1. Introduction
Metformin is a kind of biguanide drug used in the treatment of type 2 diabetes, which plays a role mainly by activating AMP-activated protein kinase (AMPK). Metformin, a biguanide-class drug and first-line therapeutic for type 2 diabetes mellitus (T2DM), exerts its primary pharmacologic action through activation of AMP-activated protein kinase (AMPK). This activation leads to inhibition of hepatic gluconeogenesis, suppression of glucagon secretion, and enhancement of insulin sensitivity, collectively contributing to effectively promoting blood glucose reduction [
1,
2,
3]. In addition to lowering blood sugar, metformin also has many functions, such as delaying the aging process and extending lifespan in both mice and nematodes [
4,
5,
6]. Moreover, it has been found to reduce the risk of Alzheimer’s disease in diabetics [
7]. Metformin can also improve obesity-induced inflammation in mice with a high-fat diet [
8]. In addition, it has shown the ability to inhibit the growth and metastasis of various types of tumor cells, including breast cancer, liver cancer, and gastric cancer, breast, hepatic, and gastric carcinomas [
9,
10]. Furthermore, metformin promotes myelin regeneration in the central nervous system. In neural systems, metformin enhances central nervous system (CNS) remyelination and restores the regenerative ability of aged oligodendrocyte progenitor cells in rats [
11]. It also facilitates heart regeneration in zebrafish by triggering autophagy [
12]. In an ischemia–reperfusion (I/R) model in mice, metformin was shown to attenuate cardiac and hypertrophic remodeling after reperfusion [
13].
Planarian
Dugesia japonica (
D. japonica) has a strong regenerative ability and possesses extraordinary regenerative capacity, serving as a key model organism for developmental biology, pharmacology, and regeneration research [
14,
15,
16,
17,
18]. The regenerative ability of planarians is mainly attributed to the abundant neoblasts in their body, which constitute approximately 20–30% of the total number of cells in the planarian [
19,
20]. Neoblasts have the ability of self-renewal, proliferation, differentiation, and migration. self-renewal, proliferative capacity, multipotent differentiation, and migratory activity. They can differentiate into cells of any tissue type, thereby providing a cellular basis for the regeneration of planarian individuals [
19,
20]. The regeneration process of planarians establishes polarity along the anterior–posterior and dorsal–ventral axes, with the anterior fragment regenerating a head and the posterior fragment regenerating a tail after amputation; the dorsal and ventral sides can be correctly distinguished and form specific tissues and structures [
14]. Planarian regeneration is related to a variety of signaling pathways, such as Wnt and EGFR [
21]. Research has shown that the Wnt signaling pathway can affect antero-posterior govern anteroposterior patterning and the proliferation of stem cells during planarian regeneration [
22,
23]. The inhibition of
Evi/Wls and
Wnt1 genes can lead to ectopic phenomena in head formation, and the suppression of the
Notum gene can result in head loss or the formation of double tails of the regenerating planarian [
24,
25]. The EGFR pathway is involved in the mediation of differentiation of various tissues such as the intestine, pharynx, and photoreceptors of planarian [
26]. There are also some genes involved in regulating the regeneration process of planarians, and abnormal expression of these genes can affect their regeneration. Knockdown of the
Djfoxk1 gene can inhibit regeneration of the cranial ganglia, resulting in the failure of eye regeneration [
27], while the degradation of the
Djequinox gene results in abnormal regeneration of planarian buds [
28]. Therefore, planarian regeneration is a complex network of regulatory processes orchestrated regulatory network influenced by various genes and microenvironments.
MicroRNA (miRNA) is an approximately 20 nt noncoding single-stranded RNA endogenous noncoding RNA molecule encoded by endogenous genes. It regulates many important biological processes, including cell proliferation, differentiation, migration, apoptosis, and tissue/organ homeostasis [
29,
30]. MiRNAs exert their biological functions mainly by binding to the 3′UTR 3′ untranslated (3′UTR) region and leading to the degradation and inhibition of expression of their target genes [
29,
30]. Further research found that miRNA can also bind to the CDS region or 5′UTR 5′ untranslated (5′UTR) region of their target genes [
31,
32]. MiRNA-regulated target genes also include tRNA, rRNA, and lncRNA (long noncoding RNA) in addition to mRNA [
31,
32]. Research on miRNAs in regeneration has found that
miR-203 hinders zebrafish fin regeneration by suppressing the Wnt signaling mediator
lef1 [
33]; the
miR-183 cluster (
miR-96,
miR-182, and
miR-183) plays a role in regulating hair cell regeneration in zebrafish ears [
34]. In planarians, the
miR-124 family is linked to brain and visual system regeneration [
35], and
miR-8b participates in brain and eye regeneration [
36]. These findings show miRNAs’ regulatory roles in regeneration biology.
Metformin can modulate the expression of various miRNAs, thereby helping to achieve its biological functions. The
let-7 microRNA family possesses tumor-suppressive activity and tumor suppressor functions. Recent studies have shown that metformin significantly increases the levels of
let-7a in cancer stem cells in mouse xenograft models, antagonizing cancer progression, inhibiting tumorigenesis [
37]. Metformin can reduce insulin resistance in human adipocytes by downregulating the expression of
miR-223 and improving the IRS/Akt/GLUT4 signaling pathway [
38]. In BALB/c wild-type mice, metformin enhances the antitumor activity of NK cells via overexpression of
miR-150 and
miR-155 [
39]. Other studies have revealed that the biological actions of metformin are mediated through the downregulation of genes specific to cancer stem cells (CSCs) and the re-expression of miRNAs (
let-7a,
let-7b,
miR-26a,
miR-101,
miR-200b, and
miR-200c) that are typically lost in pancreatic cancer, indicating that metformin may help overcome therapeutic resistance in pancreatic cancer cells [
40].
MiR-27 is a functionally diverse miRNA family; it is divided into two subtypes:
miR-27a and
miR-27b in humans.
MiR-27b is closely related to bone metabolism and tumor development. The decrease in the expression level of
miR-27b can increase the expression of its target gene
MMOL/LP-13, leading to a decrease in the expression of
COLII collagen type II (COL2A1), and causing intervertebral disc degeneration [
41]. In the field of cancer research, it has been found that high expression of
miR-27b can inhibit the expression of
PPARγ, promote the proliferation and invasion of cervical cancer cells, and increase the risk of cervical cancer [
42]. Low expression of
miR-27b may increase the occurrence and development of gastric cancer by targeting
GSPT1 or
ROR1 [
43,
44].
Pax6 is a transcription factor essential for the development of tissues like eyes, central nervous system, and endocrine glands in vertebrates (e.g., mice, zebrafish) and invertebrates (e.g., fruit flies, nematodes) [
45]. Studies in animal models (e.g., mice, frogs, chickens) showed it regulates eye development and orchestrates retinogenesis by controlling cell proliferation, differentiation, and survival in eye tissues [
46]. Mutations or deletions of the
Pax6 gene are linked to congenital eye malformations and neurological developmental abnormalities, such as anophthalmia, microphthalmia, iris defects, iridohypoplasia, and brain/retina retino-cortical dysplasia [
46,
47,
48]. It has been reported that
miR-27b is expressed in the human retinal pigment epithelial cell line ARPE-19 [
49]. In the serum and retinas of 5–10-week-old diabetic mice,
miR-27b-3p is one of the significantly dysregulated miRNAs. These miRNAs can regulate the expression of
VEGF,
CREB1,
BDNF, and
PPAR-α in diabetic retinas and may serve as potential biomarkers and therapeutic targets for early diabetic retinopathy [
50]. In early retinal development, the expression of
Pax6 is regulated by multiple signaling pathways, including BMP and TGF-β, although its exact mechanisms of action remain unclear [
51].
In this study, the function of DjmiR-27b and DjPax6 in planarian eyespot regeneration regulated by metformin was investigated. It was found that metformin can regulate planarian eyespot regeneration by modulating the expression of the target gene DjPax6 through DjmiR-27b. This study provides a theoretical basis and mechanistic insight for the research of metformin and miRNA in the field of regeneration.
