Genetic Landscape of Non-Remitting Neutropenia in Children and Chronic Idiopathic Neutropenia in Adults
Abstract
1. Introduction
2. Results
2.1. Immunological and Clinical Characteristics
2.2. Genes Potentially Associated with Neutropenia
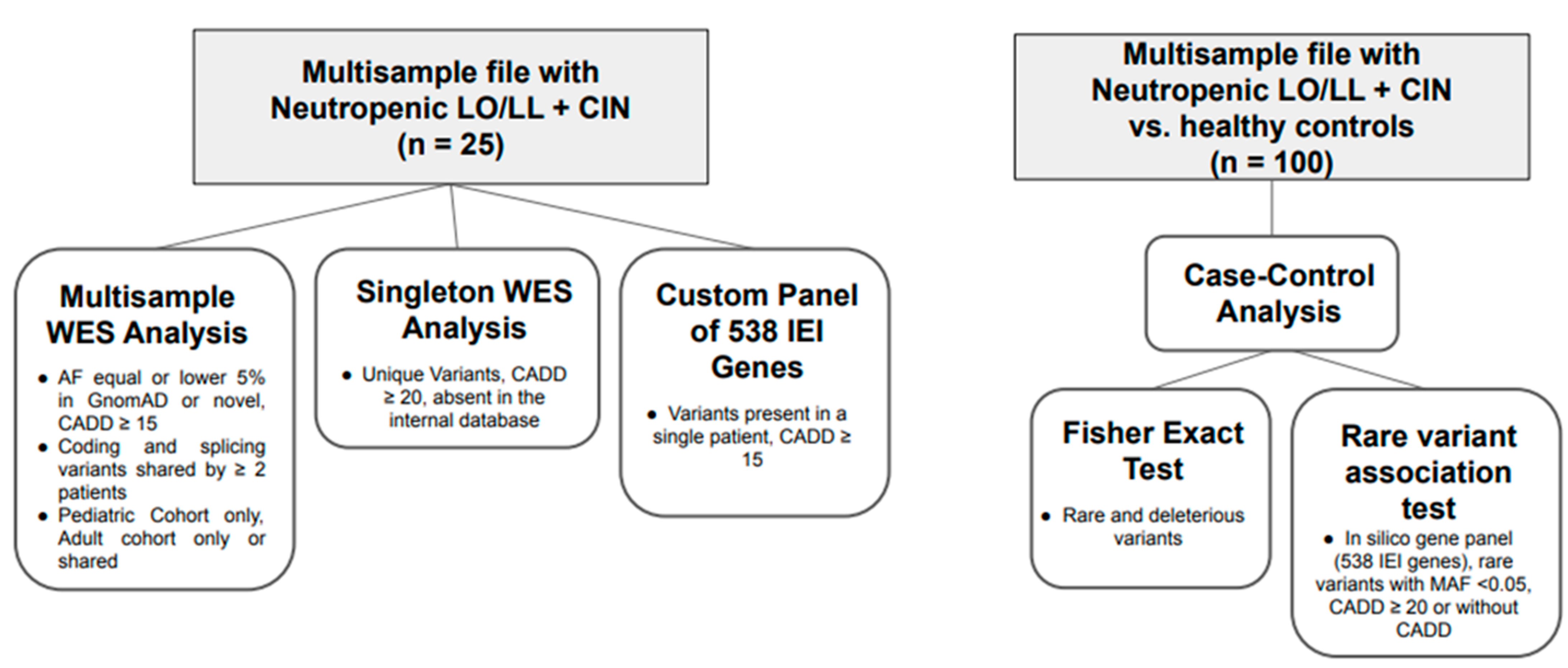
2.2.1. Multisample Analysis
2.2.2. Singleton Analysis
2.2.3. Inborn Error of Immunity Panel Analysis
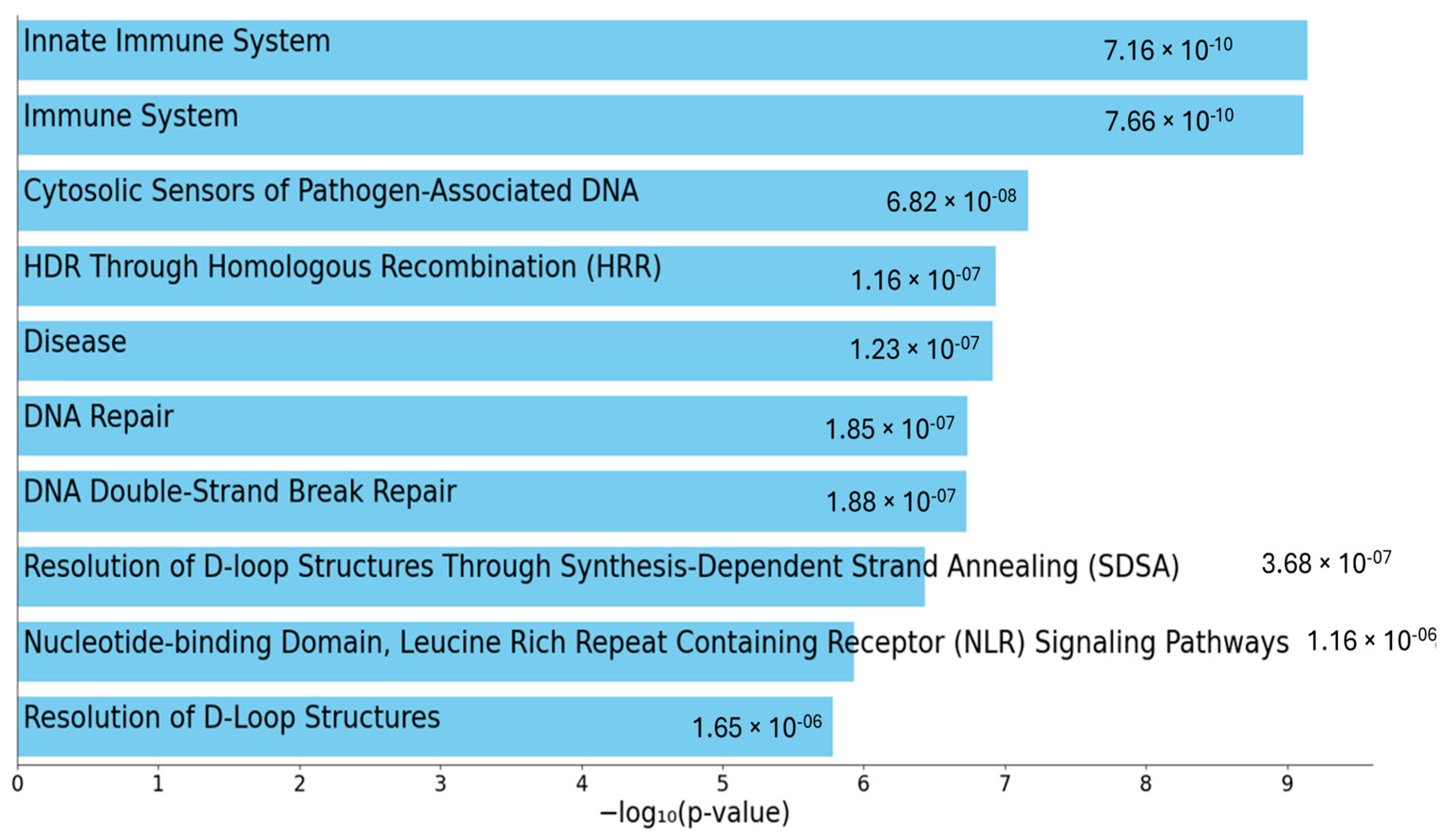
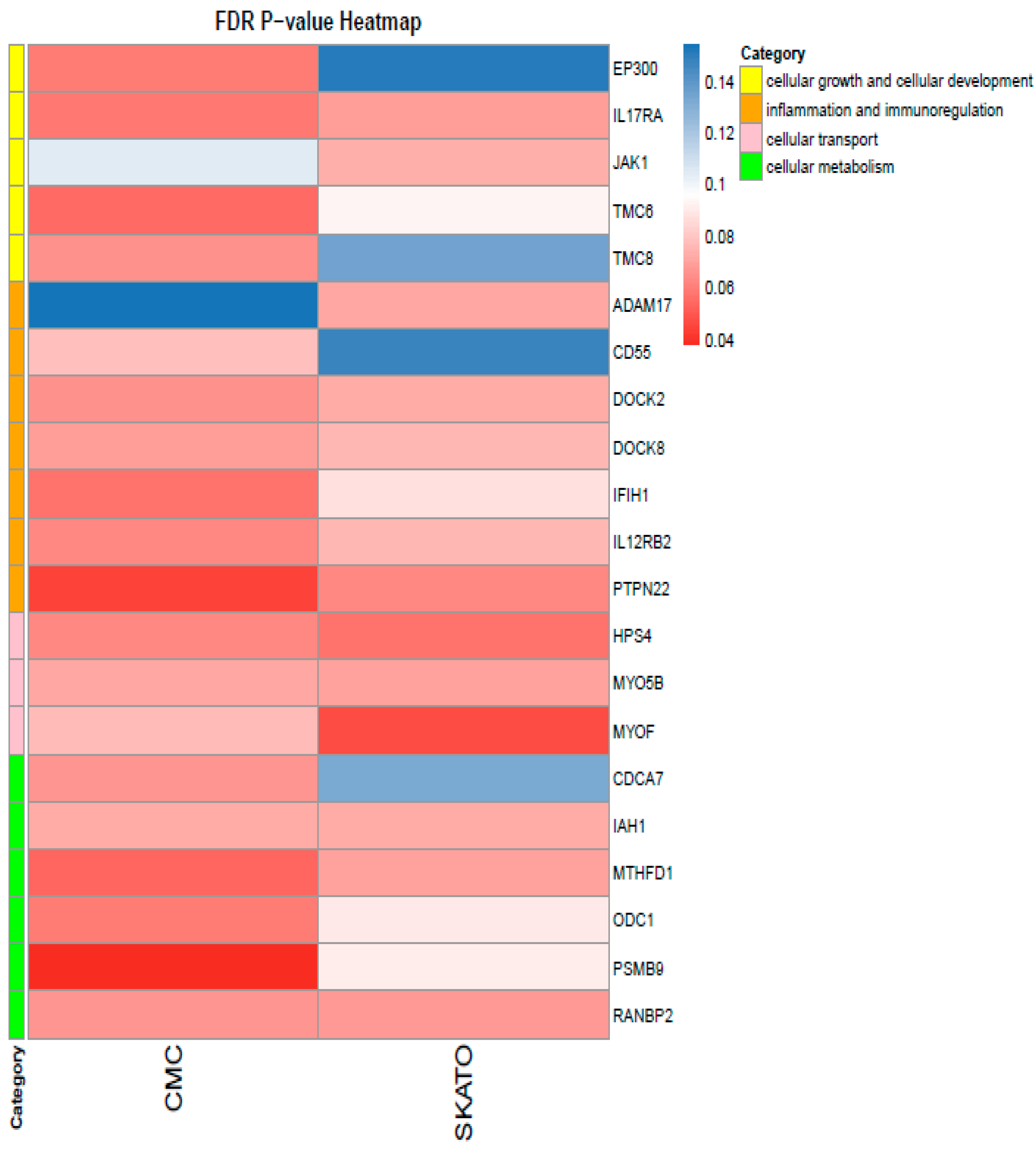
2.3. Gene Clustering
Multisample Analysis of Total Patient Group vs. Healthy Controls
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AR | Autosomal Recessive |
| ACMG | American Council of Medical Genetic |
| CADD | Combined Annotation Dependent Depletion |
| CIN | Chronic Idiopathic Neutropenia |
| CVID | Common Variable ImmunoDeficiency |
| IEI | Inborn Errors of Immunity |
| LEF1 | Lymphoid Enhancer-Binding Factor 1 |
| LL | Long Lasting |
| LO | Late Onset |
| MHC | Major Histocompatibility Complex |
| NF-κB | Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B-Cells |
| NGS | Next Generation Sequencing |
| P | Pathogenic |
| LP | Likely Pathogenic |
| TNF | Tumor Necrosis Factor |
| WES | Whole Exome Sequencing |
| PLCG1 | Phospholipase C (PLC) γ1 |
| GAB1 | GRB2-Associated Binder 1 |
| JNK | c-Jun N-Terminal Kinase |
| mTOR | Mammalian Target of Rapamycin |
| SCID | Severe Combined Immunodeficiency |
| VUS | Variant of Unknown Significance |
References
- Fioredda, F.; Skokowa, J.; Tamary, H.; Spanoudakis, M.; Farruggia, P.; Almeida, A.; Guardo, D.; Höglund, P.; Newburger, P.E.; Palmblad, J.; et al. The European Guidelines on Diagnosis and Management of Neutropenia in Adults and Children: A Consensus Between the European Hematology Association and the EuNet-INNOCHRON COST Action. Hemasphere 2023, 7, e872. [Google Scholar] [CrossRef] [PubMed]
