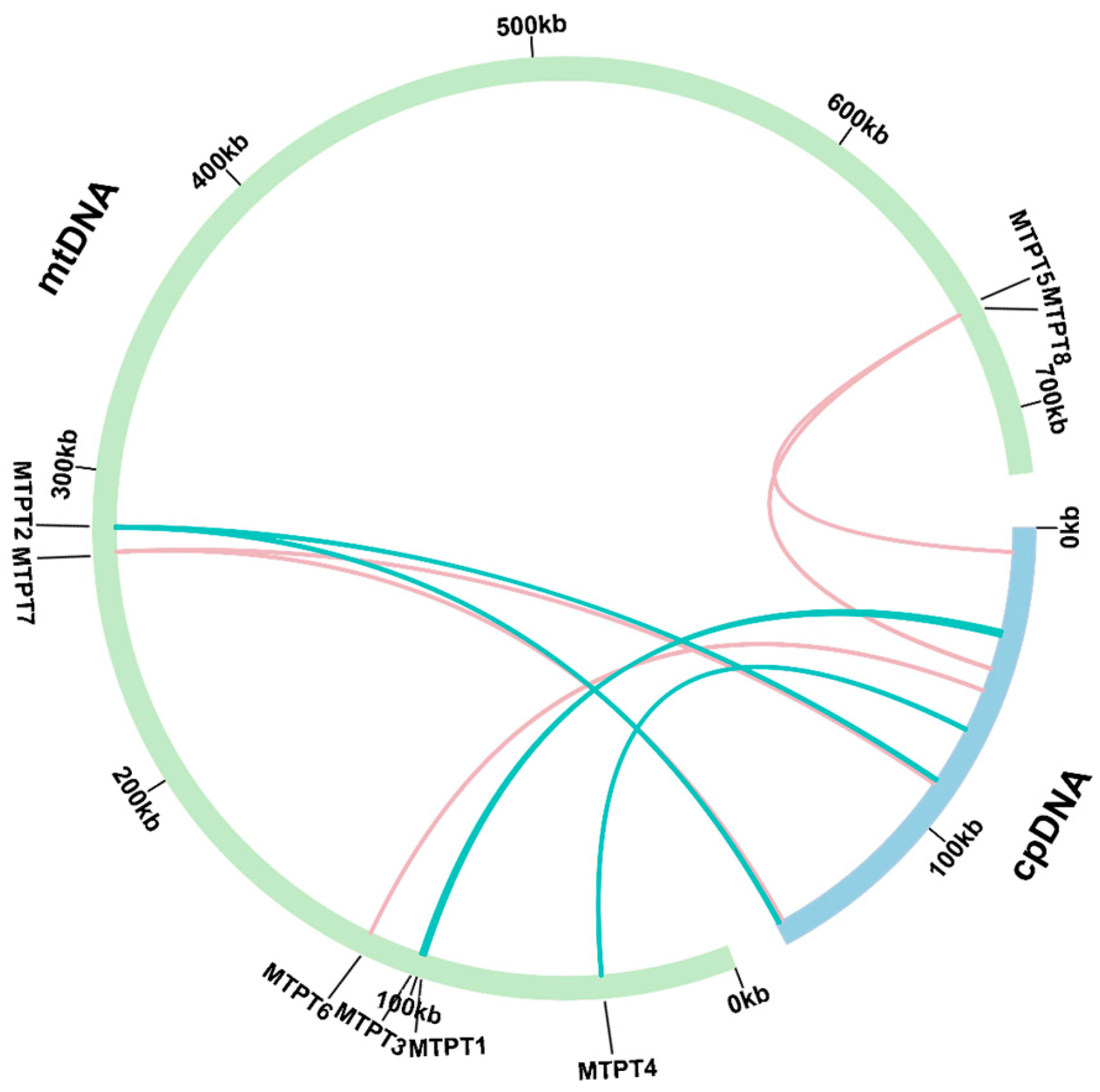
2.4. Analysis of the Homologous Sequences Between Plastids and Mitochondria
2.5. Phylogenetic Analysis
2.6. RNA-Editing Event Analysis
3. Discussion
3.1. Overview of the T. wilfordii Mitochondrial Genome
3.2. Architecture of One Molecule of the T. wilfordii Mitogenome
3.3. Research Trends for the T. wilfordii Mitogenome
4. Materials and Methods
4.1. Plant Materials, DNA and RNA Isolation, and Sequencing
4.2. Genome Assembly, Annotation, and Validation
4.3. Identification of Tandem Sequences
4.4. Identification of Repeat-Mediated Recombination
4.5. Identification of Mitochondrial Plastid Sequences (MTPTs)
4.6. Phylogenetic Tree Construction
4.7. RNA-Editing Site Identification in the PCGs of the T. wilfordii Mitogenome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PCG | protein-coding gene |
| tRNA | transfer RNA |
| rRNA | ribosomal RNA |
| SSR | simple sequence repeat |
| LTR | long tandem repeat |
| DRS | dispersed repetitive sequence |
| MTPT | mitochondrial plastid DNA |
| CDS | coding sequence |
| MVA | mevalonate |
| MEP | methylerythritol phosphate |
| TCA | tricarboxylic acid |
| HR | homologous recombination |
| BER | base excision repair |
| C | cytidines |
| U | uridines |
| DBS | double-bifurcating structure |
| SNP | single-nucleotide polymorphism |
| MC | mitochondrial chromosome |
| IR | inverted repeat |
Appendix A
| Sequencing Descriptor | Sequencing Platform | |
|---|---|---|
| Illumina | Nanopore | |
| Total number of nucleotides | 13,177,504,500 | 7,231,843,596 |
| Total number of reads | 87,850,030 | 316,356 |
| Mean read length (bp) | 150 | 22,859.8 |
| ID | SSR Type | SSR | Size | Start | End |
|---|---|---|---|---|---|
| SSR1 | p4 | (TCAT)3 | 12 | 3904 | 3915 |
| SSR2 | p3 | (TGC)4 | 12 | 14,552 | 14,563 |
| SSR3 | p2 | (GA)5 | 10 | 18,258 | 18,267 |
| SSR4 | p3 | (TTC)4 | 12 | 21,603 | 21,614 |
| SSR5 | p4 | (TTCA)3 | 12 | 22,669 | 22,680 |
| SSR6 | p1 | (T)10 | 10 | 24,476 | 24,485 |
| SSR7 | p2 | (GA)5 | 10 | 26,947 | 26,956 |
| SSR8 | p1 | (A)11 | 11 | 30,368 | 30,378 |
| SSR9 | p1 | (A)10 | 10 | 36,140 | 36,149 |
| SSR10 | p4 | (CTTA)3 | 12 | 42,128 | 42,139 |
| SSR11 | p2 | (TA)6 | 12 | 49,292 | 49,303 |
| SSR12 | p2 | (AT)5 | 10 | 55,853 | 55,862 |
| SSR13 | p1 | (A)12 | 12 | 56,245 | 56,256 |
| SSR14 | p2 | (CT)5 | 10 | 56,509 | 56,518 |
| SSR15 | p4 | (ATCA)3 | 12 | 59,470 | 59,481 |
| SSR16 | p3 | (CTT)4 | 12 | 63,294 | 63,305 |
| SSR17 | p1 | (T)12 | 12 | 97,610 | 97,621 |
| SSR18 | p2 | (TA)6 | 12 | 101,015 | 101,026 |
| SSR19 | p2 | (TA)5 | 10 | 102,475 | 102,484 |
| SSR20 | p1 | (T)10 | 10 | 102,491 | 102,500 |
| SSR21 | p1 | (A)13 | 13 | 102,503 | 102,515 |
| SSR22 | p1 | (T)10 | 10 | 103,640 | 103,649 |
| SSR23 | p1 | (A)10 | 10 | 104,561 | 104,570 |
| SSR24 | p5 | (TTTCT)3 | 15 | 108,551 | 108,565 |
| SSR25 | p4 | (ATAG)3 | 12 | 108,894 | 108,905 |
| SSR26 | p4 | (AGAA)3 | 12 | 114,292 | 114,303 |
| SSR27 | p4 | (TAAG)3 | 12 | 114,945 | 114,956 |
| SSR28 | p1 | (A)11 | 11 | 116,756 | 116,766 |
| SSR29 | p1 | (T)10 | 10 | 118,096 | 118,105 |
| SSR30 | p2 | (CT)5 | 10 | 118,358 | 118,367 |
| SSR31 | p2 | (CT)5 | 10 | 122,012 | 122,021 |
| SSR32 | p3 | (CAA)4 | 12 | 125,275 | 125,286 |
| SSR33 | p4 | (CTGT)3 | 12 | 127,624 | 127,635 |
| SSR34 | p2 | (CT)5 | 10 | 128,277 | 128,286 |
| SSR35 | p1 | (T)10 | 10 | 130,920 | 130,929 |
| SSR36 | p3 | (TAT)4 | 12 | 140,781 | 140,792 |
| SSR37 | p2 | (TA)5 | 10 | 149,059 | 149,068 |
| SSR38 | p1 | (T)12 | 12 | 149,539 | 149,550 |
| SSR39 | p1 | (T)10 | 10 | 149,968 | 149,977 |
| SSR40 | p4 | (GGCA)3 | 12 | 151,469 | 151,480 |
| SSR41 | p1 | (A)12 | 12 | 160,470 | 160,481 |
| SSR42 | p5 | (ACTAG)3 | 15 | 160,484 | 160,498 |
| SSR43 | p4 | (TTCT)3 | 12 | 162,501 | 162,512 |