3. Discussion
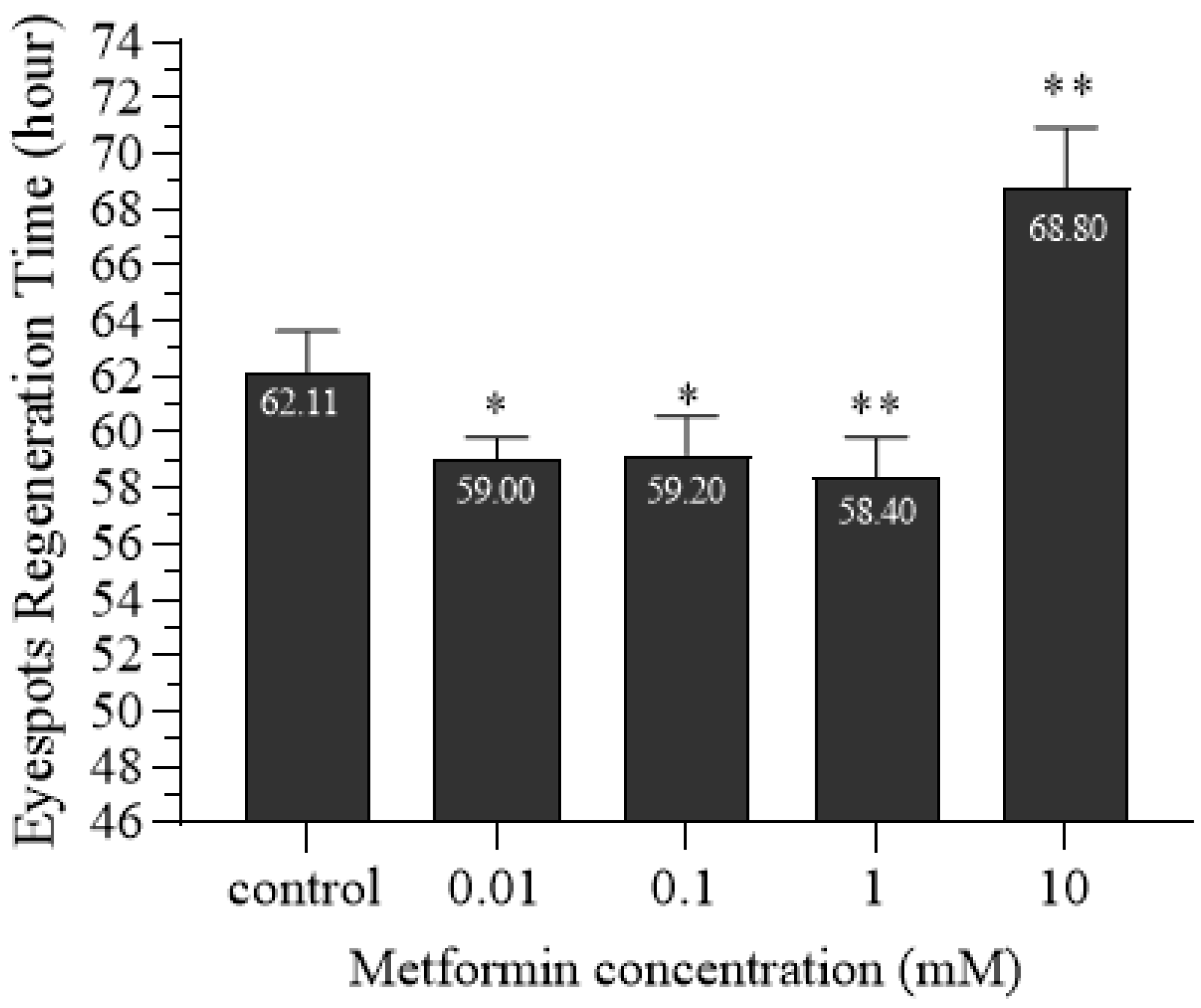
In recent years, research has found that metformin is not only a drug for the treatment of type 2 diabetes but is also associated with the regeneration of tissues and organs [
1,
11,
12,
54]. Studies have shown that 50 μM metformin can enhance autophagic flux and improve the regeneration of the epicardium, endocardium, and vascular endothelium in zebrafish, promote autophagic flux activation, accelerating regeneration of epicardial, endocardial, and vascular endothelial tissues in zebrafish [
12]. In vitro experiments with C57BL/6 mice, treatment with 100 µM metformin increases phosphorylated AMPK levels, restoring the differentiation capacity of senescent oligodendrocyte precursor cells and thereby improving central nervous system CNS remyelination [
11]. Lei et al. found that 0.1 mM metformin can promote the proliferation of human umbilical cord mesenchymal stem cells and enhance their osteogenesis and angiogenesis [
55]. These studies indicate that metformin has the effect of promoting regeneration, and the concentration of promoting regeneration varies in different tissues and organs. Similarly, the planarian eyespot regeneration experiments in this study revealed that 0.01 mM, 0.1 mM, and 1 mM metformin significantly promoted eyespot regeneration, whereas 10 mM metformin had the opposite effect. In this experiment, the regeneration time of eyespots after treatment with metformin in regenerating planarians was different from Zhao’s results [
52]. This may be due to the seasonal rhythm of planarians’ regeneration. Although the constant temperature of the incubator eliminates the influence of temperature changes as an environmental factor on planarians, planarians still have endogenous circadian rhythms and rhythmic responses to non-temperature environmental factors. The regeneration of planarians has a certain seasonal rhythm [
56,
57,
58]. Therefore, in our experiment, we indeed found that there were differences in the regeneration rate of planarian eyespots among batches from different seasons. However, for the same batch of planarians, the regeneration rate of the eyespots of planarians cultured in low concentrations of metformin is indeed significantly faster than that of those cultured in clean water, indicating that metformin can accelerate the regeneration of planarians.