- Fioredda, F.; Beccaria, A.; Casartelli, P.; Turrini, E.; Contratto, C.; Giarratana, M.C.; Bagnasco, F.; Saettini, F.; Pillon, M.; Marzollo, A.; et al. Late-onset and long-lasting neutropenias in the young: A new entity anticipating immune-dysregulation disorders. Am. J. Hematol. 2024, 99, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Kyriakou, D.; Papadaki, H.A.; Sakellariou, D.; Eliopoulos, A.G.; Kapsimali, V.; Eliopoulos, G.D. Flow-cytometric analysis of peripheral blood lymphocytes in patients with chronic idiopathic neutropenia of adults. Ann. Hematol. 1997, 75, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, H.A.; Stamatopoulos, K.; Damianaki, A.; Gemetzi, C.; Anagnostopoulos, A.; Papadaki, T.; Eliopoulos, A.G.; Eliopoulos, G.D. Activated T-lymphocytes with myelosuppressive properties in patients with chronic idiopathic neutropenia. Br. J. Haematol. 2005, 128, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Mavroudi, I.; Eliopoulos, A.G.; Pontikoglou, C.; Pyrovolaki, K.; Damianaki, A.; Koutala, H.; Zervou, M.I.; Ximeri, M.; Mastrodemou, S.; Kanellou, P.; et al. Immunoglobulin and B-cell disturbances in patients with chronic idiopathic neutropenia. Clin. Immunol. 2017, 183, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Pyrovolaki, K.; Mavroudi, I.; Papadantonakis, N.; Velegraki, M.; Ximeri, M.; Kalpadakis, C.; Gvazava, G.; Klaus, M.; Eliopoulos, G.D.; Papadaki, H.A. Transforming growth factor-beta1 affects interleukin-10 production in the bone marrow of patients with chronic idiopathic neutropenia. Eur. J. Haematol. 2007, 79, 531–538. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, S.B.; Williams, J.L. Clinical Laboratory Hematology, 3rd ed.; Pearson Clinical Laboratory Science Series; Pearson: Boston, MA, USA, 2014. [Google Scholar]
- van Gent, R.; van Tilburg, C.M.; Nibbelke, E.E.; Otto, S.A.; Gaiser, J.F.; JAnseen, P.L.; Sanders, A.M.; Borgans, J.A.M.; Wulffraat, N.M.; Bierings, M.B.; et al. Refined characterization and reference values of the pediatric T- and B-cell compartments. Clin. Immunol. 