| SSR44 | p1 | (A)10 | 10 | 163,156 | 163,165 |
| SSR45 | p4 | (TCTT)3 | 12 | 170,024 | 170,035 |
| SSR46 | p2 | (TC)5 | 10 | 172,086 | 172,095 |
| SSR47 | p2 | (AG)5 | 10 | 173,151 | 173,160 |
| SSR48 | p4 | (ATTT)3 | 12 | 177,791 | 177,802 |
| SSR49 | p2 | (AT)6 | 12 | 178,785 | 178,796 |
| SSR50 | p4 | (CTTT)3 | 12 | 179,931 | 179,942 |
| SSR51 | p1 | (T)10 | 10 | 179,992 | 180,001 |
| SSR52 | p1 | (C)11 | 11 | 182,801 | 182,811 |
| SSR53 | p2 | (CT)5 | 10 | 186,353 | 186,362 |
| SSR54 | p1 | (A)10 | 10 | 186,457 | 186,466 |
| SSR55 | p4 | (TTCT)3 | 12 | 187,055 | 187,066 |
| SSR56 | p1 | (A)10 | 10 | 188,497 | 188,506 |
| SSR57 | p1 | (T)10 | 10 | 191,228 | 191,237 |
| SSR58 | p1 | (T)11 | 11 | 192,751 | 192,761 |
| SSR59 | p4 | (GGCG)3 | 12 | 197,572 | 197,583 |
| SSR60 | p3 | (CTT)4 | 12 | 198,276 | 198,287 |
| SSR61 | p1 | (A)11 | 11 | 198,567 | 198,577 |
| SSR62 | p4 | (AGCC)3 | 12 | 200,534 | 200,545 |
| SSR63 | p4 | (AGAC)3 | 12 | 204,756 | 204,767 |
| SSR64 | p3 | (CTT)4 | 12 | 205,202 | 205,213 |
| SSR65 | p1 | (G)10 | 10 | 215,294 | 215,303 |
| SSR66 | p1 | (A)10 | 10 | 218,839 | 218,848 |
| SSR67 | p4 | (AAAG)3 | 12 | 226,751 | 226,762 |
| SSR68 | p4 | (GCCG)3 | 12 | 231,013 | 231,024 |
| SSR69 | p4 | (CAAG)3 | 12 | 234,577 | 234,588 |
| SSR70 | p2 | (GA)5 | 10 | 239,681 | 239,690 |
| SSR71 | p4 | (GCAA)3 | 12 | 250,096 | 250,107 |
| SSR72 | p4 | (TCTT)3 | 12 | 250,875 | 250,886 |
| SSR73 | p3 | (CTT)4 | 12 | 253,352 | 253,363 |
| SSR74 | p4 | (TACT)3 | 12 | 253,737 | 253,748 |
| SSR75 | p2 | (AT)8 | 16 | 261,274 | 261,289 |
| SSR76 | p4 | (GTGA)3 | 12 | 262,669 | 262,680 |
| SSR77 | p2 | (AT)7 | 14 | 265,650 | 265,663 |
| SSR78 | p1 | (A)10 | 10 | 266,208 | 266,217 |
| SSR79 | p1 | (A)11 | 11 | 266,771 | 266,781 |
| SSR80 | p3 | (TTA)4 | 12 | 275,552 | 275,563 |
| SSR81 | p1 | (A)10 | 10 | 276,586 | 276,595 |
| SSR82 | p2 | (TC)5 | 10 | 291,363 | 291,372 |
| SSR83 | p4 | (AAAG)3 | 12 | 291,575 | 291,586 |
| SSR84 | p3 | (CAA)4 | 12 | 293,222 | 293,233 |
| SSR85 | p4 | (AAAC)3 | 12 | 296,155 | 296,166 |
| SSR86 | p4 | (TCTT)3 | 12 | 307,547 | 307,558 |
| SSR87 | p2 | (TC)5 | 10 | 317,136 | 317,145 |
| SSR88 | p3 | (TCT)4 | 12 | 319,730 | 319,741 |
| SSR89 | p4 | (TCTT)3 | 12 | 320,202 | 320,213 |
| SSR90 | p4 | (TCAA)3 | 12 | 325,791 | 325,802 |
| SSR91 | p2 | (AT)5 | 10 | 329,975 | 329,984 |
| SSR92 | p2 | (CT)5 | 10 | 335,795 | 335,804 |
| SSR93 | p2 | (TC)5 | 10 | 336,109 | 336,118 |
| SSR94 | p2 | (TC)6 | 12 | 340,581 | 340,592 |
| SSR95 | p3 | (GAT)4 | 12 | 340,901 | 340,912 |
| SSR96 | p2 | (TA)6 | 12 | 342,602 | 342,613 |
| SSR97 | p4 | (TCAG)3 | 12 | 344,772 | 344,783 |
| SSR98 | p3 | (TCT)4 | 12 | 347,832 | 347,843 |
| SSR99 | p4 | (TTCT)3 | 12 | 348,543 | 348,554 |
| SSR100 | p4 | (AGAA)3 | 12 | 354,135 | 354,146 |
| SSR101 | p2 | (AG)5 | 10 | 354,778 | 354,787 |
| SSR102 | p3 | (GGA)4 | 12 | 357,969 | 357,980 |
| SSR103 | p2 | (TA)5 | 10 | 360,398 | 360,407 |
| SSR104 | p4 | (CCTT)3 | 12 | 362,330 | 362,341 |
| SSR105 | p3 | (CTT)4 | 12 | 366,171 | 366,182 |
| SSR106 | p4 | (GAGT)3 | 12 | 366,513 | 366,524 |
| SSR107 | p2 | (TA)5 | 10 | 376,772 | 376,781 |
| SSR108 | p4 | (GAAT)3 | 12 | 383,703 | 383,714 |
| SSR109 | p4 | (CTTT)3 | 12 | 384,018 | 384,029 |
| SSR110 | p2 | (CT)5 | 10 | 389,799 | 389,808 |
| SSR111 | p5 | (TTAGA)3 | 15 | 390,482 | 390,496 |
| SSR112 | p3 | (CTG)5 | 15 | 391,010 | 391,024 |
| SSR113 | p1 | (T)10 | 10 | 395,390 | 395,399 |
| SSR114 | p3 | (CTT)4 | 12 | 402,562 | 402,573 |
| SSR115 | p3 | (AGA)4 | 12 | 403,830 | 403,841 |
| SSR116 | p2 | (CT)5 | 10 | 404,825 | 404,834 |
| SSR117 | p4 | (CTAT)3 | 12 | 408,871 | 408,882 |
| SSR118 | p4 | (GCTA)3 | 12 | 409,575 | 409,586 |
| SSR119 | p1 | (A)10 | 10 | 411,626 | 411,635 |
| SSR120 | p1 | (A)13 | 13 | 412,695 | 412,707 |