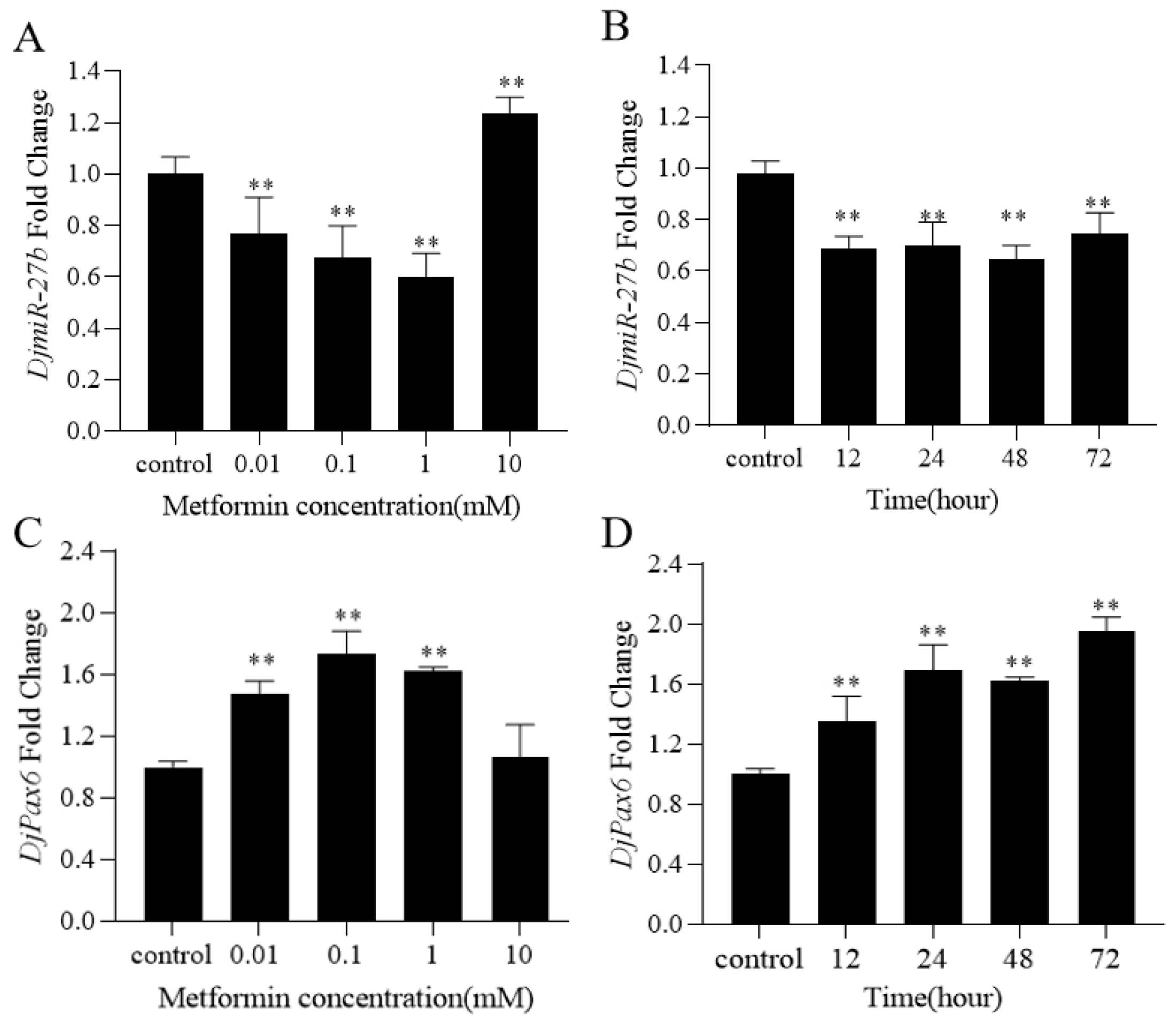
MiRNAs are involved in various physiological activities such as cell proliferation, differentiation, migration, apoptosis, and tissue metabolism in organisms [
30]. Studies have shown that metformin exerts its biological effects by regulating the expression of various miRNAs. Metformin could increase the expression of
miR-497a-5p in mouse eyes and alleviate the severity of their retinal lesions [
59]. Metformin could delay cell aging, cellular senescence in human dental pulp stem cells (DPSCs) by downregulating
miR-34a-3p in human dental pulp stem cells [
60] and reduce lipid accumulation in high glucose-induced HepG2 cells by downregulating
miR-33b expression [
61]. Research in planarians has found that
miR-8b serves as a crucial regulator involved in brain and eyespot regeneration in
D. japonica [
34]. Loss of the
miR-124 family could lead to a significant reduction in the regenerative capacity of the brain and visual system in
Schmidtea mediterranea [
62]. Therefore, we speculate that metformin might affect the regeneration of
D. japonica by regulating the expression of miRNA. Through the study of the effect of metformin on the expression of
DjmiR-27b and
DjPax6 in the regenerating planarians, we found that low concentrations of metformin significantly inhibited the expression of
DjmiR-27b and promoted the expression of
DjPax6. There is a negative regulatory relationship between
DjmiR-27b and
DjPax6. The dual-luciferase reporter assay confirmed that
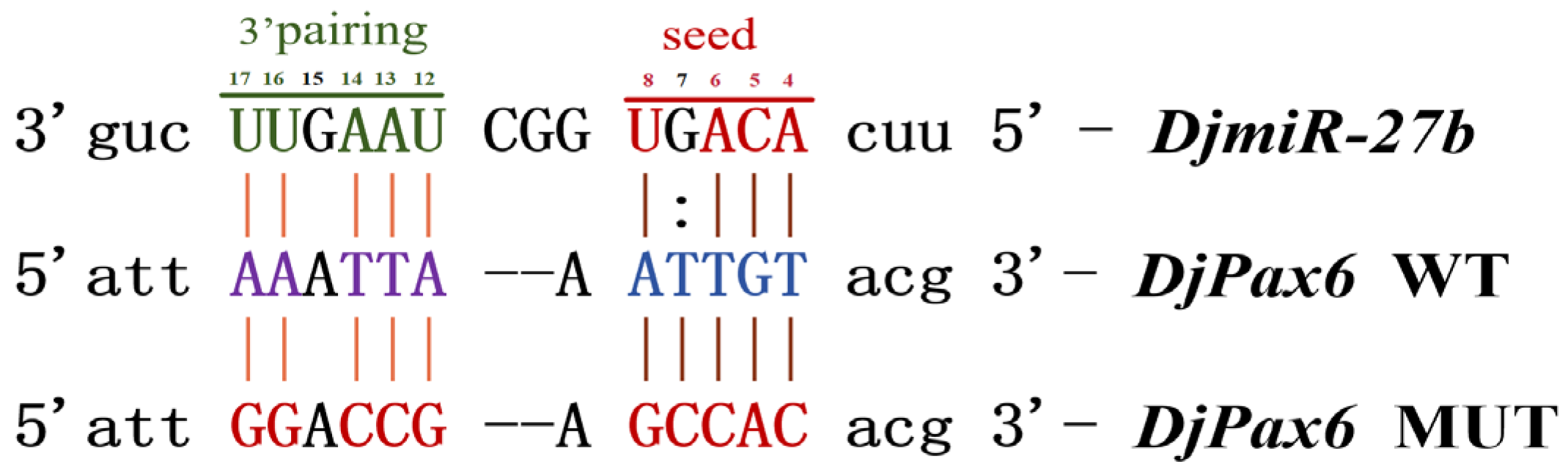
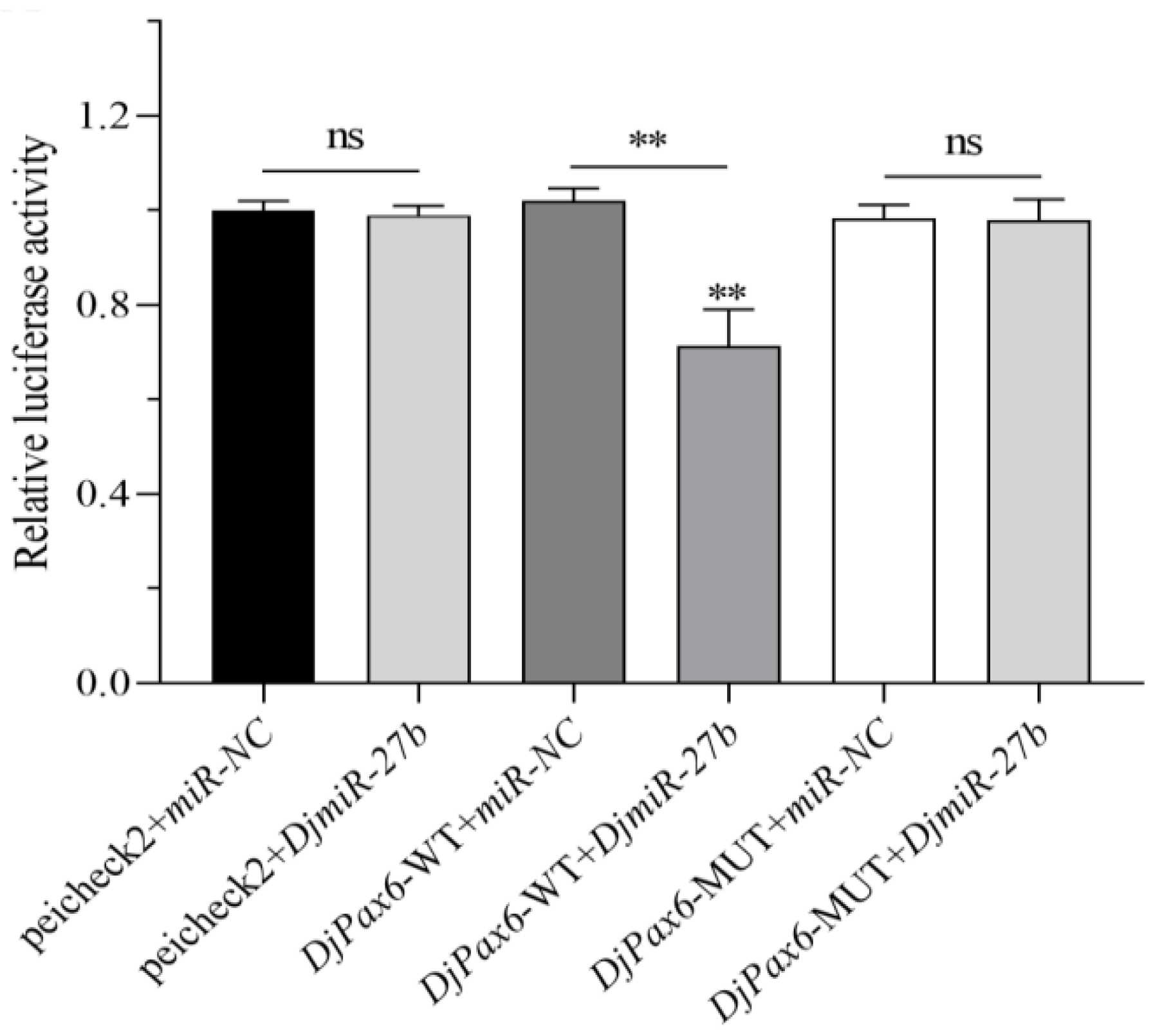
DjmiR-27b directly targets the binding site of the 3′UTR of the
DjPax6 gene, so it is speculated that metformin may regulate planarian regeneration by regulating the expression level of the target gene
DjPax6 through
DjmiR-27b.
Pax6 is a key regulator of eye development in numerous organisms, which is involved in eye development in
Drosophila,
Capitella teleta, and mammals [
63,
64,
65].