2009, 133, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Bisset, L.R.; Lung, T.L.; Kaelin, M.; Ludwig, E.; Dubs, R.W. Reference values for peripheral blood lymphocyte phenotypes applicable to the healthy adult population in Switzerland. Eur. J. Haematol. 2004, 72, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Ugazio, A.G.; Duse, M.; Notarangleo, L.D.; Plebani, A.; Porta, F. Il Bambino Immunodepresso Perche’ lo e’ e Come va Difeso, 2nd ed.; CEA: Milano, Italy, 1995. [Google Scholar]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Chavanas, S.; Garner, C.; Bodemer, C.; Ali, M.; Hamel-Teillac, D.; Wilkinson, J.; Bonafe, J.-L.; Paradisi, M.; Kelsell, D.P.; Ansai, S.; et al. Localization of the Netherton syndrome gene to chromosome 5q32, by linkage analysis and homozygosity mapping. Am. J. Hum. Genet. 2000, 66, 914–921. [Google Scholar] [CrossRef]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-H.; Ronsin, C.; Gesnel, M.-C.; Coupey, L.; Skeel, A.; Leonard, E.J.; Breathnach, R. Identification of the RON gene product as the receptor for the human macrophage stimulating protein. Science 1994, 266, 117–119. [Google Scholar] [CrossRef] [PubMed]
- Tsai, F.Y.; Keller, G.; Kuo, F.C.; Weiss, M.; Chen, J.; Rosenblatt, M.; Alt, F.W.; Orkin, S.H. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 1994, 371, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Hsu, A.P.; Sampaio, E.P.; Khan, J.; Calvo, K.R.; Lemieux, J.E.; Patel, S.Y.; Frucht, D.M.; Vinh, D.C.; Auth, R.D.; Freeman, A.F.; et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood 2011, 118, 2653–2655. [Google Scholar] [CrossRef] [PubMed]
- Bray, P.F.; Rosa, J.P.; Johnston, G.I.; Shiu, D.T.; Cook, R.G.; Lau, C.; Kan, Y.; McEver, R.P.; Shuman, M.A. Platelet glycoprotein IIb. Chromosomal localization and tissue expression. J. Clin. Investig. 1987, 80, 1812–1817. [Google Scholar] [CrossRef] [PubMed]