| SSR121 | p4 | (TTCT)3 | 12 | 413,246 | 413,257 |
| SSR122 | p1 | (A)10 | 10 | 413,768 | 413,777 |
| SSR123 | p4 | (ATAG)3 | 12 | 415,985 | 415,996 |
| SSR124 | p2 | (CT)5 | 10 | 416,396 | 416,405 |
| SSR125 | p2 | (CT)5 | 10 | 423,720 | 423,729 |
| SSR126 | p2 | (AT)9 | 18 | 429,578 | 429,595 |
| SSR127 | p2 | (GA)5 | 10 | 433,555 | 433,564 |
| SSR128 | p2 | (AG)5 | 10 | 433,863 | 433,872 |
| SSR129 | p1 | (A)11 | 11 | 437,201 | 437,211 |
| SSR130 | p1 | (T)14 | 14 | 440,277 | 440,290 |
| SSR131 | p5 | (AAAGA)3 | 15 | 450,941 | 450,955 |
| SSR132 | p4 | (GGAT)3 | 12 | 452,636 | 452,647 |
| SSR133 | p4 | (GGAG)3 | 12 | 453,553 | 453,564 |
| SSR134 | p4 | (GAAA)3 | 12 | 455,204 | 455,215 |
| SSR135 | p5 | (TCTAT)3 | 15 | 456,202 | 456,216 |
| SSR136 | p1 | (A)14 | 14 | 456,223 | 456,236 |
| SSR137 | p2 | (AT)5 | 10 | 458,251 | 458,260 |
| SSR138 | p2 | (AT)5 | 10 | 458,267 | 458,276 |
| SSR139 | p3 | (TAA)4 | 12 | 463,152 | 463,163 |
| SSR140 | p4 | (CTTT)3 | 12 | 470,520 | 470,531 |
| SSR141 | p1 | (G)10 | 10 | 472,879 | 472,888 |
| SSR142 | p1 | (A)10 | 10 | 476,424 | 476,433 |
| SSR143 | p3 | (ACT)4 | 12 | 480,790 | 480,801 |
| SSR144 | p1 | (A)10 | 10 | 481,798 | 481,807 |
| SSR145 | p3 | (TCC)4 | 12 | 485,671 | 485,682 |
| SSR146 | p1 | (A)11 | 11 | 487,453 | 487,463 |
| SSR147 | p2 | (AT)6 | 12 | 491,773 | 491,784 |
| SSR148 | p1 | (A)11 | 11 | 495,422 | 495,432 |
| SSR149 | p1 | (T)10 | 10 | 495,583 | 495,592 |
| SSR150 | p2 | (TA)5 | 10 | 497,184 | 497,193 |
| SSR151 | p1 | (T)10 | 10 | 501,990 | 501,999 |
| SSR152 | p4 | (TTTC)3 | 12 | 510,724 | 510,735 |
| SSR153 | p4 | (AAGA)3 | 12 | 514,161 | 514,172 |
| SSR154 | p1 | (T)12 | 12 | 514,486 | 514,497 |
| SSR155 | p2 | (AT)5 | 10 | 514,761 | 514,770 |
| SSR156 | p3 | (TTA)4 | 12 | 516,658 | 516,669 |
| SSR157 | p1 | (A)10 | 10 | 519,246 | 519,255 |
| SSR158 | p1 | (A)11 | 11 | 520,785 | 520,795 |
| SSR159 | p1 | (A)10 | 10 | 521,711 | 521,720 |
| SSR160 | p2 | (TC)5 | 10 | 527,030 | 527,039 |
| SSR161 | p4 | (TCTA)3 | 12 | 527,038 | 527,049 |
| SSR162 | p4 | (TAGA)3 | 12 | 527,050 | 527,061 |
| SSR163 | p3 | (CTA)4 | 12 | 527,121 | 527,132 |
| SSR164 | p1 | (A)10 | 10 | 528,215 | 528,224 |
| SSR165 | p4 | (AGCC)3 | 12 | 528,751 | 528,762 |
| SSR166 | p4 | (AAAG)3 | 12 | 537,670 | 537,681 |
| SSR167 | p1 | (T)10 | 10 | 553,153 | 553,162 |
| SSR168 | p4 | (AAGC)3 | 12 | 556,534 | 556,545 |
| SSR169 | p4 | (AACA)3 | 12 | 563,478 | 563,489 |
| SSR170 | p1 | (T)10 | 10 | 566,427 | 566,436 |
| SSR171 | p1 | (A)13 | 13 | 570,844 | 570,856 |
| SSR172 | p6 | (ATCTAT)3 | 18 | 574,227 | 574,244 |
| SSR173 | p1 | (A)11 | 11 | 578,831 | 578,841 |
| SSR174 | p1 | (A)12 | 12 | 581,826 | 581,837 |
| SSR175 | p4 | (AATG)3 | 12 | 583,827 | 583,838 |
| SSR176 | p2 | (TA)6 | 12 | 584,679 | 584,690 |
| SSR177 | p4 | (CTTT)3 | 12 | 592,151 | 592,162 |
| SSR178 | p2 | (TC)5 | 10 | 593,996 | 594,005 |
| SSR179 | p1 | (T)10 | 10 | 596,355 | 596,364 |
| SSR180 | p5 | (GTAAT)3 | 15 | 599,235 | 599,249 |
| SSR181 | p1 | (A)11 | 11 | 600,150 | 600,160 |
| SSR182 | p5 | (GCCCA)3 | 15 | 603,387 | 603,401 |
| SSR183 | p1 | (T)10 | 10 | 606,766 | 606,775 |
| SSR184 | p1 | (A)13 | 13 | 611,183 | 611,195 |
| SSR185 | p6 | (ATCTAT)3 | 18 | 614,566 | 614,583 |
| SSR186 | p4 | (ATTC)3 | 12 | 623,581 | 623,592 |
| SSR187 | p4 | (TTAG)3 | 12 | 625,006 | 625,017 |
| SSR188 | p4 | (TTTC)3 | 12 | 625,633 | 625,644 |
| SSR189 | p1 | (T)10 | 10 | 626,533 | 626,542 |
| SSR190 | p1 | (T)10 | 10 | 626,867 | 626,876 |
| SSR191 | p1 | (T)12 | 12 | 627,314 | 627,325 |
| SSR192 | p1 | (T)11 | 11 | 627,327 | 627,337 |
| SSR193 | p3 | (GCT)4 | 12 | 627,388 | 627,399 |
| SSR194 | p4 | (TCGA)3 | 12 | 630,121 | 630,132 |
| SSR195 | p4 | (TTTC)3 | 12 | 632,172 | 632,183 |
| SSR196 | p4 | (GAAT)3 | 12 | 636,280 | 636,291 |