Xenopus tropicalis Pax6 is expressed in various eye tissues, including the neural retina layers, retinal neuroepithelium, optic disc, corneal epithelium, iris, and ciliary body [
66,
67]. Abnormal expression of
Pax6 could lead to abnormal eye development, resulting in microphthalmia microphthalmos in mice [
68,
69] and anophthalmia anophthalmos in fruit flies [
70]. The research of Pineda D et al. found that
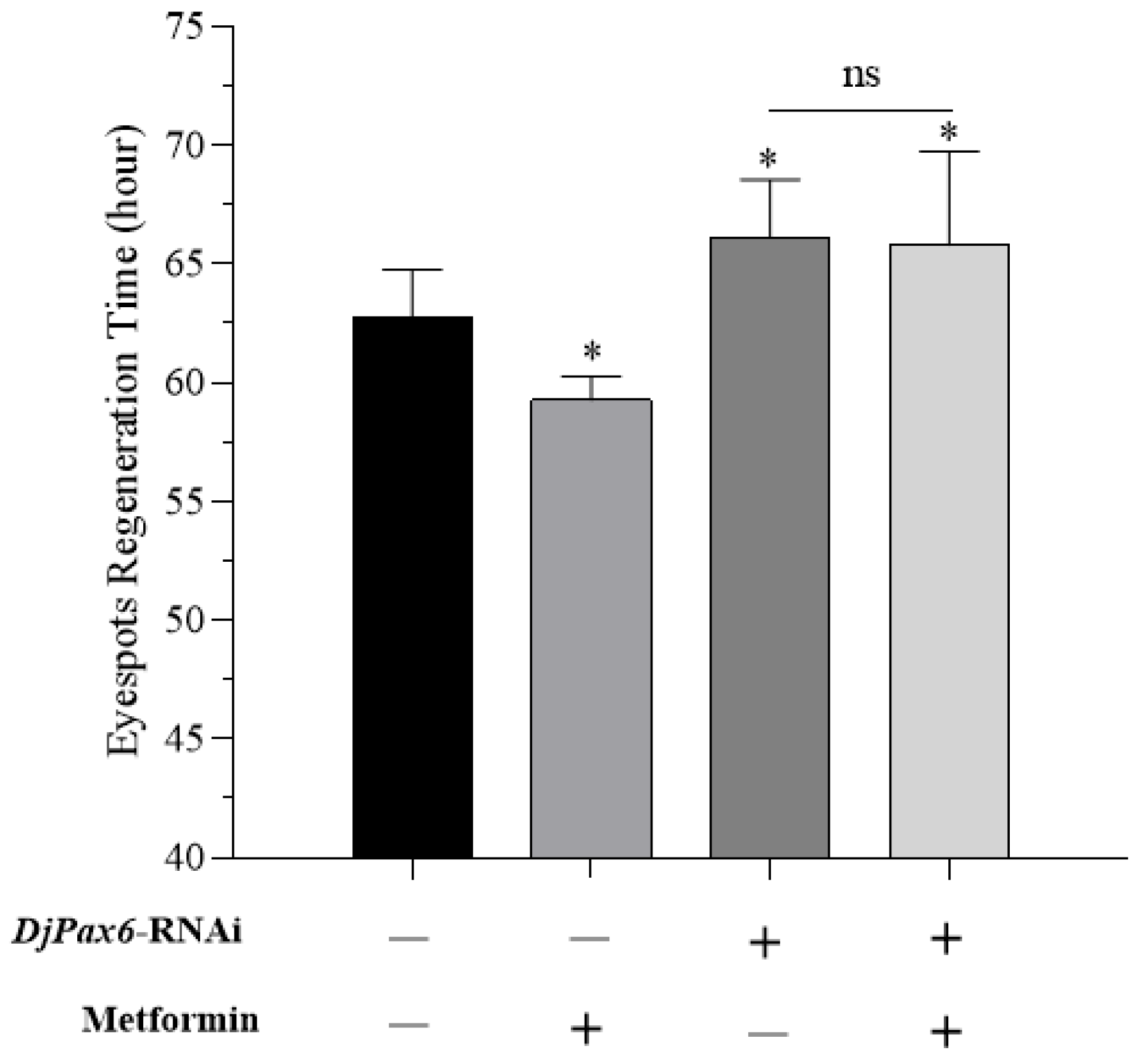
Pax6 knockdown in planarians did not disrupt normal eyespot development, leading to the conclusion that
Pax6 has no role in planarian eyespot regeneration [
71]. However, our study reveals that although
Pax6 knockdown did not cause morphological eyespot regeneration defects, it significantly prolonged the eyespot regeneration time, indicating the involvement of
Pax6 in planarian eyespot regeneration.
Studies have shown that
miR-27b is involved in tissue regeneration, metabolism, and other biological processes by regulating its target genes or related signaling pathways. Studies on human microvascular endothelial cells (HMEC)-1 have shown that downregulation of
miR-27b can activate the PI3K/AKT signaling pathway and promote angiogenesis [
72]. By targeting the regulation of
HOXC6 expression,
miR-27b could inhibit epithelial–mesenchymal transition in human retinal pigment epithelial cells, reducing retinal fibrosis fibrotic retinopathy in patients [
73]. In our work, overexpression of
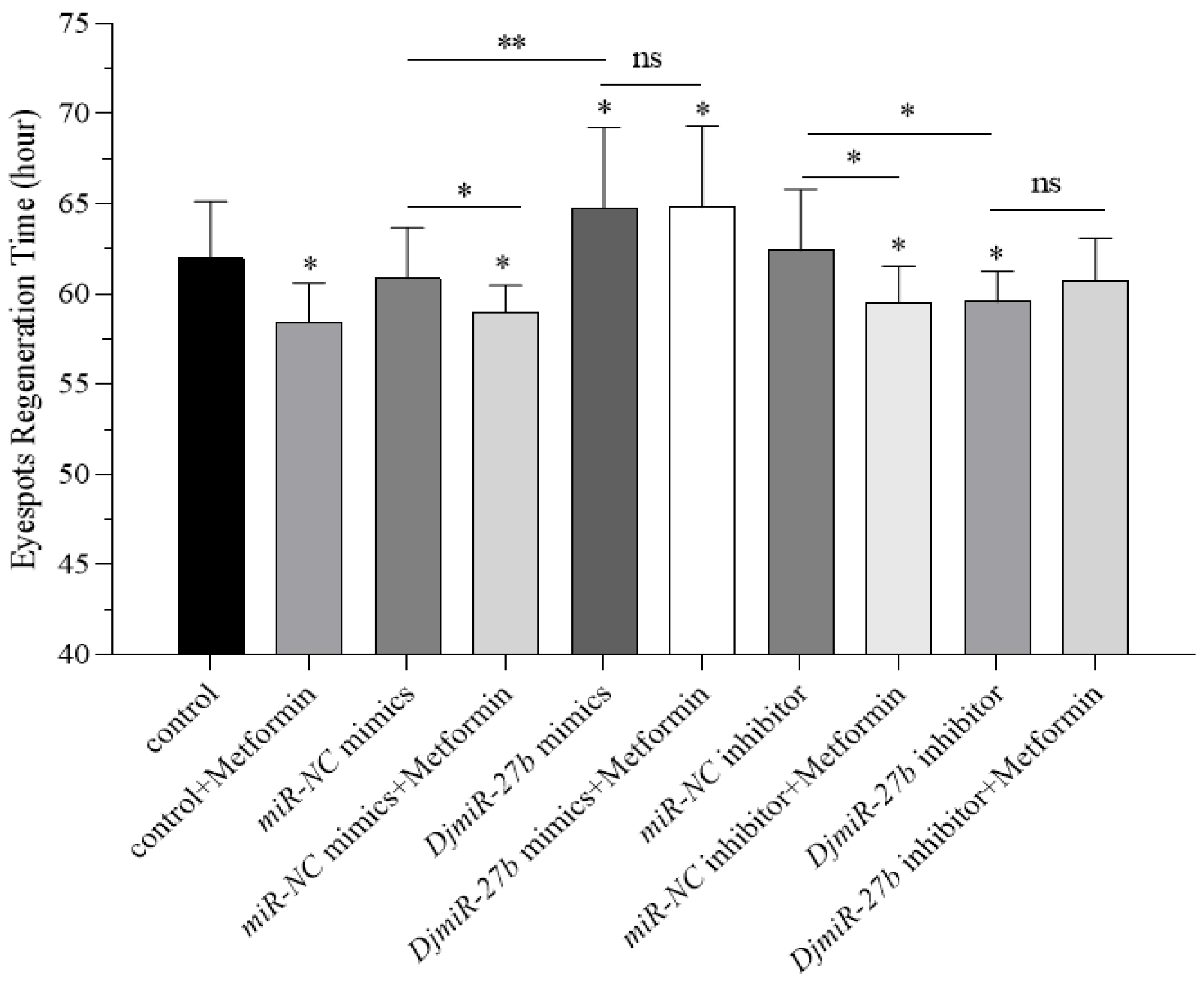
DjmiR-27b and inhibition of its target gene
DjPax6 both significantly prolonged the regeneration time of planarian eyespots, whereas knockdown of
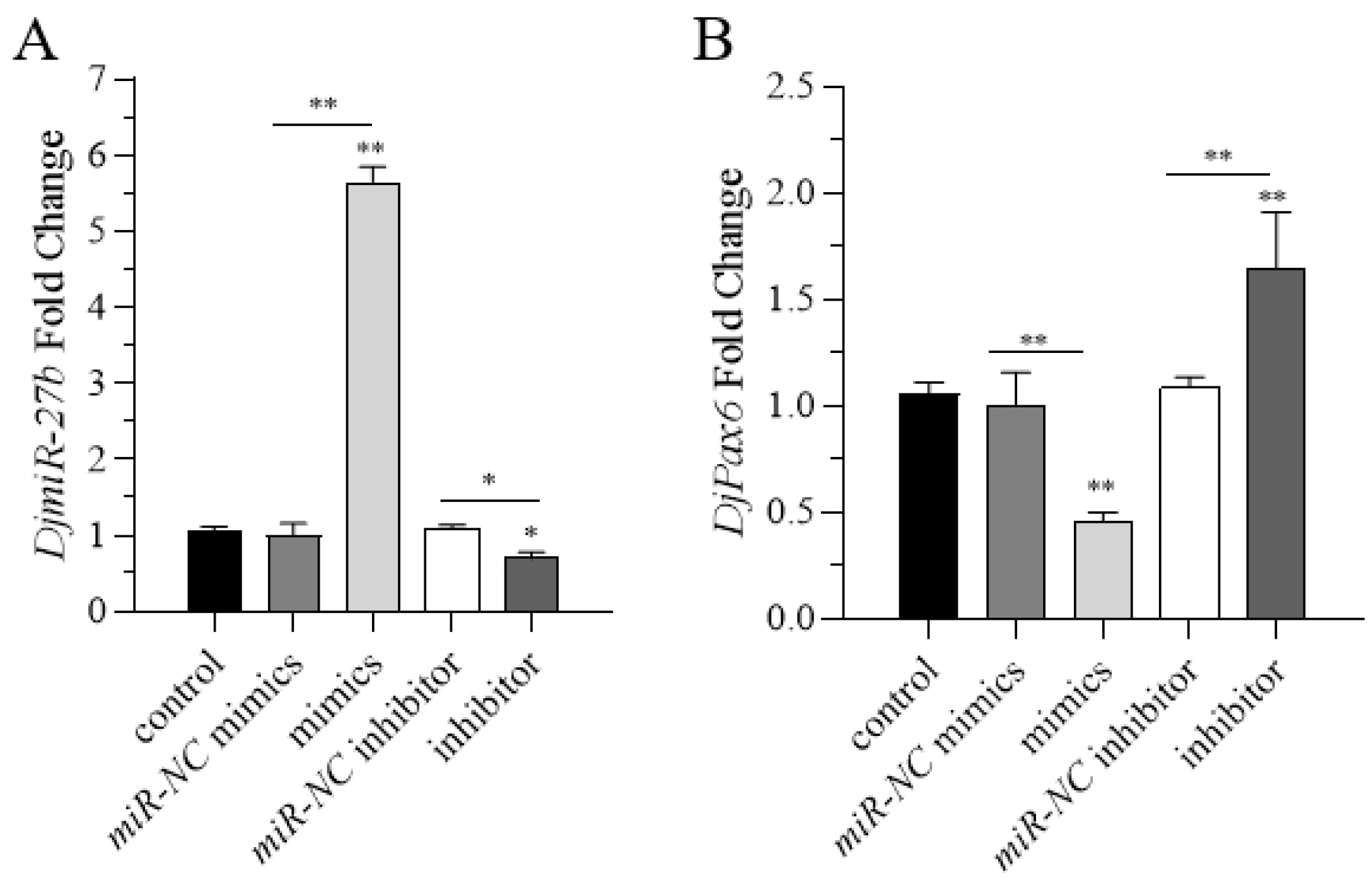
DjmiR-27b significantly shortened the regeneration time. This suggests that overexpression of