- Bajt, M.L.; Ginsberg, M.H.; Frelinger, A.L., 3rd; Berndt, M.C.; Loftus, J.C. A spontaneous mutation of integrin alpha IIb beta 3 (platelet glycoprotein IIb-IIIa) helps define a ligand binding site. J. Biol Chem. 1992, 267, 3789–3794. [Google Scholar] [CrossRef] [PubMed]
- Badran, Y.R.; Dedeoglu, F.; Leyva Castillo, J.M.; Bainter, W.; Ohsumi, T.K.; Bousvaros, A.; Goldsmith, J.D.; Geha, R.S.; Chou, J. Human RELA haploinsufficiency results in autosomal-dominant chronic mucocutaneous ulceration. J. Exp. Med. 2017, 214, 1937–1947. [Google Scholar] [CrossRef] [PubMed]
- Batu, E.D.; Basaran, O.; Bilginer, Y.; Ozen, S. Familial Mediterranean Fever: How to Interpret Genetic Results? How to Treat? A Quarter of a Century After the Association with the Mefv Gene. Curr. Rheumatol. Rep. 2022, 24, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.M.; Mathialagan, N.; Duffy, J.Y.; Smith, G.W. Regulation and regulatory role of proteinase inhibitors. Crit. Rev. Eukaryot. Gene Expr. 1995, 4, 385–436. [Google Scholar] [CrossRef] [PubMed]
- Wlodarski, M.W.; Collin, M. Horwitz MS GATA2 defciency and related myeloid neoplasms. Semin. Hematol. 2017, 54, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Lecerf, K.; Koboldt, D.C.; Kuehn, H.S.; Jayaraman, V.; Lee, K.; Mosher, T.M.; Yonkof, J.R.; Mori, M.; Hickey, S.E.; Franklin, S.; et al. Case report and review of the literature: Immune dysregulation in a large familial cohort due to a novel pathogenic RELA variant. Rheumatology 2023, 62, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Comrie, W.A.; Faruqi, A.J.; Price, S.; Zhang, Y.; Rao, V.K.; Su, H.C.; Lenardo, M.J. RELA haploinsufficiency in CD4 lymphoproliferative disease with autoimmune cytopenias. J. Allergy Clin. Immunol. 2018, 141, 1507–1510. [Google Scholar] [CrossRef] [PubMed]
- Turvey, S.E.; Durandy, A.; Fischer, A.; Fung, S.Y.; Geha, R.S.; Gewies, A.; Giese, T.; Greil, J.; Keller, B.; McKinnon, M.L.; et al. The CARD11-BCL10-MALT1 (CBM) signalosome complex: Stepping into the limelight of human primary immunodeficiency. J. Allergy Clin. Immunol. 