| SSR197 | p4 | (AGAA)3 | 12 | 647,143 | 647,154 |
| SSR198 | p2 | (AG)5 | 10 | 656,323 | 656,332 |
| SSR199 | p1 | (A)10 | 10 | 656,397 | 656,406 |
| SSR200 | p3 | (GGT)4 | 12 | 657,497 | 657,508 |
| SSR201 | p2 | (GA)6 | 12 | 657,832 | 657,843 |
| SSR202 | p3 | (GTT)5 | 15 | 658,242 | 658,256 |
| SSR203 | p1 | (T)11 | 11 | 658,267 | 658,277 |
| SSR204 | p4 | (CTTG)3 | 12 | 668,106 | 668,117 |
| SSR205 | p1 | (T)10 | 10 | 670,382 | 670,391 |
| SSR206 | p4 | (AAGA)3 | 12 | 670,430 | 670,441 |
| SSR207 | p3 | (CTT)4 | 12 | 676,856 | 676,867 |
| SSR208 | p2 | (CT)5 | 10 | 684,284 | 684,293 |
| SSR209 | p4 | (CTTT)3 | 12 | 686,396 | 686,407 |
| SSR210 | p4 | (ATTC)3 | 12 | 689,697 | 689,708 |
| SSR211 | p4 | (AAAG)3 | 12 | 694,379 | 694,390 |
| SSR212 | p1 | (C)11 | 11 | 697,246 | 697,256 |
| SSR213 | p2 | (CT)5 | 10 | 697,892 | 697,901 |
| SSR214 | p1 | (A)10 | 10 | 698,667 | 698,676 |
| SSR215 | p1 | (T)10 | 10 | 699,203 | 699,212 |
| SSR216 | p4 | (TTAA)3 | 12 | 699,408 | 699,419 |
| SSR217 | p4 | (AAGA)3 | 12 | 700,384 | 700,395 |
| SSR218 | p4 | (GATA)3 | 12 | 700,546 | 700,557 |
| SSR219 | p3 | (TCT)4 | 12 | 701,847 | 701,858 |
| SSR220 | p3 | (TCC)4 | 12 | 706,852 | 706,863 |
| SSR221 | p4 | (AAGC)3 | 12 | 709,507 | 709,518 |
| SSR222 | p2 | (CT)5 | 10 | 709,834 | 709,843 |
| SSR223 | p2 | (TC)5 | 10 | 715,387 | 715,396 |
Appendix B

References
- Zhong, Y.; Zhang, J.Z.; Bao, Z.Z. The complete chloroplast genome of Tripterygium wilfordii Hook. f.(Celastraceae). Mitochondrial DNA Part B 2022, 7, 1696–1698. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.F.; Tu, S.H.; Gao, W.N.; Wang, Y.; Liu, P.; Hu, Y.H.; Dong, H. Extracts of Tripterygium wilfordii Hook F in the treatment of rheumatoid arthritis: A systemic review and meta-analysis of randomised controlled trials. Evid.-Based Complement. Altern. Med. 2013, 2013, 41079. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.Y.; Liu, J.T.; Zhao, Z.M.; Li, Z.P.; Wu, K.Y. Tripterygium and its plant extraction for systemic lupus erythematosus: A protocol for systematic review and meta analysis. Medicine 2020, 99, e21909. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Lin, G.J.; Chia, W.T.; Lin, C.K.; Chuang, Y.P.; Sytwu, H.K. Triptolide exerts anti-tumor effect on oral cancer and KB cells in vitro and in vivo. Oral Oncol. 2009, 45, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Kuchta, K.; Xiang, Y.; Huang, S.; Tang, Y.; Peng, X.; Wang, X.; Zhu, Y.; Li, J.; Xu, J.; Lin, Z.; et al. Celastrol, an active constituent of the TCM plant Tripterygium wilfordii Hook. f., inhibits prostate cancer bone metastasis. Prostate Cancer Prostatic Dis. 2017, 20, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Hao, J.J. Treatment of neurodegenerative diseases with bioactive components of Tripterygium wilfordii. Am. J. Chin. Med. 2019, 47, 769–785. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Su, C.; Iyaswamy, A.; Krishnamoorthi, S.K.; Zhu, Z.; Yang, S.; Tong, B.C.; Liu, J.; Sreenivasmurthy, S.G.; Guan, X.; et al. Celastrol enhances transcription factor EB (TFEB)-mediated autophagy and mitigates Tau pathology: Implications for Alzheimer’s disease therapy. Acta Pharm. Sin. B 2022, 12, 1707–1722. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.H.; Feng, Y.Q.; He, W.S.; Xu, W.; Xu, W.; Yang, H.J.; Li, X.Y. Celastrol in metabolic diseases: Progress and application prospects. Pharmacol. Res. 2021, 167, 105572. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.D.; Guan, D.X.; Auen, T.; Choi, J.W.; Hernández, M.A.S.; Lee, J.; Chun, H.; Faruk, F.; Kaplun, E.; Herbert, Z.; et al. IL1R1 is required for celastrol’s leptin-sensitization and antiobesity effects. Nat. Med. 2019, 25, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.B.; Zhang, J.; Zhang, B.; Chen, L.; Zhang, X.; Zhu, C.S. MYC2 transcription factors TwMYC2a and TwMYC2b negatively regulate triptolide biosynthesis in Tripterygium wilfordii hairy roots. Plants 2021, 10, 679. [Google Scholar] [CrossRef] [PubMed]