DjmiR-27b downregulates its target gene
DjPax6, thereby inhibiting eyespot regeneration in planarians. Conversely, inhibition of
DjmiR-27b upregulates
DjPax6 expression and promotes eyespot regeneration. Combining the regulatory relationship between miRNA and target genes, it is speculated that metformin may inhibit the expression of
DjmiR-27b, thereby promoting the expression of the target gene
DjPax6 and regulating the process of regeneration in regenerating planarians. Previous studies have demonstrated that
miR-27b could promote muscle regeneration in mice after injury by targeting the target gene
MDFI, and the expression of type II collagen during the differentiation of rat articular chondrocytes by regulating the expression of the target gene
Pparγ2 [
74,
75]. Therefore, as an important regulatory factor,
miR-27b can regulate cell proliferation, differentiation, and metabolism by modulating different target genes, thereby affecting tissue development and repair.
In situ hybridization has been utilized to detect the spatial expression patterns of genes in organisms. So far, only a few studies have been reported for the expression localization of miRNAs in planarians [
34,
62].
DjmiR-124 and
DjmiR-8b are involved in the regeneration of the eye or brain in planarians.
DjmiR-8b is specifically expressed in the head and eyespots of planarians [
34], and
DjmiR-124 is predominantly localized to the cephalic ganglia, ventral nerve cords, and visual system [
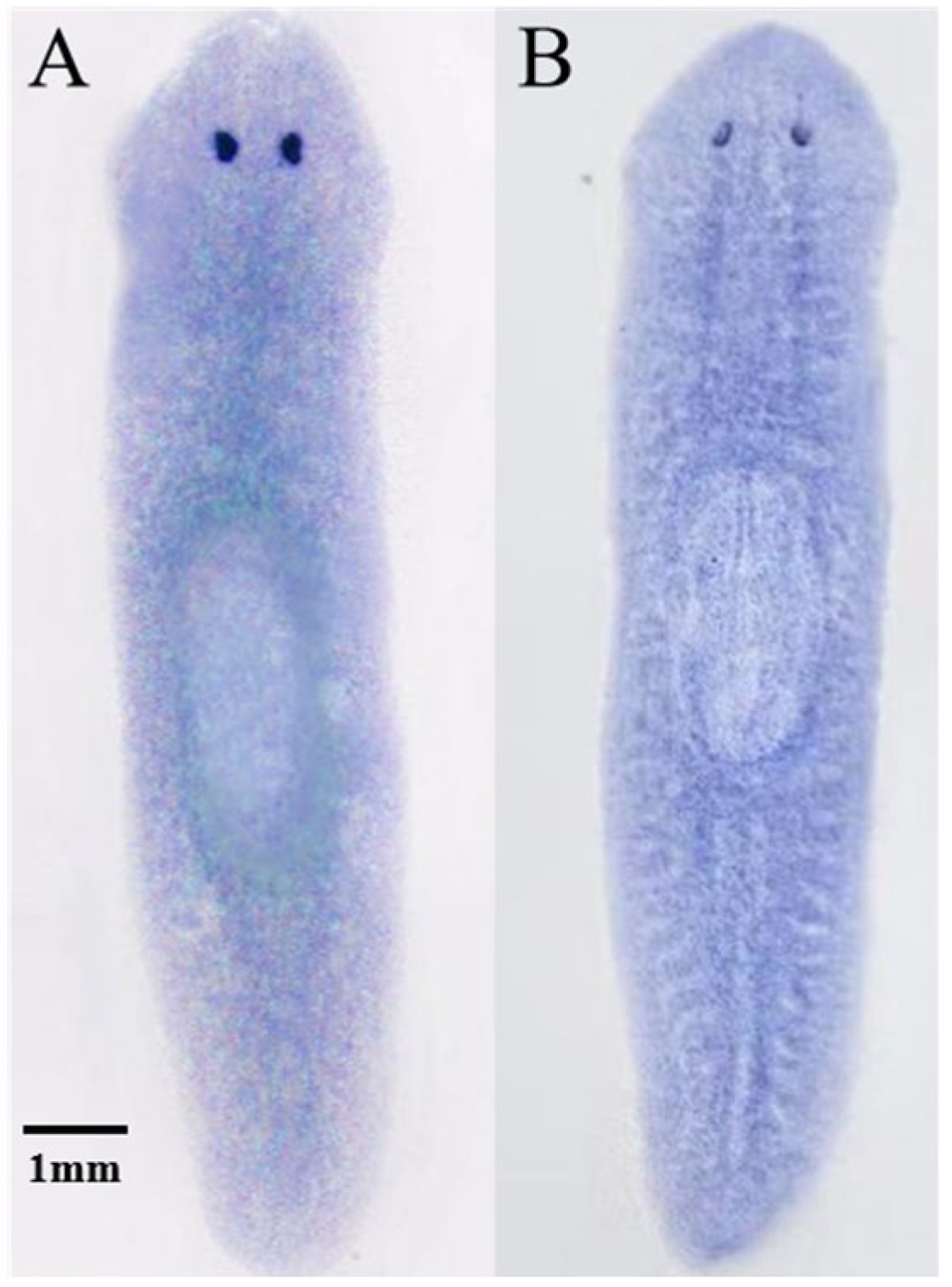
12]. The morphogenesis of eyespots is crucial for head regeneration in planarians. Results of in situ hybridization showed that
DjmiR-27b and its target gene
DjPax6 are widely expressed in planarian head tissue.