2014, 134, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Yokosuka, T.; Hirakawa, H.; Ishihara, C.; Yasukawa, S.; Yamazaki, M.; Koseki, H.; Hiroki, H.; Yoshida, H.; Saito, T. Clustering of CARMA1 through SH3-GUK domain interactions is required for its activation of NF-κB signalling. Nat. Commun. 2015, 6, 5555. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dorjbal, B.; Stinson, J.R.; Ma, C.A.; Weinreich, M.A.; Miraghazadeh, B.; Hartberger, J.M.; Frey-Jakobs, S.; Weidinger, S.; Moebus, L.; Franke, A. AHypomorphic caspase activation and recruitment domain 11 (CARD11) mutations associated with diverse immunologic phenotypes with or without atopic disease. J. Allergy Clin. Immunol. 2019, 143, 1482–1495. [Google Scholar] [CrossRef] [PubMed]
- Masters, S.L.; Lagou, V.; Jeru, I.; Baker, P.J.; Van Eyck, L.; Parry, D.A.; Lawless, D.; De Nardo, D.; Grcia-Perez, J.E.; Dagley, L.F.; et al. Familial autoinflammation with neutrophilic dermatosis reveals a regulatory mechanism of pyrin activation. Sci. Transl. Med. 2016, 8, 332ra45. [Google Scholar] [CrossRef] [PubMed]
- Touitou, I. The spectrum of Familial Mediterranean fever (FMF) mutations. Eur. J. Hum. Genet. 2001, 9, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Aksentijevich, I.; Centola, M.; Deng, Z.; Sood, R.; Balow, J.E., Jr.; Wood, G.; Zaks, N.; Mansfield, E.; Chen, X.; Eisenberg, S.; et al. Missense Mutations in a New Member of the RoRet Gene Family Are Likely to Cause Familial Mediterranean Fever. Cell 1997, 90, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, J. Unraveling the mystery of oligogenic inheritance under way? Mol. Cells 2023, 4, 100003. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.; Wang, Y.; Meng, X.; Yang, H.; Liu, Z.; Qian, J.; Zhou, W.; Li, J. Whole Exome Sequencing of Ulcerative Colitis–associated Colorectal Cancer Based on Novel Somatic Mutations Identified in Chinese Patients. Inflamm. Bowel Dis. 2019, 25, 1293–1300. [Google Scholar] [CrossRef] [PubMed]