- Tu, L.C.; Su, P.; Zhang, Z.R.; Gao, L.H.; Wang, J.D.; Hu, T.Y.; Zhou, J.W.; Zhang, Y.F.; Zhao, Y.J.; Liu, Y.; et al. Genome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis. Nat. Commun. 2020, 11, 971. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Chevigny, N.; Schatz-Daas, D.; Lotfi, F.; Gualberto, J.M. DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci. 2020, 21, 328. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.; Jörg, A.; Burger, M.; Haag, S. RNA editing mutants as surrogates for mitochondrial SNP mutants. Plant Physiol. Biochem. 2019, 135, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Chen, H.; Shao, J.; Zhang, H.; Wu, K.; Liu, C. Identification of symmetrical RNA editing events in the mitochondria of Salvia miltiorrhiza by strand-specific RNA sequencing. Sci. Rep. 2017, 7, 42250. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Jian, Y.Q.; Yan, W.Y.; Xu, J.F.; Duan, S.G.; Li, G.C.; Jin, L.P. Genome-wide simple sequence repeat markers in potato: Abundance, distribution, composition, and polymorphism. DNA Res. 2021, 28, dsab020. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Hannan, A.J. Tandem repeat polymorphisms: Mediators of genetic plasticity, modulators of biological diversity and dynamic sources of disease susceptibility. Adv. Exp. Med. Biol. 2012, 769, 1–9. [Google Scholar] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Carrington, M.; Roditi, I.; Williams, R.O. The structure and transcription of an element interspersed between tandem arrays of mini-exon donor RNA genes in Trypanosoma brucei. Nucleic Acids Res. 1987, 15, 10179–10198. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H. Mobile elements: Drivers of genome evolution. Science 2004, 303, 1626–1632. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B. One ring to rule them all? Genome sequencing provides new insights into the master circle model of plant mitochondrial DNA structure. New Phytol. 2013, 200, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Wynn, E.L.; Christensen, A.C. Repeats of unusual size in plant mitochondrial genomes: Identification, incidence and evolution. G3 Genes Genomes Genet. 2019, 9, 549–559. [Google Scholar] [CrossRef]
- Wang, X.C.; Chen, H.M.; Yang, D.; Liu, C. Diversity of mitochondrial plastid DNAs (MTPTs) in seed plants. Mitochondrial DNA Part A 2018, 29, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Q.; Ni, Y.; Li, J.L.; Huang, L.F.; Li, H.S.; Chen, H.M.; Liu, C. Multiple configurations of the plastid and mitochondrial genomes of Caragana spinosa. Planta 2023, 258, 98. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Y.; Chen, H.M.; Ni, Y.; Li, J.L.; Cai, Y.S.; Ma, B.X.; Yu, J.; Wang, J.H.; Liu, C. De novo hybrid assembly of the Salvia miltiorrhiza mitochondrial genome provides the first evidence of the multi-chromosomal mitochondrial DNA structure of salvia species. Int. J. Mol. Sci. 2022, 23, 14267. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Liu, G.X.; Wang, W.P.; Shen, W.; Zhao, Y.P.; Sun, J.L.; Yang, Q.Y.; Zhang, Y.X.; Fan, W.J.; Pei, S.S.; et al. RNA editing and its roles in plant organelles. Front. Genet. 2021, 12, 757109. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, M.; Sugita, M. RNA editing and its molecular mechanism in plant organelles. Genes 2016, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.W.; Jiang, L.P.; Zhu, M.D.; Li, Y.X.; Luo, M.; Jiang, P.H.; Tong, S.Q.; Zhang, H.J.; Yan, J.Z. The genus Tripterygium: A phytochemistry and pharmacological review. Fitoterapia 2019, 137, 104190. [Google Scholar] [CrossRef] [PubMed]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef] [PubMed]