DjmiR-27b likely regulates eyespot regeneration by targeting
DjPax6 in the early stage of head regeneration.
The results of this study reveal the regulatory effects of metformin on DjmiR-27b and DjPax6, as well as their targeting relationship, indicating that metformin may modulate planarian eyespot regeneration by regulating DjPax6 expression via DjmiR-27b. This research sheds light on the potential molecular mechanisms by which metformin regulates planarian regeneration, providing a theoretical basis for studying metformin in regenerative medicine. However, the regeneration of planarian eyespots involves complex molecular regulatory mechanisms. As a drug, metformin does not regulate planarian regeneration in a single-targeted manner. DjmiR-27b is just one of the many molecules involved in regulating regeneration. Further studies beyond the DjmiR-27b/Pax6 axis will enhance our understanding of the molecular mechanisms by which metformin regulates planarian regeneration.
4. Materials and Methods
4.1. Experimental Animal
The planarian used in this study was an asexual strain
Dugesia ZB-1 kept in our laboratory [
76]. The animals were cultured in
Montjuïc water (1.6 mM NaCl, 1 mM CaCl
2, 1 mM MgSO
4, 0.1 mM MgCl
2, 0.1 mM KCl, and 1.2 mM NaHCO
3) (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) in an incubator at 20 °C. Planarians were fed with nematode homogenate once a week. Before the experiments, planarians were subjected to a week-long period of starvation.
4.2. Experimental Material
E. coli HT115 and L4440 vectors were preserved by our laboratory.
E. coli DH5α competent cells were purchased from Shanghai Angyu Biotechnology Co., Ltd. (Shanghai, China). The restriction enzymes
XhoI and
PstI, along with T4 DNA ligase, were purchased from Takara Bio Inc. (Kusatsu, Shiga, Japan). Revertra Ace qPCR RT Kit was obtained from Toyobo Co., Ltd. (Osaka, Japan). Dual-luciferase reporter vector psiCHECK2 and 293T cells were provided by Biotech Primer Co., Ltd. (Beijing, China). LipoFiter Transfection Reagent was purchased from Shanghai Hanbiotechnology Co., Ltd. (Shanghai, China).
DjmiR-27b mimics,
DjmiR-27b inhibitor, and
DjmiR-27b-specific probe were synthesized by Sangon Biotech (Shanghai) Co., Ltd. (China). All primers were commercially synthesized by Beijing Genomics Institute (BGI), and their sequences are listed in
Appendix A,
Table A1,
Table A2,
Table A3,
Table A4,
Table A5,
Table A6,
Table A7 and
Table A8. In addition, 4% paraformaldehyde (PFA) was purchased from Wuhan Service Biotechnology Co., Ltd. (Wuhan, China). T7 RNA Polymerase and RNase Inhibitor were purchased from Beijing GenStar Biotechnology Co., Ltd. (Beijing, China). DIG, DIG antibody, and NBT/BCIP were purchased from F. Hoffmann-La Roche Co., Ltd. (Basel, Switzerland). Maleic acid, sodium heparin, and Tween 20 were obtained from Sigma-Aldrich Co. LLC (St. Louis, MO, USA). In addition, 20 × SSC were purchased from Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China). Yeast RNA and 50×Denhardt’s solution were obtained from Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China).
4.3. Determination of DjmiR-27b and DjPax6 Gene as Research Subjects
Two databases generated from our previous work were analyzed: the miRNA database of the regenerating planarians [
53] and the transcriptome database of metformin-treated regenerating planarians [
52]. Downregulated miRNAs were selected from the miRNA database, and upregulated genes were identified from the transcriptome database. Target genes for the selected miRNAs were predicted using TargetScan (
https://www.targetscan.org/vert_72/) (accessed on 19 July 2023). Consequently,
DjmiR-27b and its target gene
DjPax6 were chosen as the research targets.
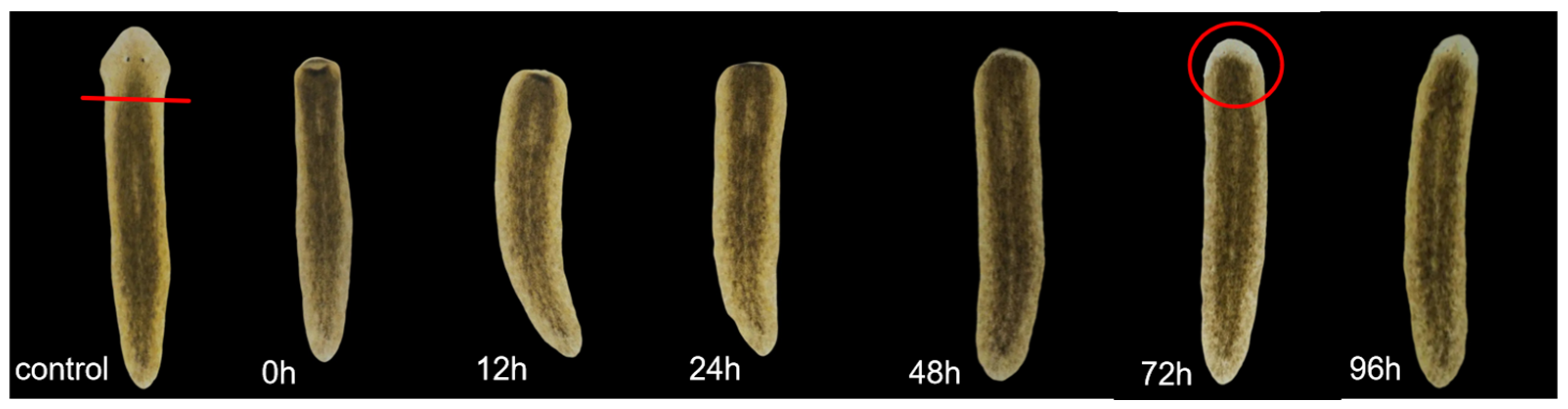
4.4. Planarian Eyespot Regeneration Experiment
A total of 50 planarians with a body length of approximately 0.8–1 cm were selected. Eyespots were surgically removed posterior to the auricles under SZ650 stereomicroscopy (Chongqing Aote Optical Instruments Co., Ltd., Chongqing, China). The regenerating planarians were divided into five groups, with ten individuals in each group. Four experimental groups were placed in metformin solutions of 0.01 mM, 0.1 mM, 1 mM, and 10 mM, respectively, while the control group was placed in clear water. Solutions were replaced daily to ensure consistent drug concentration. Eyespot regeneration was observed every 12 h within the first 48 h, then hourly after 48 h. Photos were taken at 0 h, 12 h, 24 h, 48 h, 72 h, and 96 h. When two distinct and symmetrical black eyespots could be clearly observed under the stereomicroscope, the corresponding time was recorded as the eyespots’ regeneration time. The experiment was repeated three times.
4.5. RT-qPCR
Total RNA was extracted from planarians using Trizol reagent (Thermo Fisher Scientific Inc., Waltham, MA, USA). Reverse transcription was performed with Oligo (dT)15 primers (Beijing Genomics Institute, Shenzhen, Guangdong, China) using ReverTra Ace qPCR RT Kit (Toyobo Co., Ltd., Osaka, Japan) according to the manufacturer’s protocol. Quantitative PCR (qPCR) reaction mixture (10 µL) contained 1 µL cDNA of 10 ng/µL, 0.4 µL 10 µM forward primer, 0.4 µL 10 µM reverse primer, 5 µL TB Green Premix Ex Taq II (2×), and 3.2 µL ddH2O. Amplification was performed on a Light Cycler 480 system (Roche Diagnostics, Basel, Switzerland) with the following protocol: initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The experiment was repeated three times. DjActin gene was used as an endogenous reference, and the expression levels of genes were calculated using the 2−ΔΔCt method.