- Tangye, S.G.; Al-Herz, W.; Bousfiha, A.; Cunningham-Rundles, C.; Franco, J.L.; Holland, S.M.; Klein, C.; Morio, T.; Oksenhendler, E.; Picard, C.; et al. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 2022, 42, 1473–1507. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Hu, Y.; Li, B.; Abecasis, G.R.; Liu, D.J. RVTESTS: An efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 2016, 32, 1423–1426. [Google Scholar] [CrossRef] [PubMed]
| PT | Gender | Ethnicity | Consanguinity | Date of Birth | Age at Onset (y) | Age at Diagnosis of NP (y) | Duration FUP (mo) | Main Clinical | Family History | AutoimmunityIncluding AbAN | WBC (×109/L) | Neu (×109/L) | Lymph (×109/L) | Mono (×109/L) | IgA (48–368 mg/dL) | IgG (701–1600 mg/dL) | IgM (25–170 mg/dL) | IgG1 (490–1140 mg/dL) | IgG2 (150–640 mg/dL) | IgG3 (20–110 mg/dL) | IgG4 (8–140 mg/dL) | CD3% | CD3/CD4% | CD3/CD8% | CD19% | CD3-(CD16/56)+% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pt1 | F | C * | N° | 1991 | 16 | 27 | 41 | Fatigue/Apthae/ Arthralgias | NR # | ANA+ Ab AN+ | 2.3 | 0.5 | 1.3 | 0.3 | 153 | 1171 | 193 | 752 | 369 | 38.4 | 9.2 | 82.7 | 50.3 | 18.4 | 11.6 | 4.0 |
| Pt2 | F | C * | N° | 2003 | 2 | 16 | 32 | Fatigue/Apthae/ Arthralgias | NR # | Ab AN+ | 1.7 | 0.5 | 0.97 | 0.15 | 220 | 1028 | 67 | 546 | 435 | 19.3 | 5.3 | 84.9 | 57.0 | 20.1 | 5.4 | 8.3 |
| Pt3 | M | C * | N° | 2002 | 12 | 13 | 72 | Apthae | Early Cancer a | ANA+ Ab AN+ | 3.5 | 0.4 | 2.2 | 0.3 | 222 | 1196 | 156 | 949 | 328 | 109 | 84 | 78.0 | 45.0 | 27.7 | 8.6 | 13.4 |
| Pt4 | F | C * | N° | 2003 | 11 | 13 | 66 | Fatigue/Apthae | Neurodegenerative disease b | ANA+ Ab AN+ | 2.7 | 0.45 | 1.6 | 0.4 | 136 | 1436 | 305 | 881 | 437 | 37.2 | 112 | 78.0 | 58.7 | 2.4 | 14.3 | 5.0 |
| Pt5 | M | C * | N° | 2011 | 2 | 4 | 64 | Nothing | NR # | Ab AN+ | 2.9 | 0.95 | 1.5 | 0.24 | 116 | 1207 | 67 | 1060 | 189 | 74 | 80 | 77.4 | 43.4 | 25.2 | 12.1 | 6.6 |