- Jackman, S.D.; Coombe, L.; Warren, R.L.; Kirk, H.; Trinh, E.; MacLeod, T.; Pleasance, S.; Pandoh, P.; Zhao, Y.J.; Coope, R.J. Complete mitochondrial genome of a gymnosperm, Sitka spruce (Picea sitchensis), indicates a complex physical structure. Genome Biol. Evol. 2020, 12, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Yu, R.X.; Chen, J.F.; Liu, Y.; Zhou, R.C. Highly active repeat-mediated recombination in the mitogenome of the holoparasitic plant Aeginetia indica. Front. Plant Sci. 2022, 13, 988368. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.J.; Ni, Y.; Lin, Z.B.; Yang, L.B.; Chen, G.T.; Nijiati, N.; Hu, Y.Z.; Chen, X.Y. De novo assembly of the complete mitochondrial genome of sweet potato (Ipomoea batatas [L.] Lam) revealed the existence of homologous conformations generated by the repeat-mediated recombination. BMC Plant Biol. 2022, 22, 285. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.S.; Zhao, C.X.; Chen, F.; Liu, Y.H.; Zhang, S.Z.; Wu, H.; Zhang, L.S.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef] [PubMed]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Luo, C. Molecular and functional diversity of RNA editing in plant mitochondria. Mol. Biotechnol. 2018, 60, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Chateigner-Boutin, A.L.; Small, I. Organellar RNA editing. Adv. Rev. 2011, 2, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate denovo assembly of organelle genomes. Genome Biol. 2020, 21, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Richly, E.; Leister, D. NUMTs in sequenced eukaryotic genomes. Mol. Biol. Evol. 2004, 21, 1081–1084. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap and miniasm: Fast mapping and de novo assembly for noisy long sequences. Bioinformatics 2016, 32, 2103–2110. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genomeassemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.C.; Chen, H.M.; Jiang, M.; Wang, L.Q.; Wu, X.; Huang, L.F.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator andanalyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Ni, Y.; Li, J.L.; Zhang, X.Y.; Yang, H.Y.; Chen, H.M.; Liu, C. CPGView: A package for visualizing detailed chloroplast genome structures. Mol. Ecol. Resour. 2023, 23, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.E.; Searle, S.M.J.; Harris, N.; Gibson, M.; Iyer, V.; Richter, J.; Wiel, C.; Bayraktaroglu, L.; Birney, E.; Crosby, M.A.; et al. Apollo: A sequence annotation editor. Genome Biol. 2002, 3, research0082.1. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primersfor polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence align ment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data vis ualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Swindell, S.R.; Plasterer, T.N. SEQMAN: Contig assembly. In Sequence Data Analysis Guidebook; Springer: Berlin/Heidelberg, Germany, 1997; pp. 75–89. [Google Scholar] [CrossRef]
- Park, H.S.; Jayakodi, M.; Lee, S.H.; Jeon, J.-H.; Lee, H.-O.; Park, J.Y.; Moon, B.C.; Kim, C.-K.; Wing, R.A.; Newmaster, S.G. Mitochondrial plastid DNA can cause DNA barcoding paradox in plants. Sci. Rep. 2020, 10, 6112. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ye, W.C.; Zhang, Y.D.; Xu, Y.S. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovli’c, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; VonHaeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Pesole, G. REDItools: High-throughput RNA editing detection made easy. Bioinformatics 2013, 29, 1813–1814. [Google Scholar] [CrossRef] [PubMed]



| No. | Name | Length (bp) | Bifurcation Structures |
|---|---|---|---|
| 1 | contig1 | 71,533 | No |