4.6. Dual-Luciferase Reporter Assay
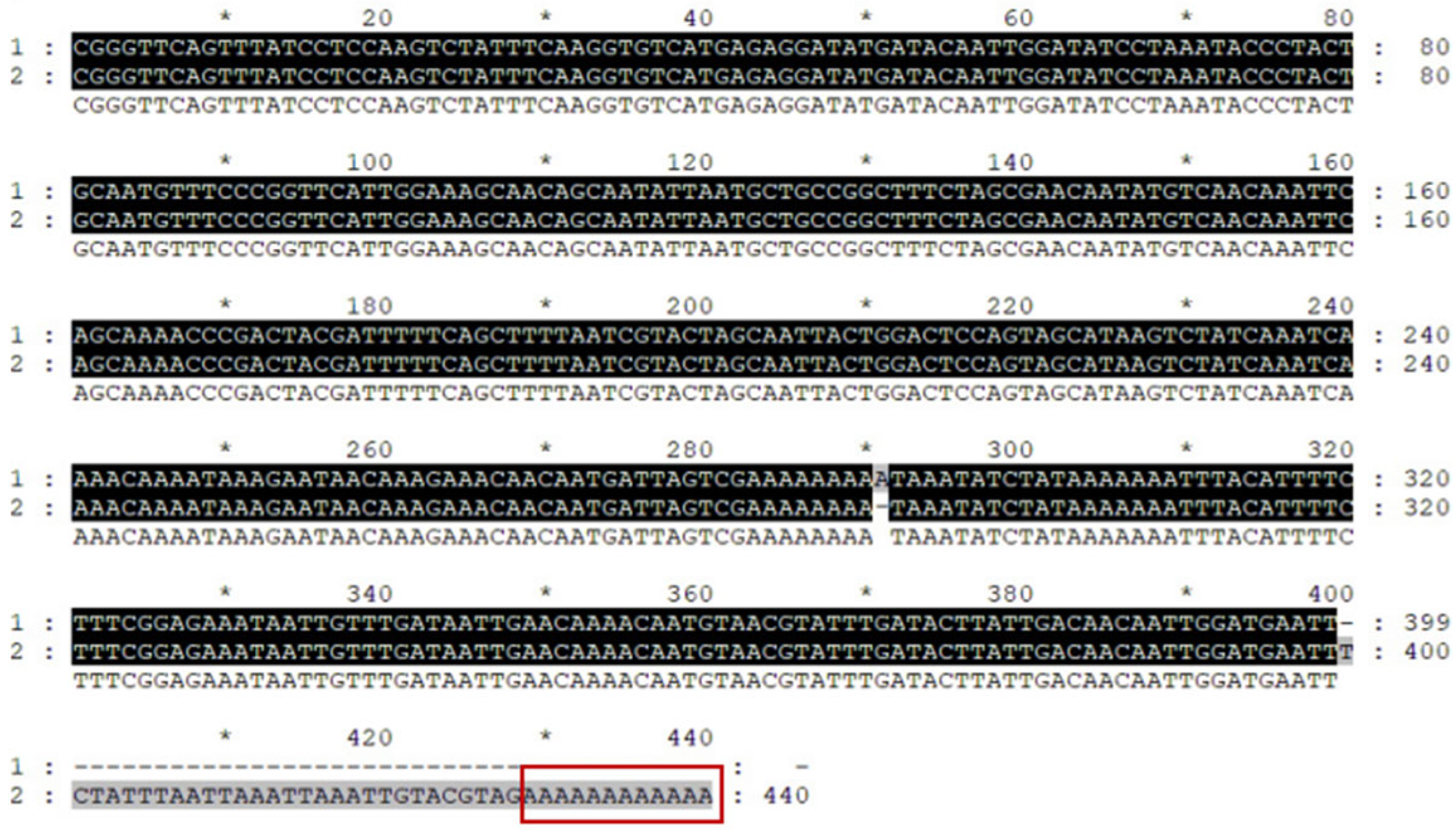
The binding sites and energy between miRNA and target genes were predicted using the Bioinformatics Platform (
http://www.bioinformatics.com.cn) (accessed on 19 July 2023). Site-directed mutagenesis was performed on the miRNA-binding regions with A→G and T→C substitutions. About 200 bp sequences with
XhoI and
NotI restriction sites flanking the wild-type (WT) and mutant (MUT) binding sites of
DjPax6 were synthesized by Hunan Pulettech Biotechnology Co., Ltd. (Changsha, Hunan, China). These fragments were cloned into the psiCHECK-2 dual-luciferase reporter vector using
XhoI/NotI digestion and T4 DNA Ligase, then transformed into DH5α cells. Positive clones were sequenced by Suzhou Genewiz Biotechnology Co., Ltd. The validated plasmids were transfected into 293T cells, incubated for 24 h at 37 °C with 5% CO
2, and lysed with Passive Lysis Buffer. Firefly and Renilla luciferase activities were measured sequentially using the Lux-T020 Detector (Guangdong Biolight Medical Equipment Co., Ltd., Zhuhai, China). The instrument settings were: 2 s interval, 10 s measurement time, and 40–100 µL sample volume. Firefly luciferase activity was measured first, followed by Renilla luciferase activity as an internal reference.
4.7. DjmiR-27b Overexpression and Inhibition Experiments
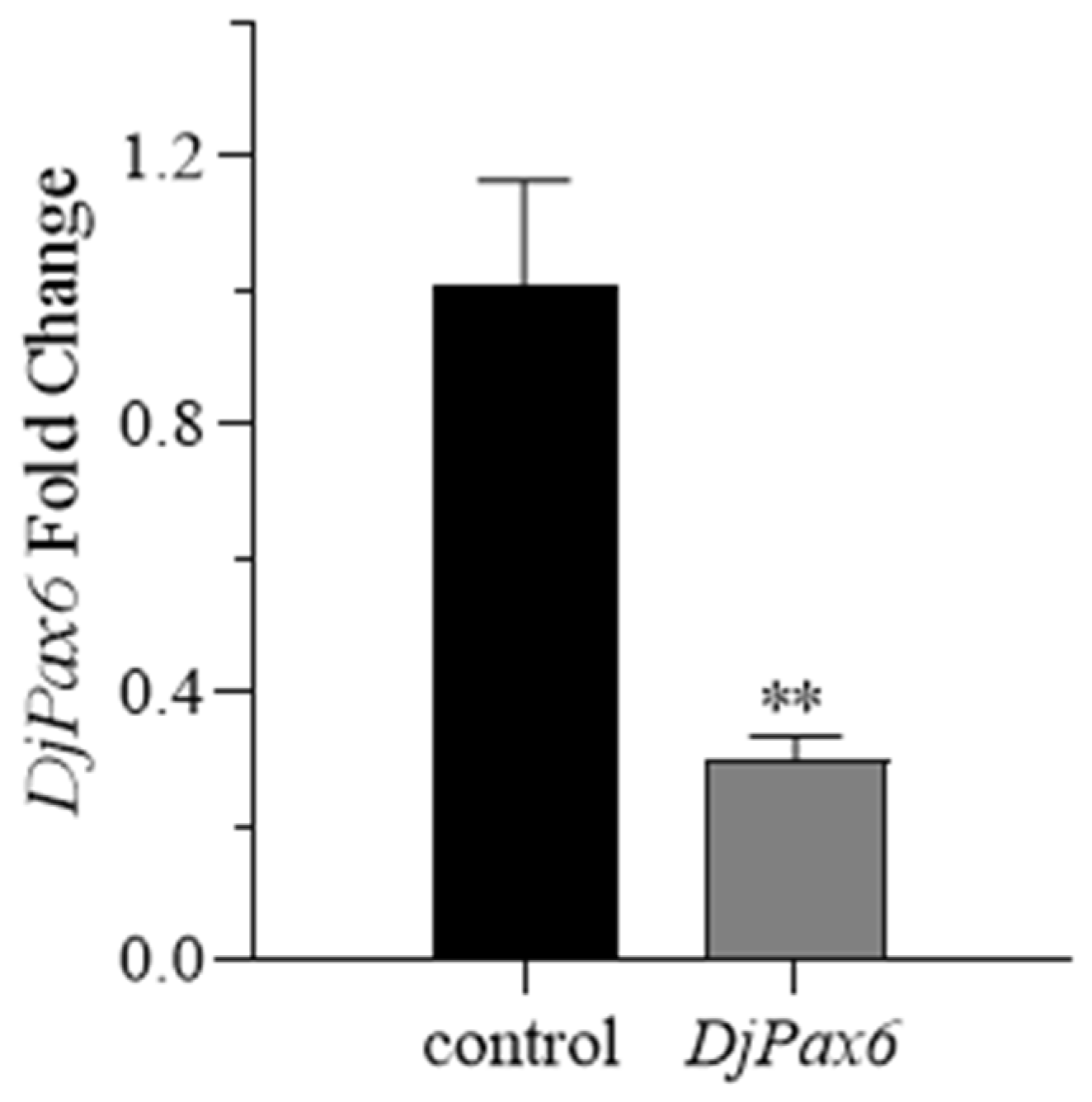
The experiments of overexpression and knockdown of DjmiR-27b in planarians were performed through the feeding method. Each food homogenate contained 15 µL nematode homogenate mixed with 5 µL 2% low-melting-point agarose (Biosharp, Hefei, Anhui, China). A total of 30 planarians with a body length of 0.6–0.8 cm were selected for each experimental group. The overexpression group was fed with 8 µL of DjmiR-27b mimics mixed with 20 µL of food homogenate. The knockdown group was fed with 8 µL of DjmiR-27b inhibitor mixed with 20 µL of food homogenate. The negative control group was fed with 8 µL DjmiR-27b negative control (NC) mixed with 20 µL food homogenate, and the blank control group was fed with 8 µL RNase-free water mixed with 20 µL food homogenate. Planarians were fed once a day for 10 consecutive days. Total RNAs were then extracted from the planarians, and RT-qPCR was performed to assess the efficiency of DjmiR-27b overexpression and knockdown. The experiment was repeated three times.