| Pt6 | F | C * | N° | 2015 | 0 | 0.3 | 66 | Nothing | Early Cancer c | Ab AN+ | 4.8 | 0.48 | 1.9 | 0.35 | 54 | 905 | 75 | 677 | 221 | 36 | 24 | 67.0 | 39.0 | 19.0 | 11.0 | 18.4 |
| Pt7 | F | C * | N° | 2001 | 17 | 19 | 12 | Nothing | NR # | Anti Thy Ab+ | 2.2 | 0.32 | 1.4 | 0.4 | 58 | 1322 | 223 | 873 | 313 | 37.1 | 12.2 | 82.1 | 54.4 | 20.8 | 13.0 | 4.0 |
| Pt8 | F | C * | N° | 2000 | 17 | 18 | 41 | Nothing | Cancer in the family d | Ab AN+ | 1.9 | 0.22 | 1.1 | 0.4 | 185 | 1308 | 68 | 748 | 692 | 52 | 12 | 78.4 | 39.5 | 22.3 | 12.0 | 5.6 |
| Pt9 | M | C * | N° | 2003 | 15 | 15 | 34 | Fatigue | NR # | Anti Thy Ab+ Ab AN+ | 1.9 | 0.35 | 1.1 | 0.3 | 165 | 959 | 75 | 750 | 189 | 22 | 118 | 81.3 | 34.4 | 31.1 | 9.9 | 6.9 |
| Pt10 | M | C * | N° | 1993 | 25 | 26 | 48 | Mild psoriasis | Neutropenic sibling | Ab AN+ | 2.9 | 0.8 | 1.3 | 0.6 | 245 | 1340 | 166 | 785 | 417 | 29.2 | 19.2 | 76.5 | 48.0 | 25.3 | 14.7 | 7.8 |
| Pt11 | F | C * | N° | 1968 | 50 | 50 | 60 | Arthralgias | NR # | Neg | 3.1 | 1.5 | 1.1 | 0.4 | 117 | 1230 | 130 | 822 | 393 | 52.1 | 0.7 | 66.3 | 44.0 | 19.4 | 16.6 | 16.8 |
| Pt12 | F | C * | N° | 1964 | 36 | 38 | 240 | Arthralgias | NR # | Neg | 3.0 | 1.2 | 1.5 | 0.3 | 144 | 1040 | 82 | 615 | 391 | 25.4 | 2.5 | 68.6 | 52.0 | 15.0 | 14.4 | 14.3 |
| Pt13 | F | C * | N° | 1979 | 18 | 18 | 372 | Arthralgias | Neutropenic sibling | Neg | 3.3 | 1.2 | 1.6 | 0.3 | 253 | 1100 | 135 | 651 | 598 | 34.0 | 41.1 | 71.0 | 38.0 | |||
| Pt14 | M | C * | N° | 1957 | 32 | 32 | 156 | Nothing | NR # | Neg | 2.8 | 0.7 | 1.4 | 0.5 | 307 | 720 | 45 | 367 | 355 | 13.5 | 35.6 | 74.7 | 30.9 | 41.2 | 9.6 | 14.8 |
| Pt15 | F | C * | N° | 1963 | 54 | 56 | 36 | Ankylosis spondylitis | NR # | Neg | 2.4 | 1.1 | 1.0 | 0.3 | 222 | 930 | 187 | 610 | 340 | 35.3 | 48.4 | 79.7 | 52.4 | 22.5 | 11.6 | 8.8 |
| Pt16 | F | C * | N° | 1951 | 35 | 35 | 180 | Arthralgias | NR # | ANA+ | 3.3 | 1.3 | 1.5 | 0.4 | 189 | 1330 | 37 | 925 | 226 | 67.4 | 3.0 | 78.0 | 58.2 | 23.8 | 6.4 | |