| 2 | contig2/DBS3 | 5483 | Yes |
| 3 | contig3 | 151,728 | No |
| 4 | contig4 | 17,421 | No |
| 5 | contig5/DBS1 | 5937 | Yes |
| 6 | contig6 | 10,857 | No |
| 7 | contig7 | 51,730 | No |
| 8 | contig8/DBS4 | 1728 | Yes |
| 9 | contig9 | 18,187 | No |
| 10 | contig10 | 140,923 | No |
| 11 | contig11 | 18,808 | No |
| 12 | contig12 | 86,897 | No |
| 13 | contig13/DBS2 | 5600 | Yes |
| 14 | contig14 | 55,721 | No |
| 15 | contig15 | 49,130 | No |
| 16 | contig16 | 9875 | Yes |
| Group of Genes | Names of Genes |
|---|---|
| Subunit of ATPase | atp1, atp4, atp6, atp8, atp9 |
| Cytochrome c biogenesis | ccmB, ccmC, ccmFN |
| Apocytochrome b | cob |
| Subunit of cytochrome c oxidase | cox1, cox2, cox3 |
| Maturase R | matR |
| Transport membrane protein | mttB |
| Subunit of NADH dehydrogenase | nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9 |
| Small subunit of ribosome | rps1, rps10, rps12, rps13, rps19, rps3, rps4 |
| Large subunit of ribosome | rpl10, rpl16, rpl2, rpl5 |
| Subunit of succinate dehydrogenase | sdh4 |
| Transfer RNAs | trnF–GAA(×2), trnN–GUU, trnQ–UUG, trnT–GGU, trnC–GCA, trnE–UUC, trnG–GCC(×2), trnK–UUU, tr nM–CAU, trnI–CAU, trnM–CAU (×2), trnP–UGG ×3), trnS–GCU, trnS–GGA, trnS–UGA, trnW–CCA, trnY–GUA |
| Ribosomal RNAs | rrn18, rrn26(×2), rrn5 |
| ID | Repeat Length | Repeat Type | Start1 | End1 | Start2 | End2 |
|---|---|---|---|---|---|---|
| DRS1 | 9875 | F | 565,608 | 575,482 | 605,947 | 615,821 |
| DRS2 | 5937 | F | 213,806 | 219,742 | 471,391 | 477,327 |
| DRS3 | 5600 | F | 86,898 | 92,497 | 141,628 | 147,227 |
| DRS4 | 5483 | P | 237,164 | 242,646 | 319,662 | 314,180 |
| DRS5 | 1728 | P | 529,058 | 530,785 | 550,700 | 548,973 |
| DRS6 | 388 | P | 250,458 | 250,845 | 593,826 | 593,439 |
| DRS7 | 295 | F | 289,376 | 289,670 | 341,859 | 342,153 |
| DRS8 | 288 | F | 260,811 | 261,098 | 301,192 | 301,479 |
| DRS9 | 241 | P | 73,190 | 73,430 | 252,355 | 252,115 |
| DRS10 | 225 | P | 380,076 | 380,300 | 455,856 | 455,632 |
| DRS11 | 200 | F | 338,753 | 338,952 | 536,718 | 536,917 |
| DRS12 | 165 | F | 523,613 | 523,777 | 552,149 | 552,313 |
| DRS13 | 138 | F | 444,909 | 445,046 | 682,863 | 683,000 |
| DRS14 | 130 | F | 11,917 | 120,46 | 197,372 | 197,501 |
| DRS15 | 125 | F | 19,468 | 19,592 | 163,717 | 163,841 |
| DRS16 | 120 | P | 409,541 | 409,422 | 250,339 | 250,458 |
| DRS17 | 111 | F | 503,030 | 503,140 | 716,487 | 716,377 |
| DRS18 | 106 | F | 362,472 | 362,577 | 394,369 | 394,474 |
| DRS19 | 101 | F | 159,986 | 160,086 | 700,671 | 700,771 |
| DRS20 | 98 | P | 227,118 | 227,215 | 716,057 | 715,960 |
| DRS21 | 85 | P | 701,099 | 701,115 | 300,221 | 300,305 |
| DRS22 | 85 | F | 162,454 | 162,538 | 413,199 | 413,283 |
| DRS23 | 73 | P | 553,929 | 554,001 | 586,127 | 586,055 |
| DRS24 | 73 | P | 226,954 | 227,026 | 716,254 | 716,182 |
| DRS25 | 73 | P | 61,726 | 61,798 | 590,179 | 590,107 |
| DRS26 | 71 | F | 410,796 | 410,866 | 700,746 | 700,816 |
| DRS27 | 70 | F | 248,538 | 248,607 | 482,163 | 482,232 |
| DRS28 | 70 | P | 244,244 | 244,313 | 576,606 | 576,537 |
| DRS29 | 67 | F | 87,186 | 87,252 | 141,916 | 141,982 |
| DRS30 | 67 | F | 87,186 | 87,252 | 339,733 | 339,799 |
| DRS31 | 67 | F | 141,916 | 141,982 | 339,733 | 339,799 |
| DRS32 | 65 | P | 195,478 | 195,542 | 657,450 | 657,386 |
| DRS33 | 64 | F | 91,720 | 91,783 | 146,450 | 146,513 |
| DRS34 | 64 | F | 91,720 | 91,783 | 179,993 | 180,056 |
| DRS35 | 64 | F | 146,450 | 146,513 | 179,993 | 180,056 |
| DRS36 | 63 | F | 182,246 | 182,308 | 216,152 | 216,214 |
| DRS37 | 63 | F | 182,246 | 182,308 | 473,737 | 473,799 |
| DRS38 | 63 | F | 216,152 | 216,214 | 473,737 | 473,799 |
| DRS39 | 61 | P | 291,156 | 291,216 | 504,253 | 504,193 |
| DRS40 | 61 | F | 57,622 | 57,682 | 30,6075 | 30,6135 |