4.8. RNA Interference (RNAi) of DjPax6 Gene
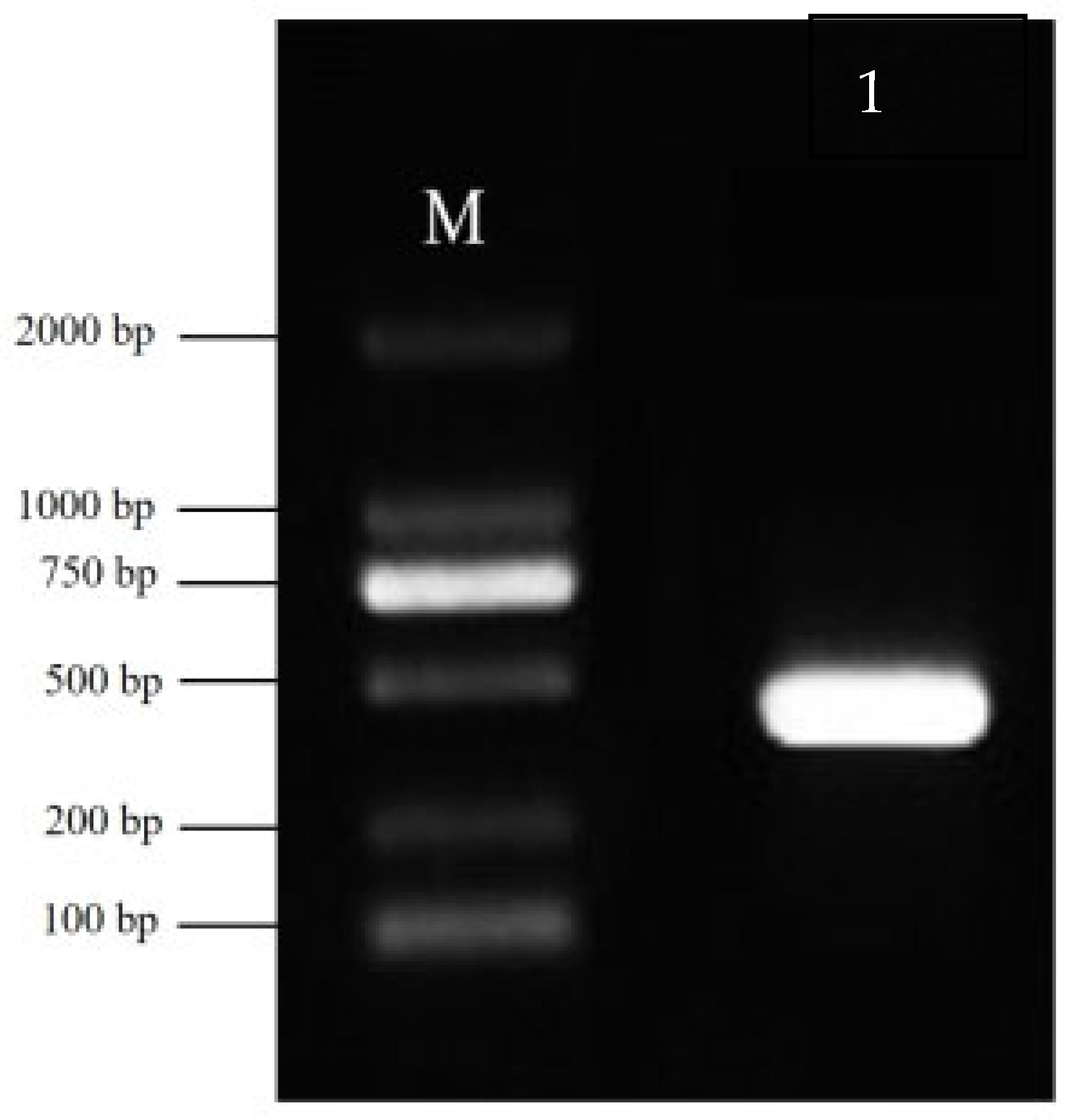
Total RNA was extracted from planarians and reverse-transcribed into cDNA. The amplified
DjPax6 gene fragment and plasmid L4440 were digested with
XhoI and
PstI and then ligated using T4 DNA Ligase (Takara Bio Inc. Kusatsu, Shiga, Japan). Sequences of
DjPax6 interference primers are listed in
Appendix A,
Table A6. L4440-
DjPax6 recombinant plasmid and empty L4440 plasmid were, respectively, transformed into HT115 competent cells and induced with 1 mmol/L IPTG (Beyotime Biotechnology Co., Ltd., Shanghai, China). After induction, the bacterial cells were mixed with nematode homogenate at a 1:2 weight ratio and fed to planarians for five consecutive days. A total of 200 planarians were evenly divided into two groups. The RNAi-treated group was fed with a mixture of bacteria expressing the recombinant plasmid L4440-
DjPax6 and nematode homogenate, while those fed with mixture of bacteria expressing the empty L4440 plasmid and nematode homogenate served as control group. Total RNAs were extracted from the planarians and reverse transcribed into cDNAs on the next day after the final feeding. RNAi efficiency was assessed by RT-qPCR. After the amputation of eyespots, 100 planarians of each group were cultured in fresh water and metformin solution, respectively. A total of 200 planarians were divided into four groups: (1) freshwater-cultured control planarians, (2) freshwater-cultured RNAi-treated planarians, (3) metformin-treated non-RNAi planarians, and (4) metformin-treated RNAi-treated planarians. The regeneration time of planarian eyespots was recorded.
4.9. In Situ Hybridization
Ten planarians with a body length of approximately 3–5 mm were selected for the experiment. DIG-labeled antisense probes for DjmiR-27b and Djpax6 were synthesized by BGI Genomics Co., Ltd. (Shenzhen, Guangdong, China). Planarians were fixed with 4% PFA (Wuhan Service Biotechnology Co., Ltd., China). at room temperature for 1 h, followed by sequential dehydration with 50% and 100% methanol, and then bleached under strong light with 6% Bleach Solution overnight. Rehydration was performed with 100% and 50% methanol. Subsequently, the samples were washed for 10 min with a solution of PBST (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) and PreHyb mixed in a 1:1 ratio, pre-hybridized for 1.5 h in pre-hybridization solution, and then hybridized for more than 16 h in hybridization solution containing a final concentration of 0.1 to 1 ng/µL of the probe, with temperature maintained at 56 °C. Planarians were blocked with Blocking Solution at room temperature for 2 h and then incubated overnight at 4 °C in Blocking Solution containing anti-DIG antibody (F. Hoffmann-La Roche Co., Ltd., Basel, Switzerland). They were washed with Alkaline Phosphatase Buffer (F. Hoffmann-La Roche Co., Ltd., Basel, Switzerland). and developed in the dark on a shaker with a solution mixed in a 1:50 ratio of NBT/BCIP and AP Buffer until they turned purple-red. Samples were imaged by a stereomicroscope (Nikon SMZ 1500, Tokyo, Japan).
4.10. Bioinformatics Analysis
Nucleotide sequences were translated into protein sequences using the ExPASy Translate tool (
https://web.expasy.org/translate/) (accessed on 19 July 2023). Multiple sequence alignment of sequencing results was performed using GeneDoc software 2.7. Statistical analyses were conducted with SPSS 26.0. Group comparisons were analyzed by one-way ANOVA, with significance levels set at
p < 0.05 for statistical significance and
p < 0.01 for extremely significant differences. Data analysis and plotting were carried out using GraphPad Prism 5.