| DRS41 | 59 | F | 495,093 | 495,151 | 528,531 | 528,589 |
| DRS42 | 58 | F | 47,031 | 47,088 | 693,744 | 693,801 |
| DRS43 | 55 | P | 167,131 | 167,185 | 710,277 | 710,223 |
| DRS44 | 53 | F | 195,516 | 195,568 | 312,661 | 312,713 |
| DRS45 | 51 | F | 244,667 | 244,717 | 576,983 | 577,033 |
| DRS46 | 50 | P | 442,609 | 442,560 | 248,466 | 248,515 |
| DRS47 | 50 | P | 226,067 | 226,116 | 441,070 | 441,021 |
| Region | Position | Reference | Coverage | Base Count [A, C, G, T] | AllSubs | Frequency |
|---|---|---|---|---|---|---|
| atp1 | 69 | G | 439 | [56, 0, 383, 0] | GA | 0.13 |
| atp1 | 71 | A | 415 | [352, 63, 0, 0] | AC | 0.15 |
| atp1 | 74 | T | 469 | [0, 62, 0, 407] | TC | 0.13 |
| atp1 | 75 | G | 472 | [0, 0, 410, 62] | GT | 0.13 |
| atp1 | 76 | A | 448 | [386, 62, 0, 0] | AC | 0.14 |
| atp1 | 79 | G | 453 | [59, 0, 394, 0] | GA | 0.13 |
| cox2 | 698 | C | 302 | [0, 255, 0, 47] | CT | 0.16 |
| cox2 | 721 | C | 205 | [0, 141, 0, 64] | CT | 0.31 |
| matR | 1753 | G | 450 | [43, 0, 407, 0] | GA | 0.1 |
| matR | 1760 | A | 438 | [396, 0, 42, 0] | AG | 0.1 |
| nad5 | 83 | C | 253 | [0, 225, 28, 0] | CG | 0.11 |
| nad5 | 84 | C | 254 | [28, 226, 0, 0] | CA | 0.11 |
| nad5 | 1295 | A | 441 | [374, 0, 0, 67] | AT | 0.15 |
| nad5 | 1301 | C | 428 | [62, 366, 0, 0] | CA | 0.14 |
| nad5 | 1312 | C | 417 | [0, 366, 0, 51] | CT | 0.12 |
| nad5 | 1351 | C | 376 | [0, 322, 0, 54] | CT | 0.14 |
| nad5 | 1353 | G | 378 | [53, 0, 325, 0] | GA | 0.14 |
| nad5 | 1361 | T | 356 | [0, 49, 0, 307] | TC | 0.14 |
| nad5 | 1363 | C | 353 | [0, 304, 0, 49] | CT | 0.14 |
| nad5 | 1377 | G | 349 | [58, 0, 291, 0] | GA | 0.17 |
| nad5 | 1509 | T | 279 | [0, 0, 40, 239] | TG | 0.14 |
| nad5 | 1510 | G | 278 | [40, 3, 235, 0] | GA GC | 0.15 |
| nad6 | 26 | C | 425 | [0, 333, 0, 92] | CT | 0.22 |
| nad6 | 55 | C | 520 | [0, 376, 0, 144] | CT | 0.28 |
| nad6 | 154 | A | 727 | [487, 0, 240, 0] | AG | 0.33 |
| nad6 | 185 | C | 764 | [0, 502, 261, 1] | CG CT | 0.34 |
| rps3 | 74 | A | 1 | [0, 0, 0, 1] | AT | 1 |
| rps4 | 63 | G | 570 | [0, 0, 448, 122] | GT | 0.21 |
| rps4 | 75 | T | 550 | [0, 102, 0, 448] | TC | 0.19 |
| rps4 | 78 | A | 560 | [467, 0, 0, 93] | AT | 0.17 |
| rps4 | 87 | A | 525 | [437, 88, 0, 0] | AC | 0.17 |
| rps4 | 93 | C | 553 | [0, 472, 0, 81] | CT | 0.15 |
| rps4 | 100 | C | 540 | [75, 465, 0, 0] | CA | 0.14 |
| rps4 | 159 | T | 646 | [0, 0, 121, 525] | TG | 0.19 |
| rps4 | 185 | C | 652 | [109, 542, 0, 1] | CA CT | 0.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Y.; Yang, S.; Chen, H.; Ni, Y.; Li, J.; Zhang, J.; Liu, C. De Novo Hybrid Assembly of the Tripterygium wilfordii Mitochondrial Genome Provides the Chromosomal Mitochondrial DNA Structure and RNA Editing Events. Int. J. Mol. Sci. 2025, 26, 7093. https://doi.org/10.3390/ijms26157093
Cai Y, Yang S, Chen H, Ni Y, Li J, Zhang J, Liu C. De Novo Hybrid Assembly of the Tripterygium wilfordii Mitochondrial Genome Provides the Chromosomal Mitochondrial DNA Structure and RNA Editing Events. International Journal of Molecular Sciences. 2025; 26(15):7093. https://doi.org/10.3390/ijms26157093
Chicago/Turabian StyleCai, Yisha, Suxin Yang, Haimei Chen, Yang Ni, Jingling Li, Jinghong Zhang, and Chang Liu. 2025. "De Novo Hybrid Assembly of the Tripterygium wilfordii Mitochondrial Genome Provides the Chromosomal Mitochondrial DNA Structure and RNA Editing Events" International Journal of Molecular Sciences 26, no. 15: 7093. https://doi.org/10.3390/ijms26157093
APA StyleCai, Y., Yang, S., Chen, H., Ni, Y., Li, J., Zhang, J., & Liu, C. (2025). De Novo Hybrid Assembly of the Tripterygium wilfordii Mitochondrial Genome Provides the Chromosomal Mitochondrial DNA Structure and RNA Editing Events. International Journal of Molecular Sciences, 26(15), 7093. https://doi.org/10.3390/ijms26157093