| Pt17 | M | C * | N° | 1973 | 27 | 27 | 209 | Nothing | NR # | Neg | 3.2 | 0.5 | 2.0 | 0.4 | 252 | 1340 | 647 | 703 | 570 | 31.0 | 57.5 | 79.7 | 38.9 | 38.5 | 13.6 | 5.3 |
| Pt18 | M | C * | N° | 1973 | 26 | 30 | 96 | Psoriasis | NR # | Neg | 2.7 | 1.4 | 1.0 | 0.3 | 265 | 1420 | 63 | 895 | 436 | 13.9 | 50.0 | 51.2 | 29.2 | 26.5 | 10.7 | 28.2 |
| Pt19 | F | C * | N° | 1953 | 60 | 66 | 60 | Nothing | NR # | Neg | 2.5 | 0.4 | 1.5 | 0.5 | 120 | 1380 | 79 | 949 | 423 | 8.3 | 108.0 | 65.0 | 39.0 | 35.0 | 16.3 | 13.7 |
| Pt20 | F | C * | N° | 1954 | 35 | 35 | 276 | Nothing | NR # | ANA+ | 3.8 | 1.6 | 1.7 | 0.4 | 381 | 1120 | 48 | 872 | 185 | 18.1 | 0.3 | 71.0 | 37.0 | 34.0 | 18.0 | 8.0 |
| Pt21 | F | C * | N° | 1968 | 32 | 32 | 288 | Nothing | NR # | Neg | 3.7 | 1.5 | 1.7 | 0.3 | 155 | 1200 | 193 | 717 | 260 | 24.0 | 107.0 | |||||
| Pt22 | F | C * | N° | 1958 | 23 | 25 | 276 | Nothing | NR # | Neg | 4.0 | 1.5 | 1.6 | 0.5 | 145 | 790 | 120 | 363 | 370 | 16.7 | 16.9 | 72.2 | 44.2 | 27.7 | 16.3 | 11.4 |
| Pt23 | F | C * | N° | 1968 | 41 | 50 | 72 | Arthralgias | NR # | Neg | 3.0 | 1.0 | 1.6 | 0.3 | 244 | 981 | 121 | 630 | 388 | 16.3 | 94.7 | 63.8 | 35.0 | 23.0 | 12.3 | 13.8 |
| Pt24 | M | C * | N° | 1999 | 18 | 18 | 120 | Nothing | NR | Neg | 2.8 | 1.0 | 1.5 | 0.2 | 62 | 1150 | 183 | 721 | 269 | 28.7 | 59.4 | 77.0 | 57.4 | 19.6 | 8.1 | |
| Pt25 | F | C * | N° | 1966 | 40 | 40 | 216 | Nothing | NR | Neg | 4.0 | 1.6 | 1.8 | 0.5 | 104 | 836 | 29 | 440 | 201 | 19.1 | 108.0 | 84.9 | 54.9 | 28.8 | 9.1 | 11.7 |
| Pt_ID | MULTISAMPLES Test | SINGLETON Test | Inborn Errors of Immunity (IEI) PANEL (538 Genes) |
|---|---|---|---|
| Pt1 | MST1R | MPEG1 | CARD10 |
| CASP8 | LAT | ||
| Pt2 | DNM2/MRE11/IRF7/CD36 | ||
| XRCC2/NBAS | TGFBR2 | ||
| SIGLEC1/MPO/ZNFX1 | RELA/TRNT1 | ||
| Pt3 | BRCA1/MPO/CASP5 | IFNAR1/SLC7A7 | |
| EPX/RIF1 | |||
| Pt4 | RANBP2/DOCK1/DOCK8/PLCH2/PIEZO1 | ||
| SPINK5 | |||
| Pt5 | EP300/VPS13B/ZNFX1 | AP3D1 | |
| ATP7B/PLCL2 | |||
| Pt6 | BCLAF1/EPX/MYO9B/PDGFRB/PIK3C2G | CASP4 | ADAM17/IL18BP/IPO8/LRRC32 |
| Pt7 | FANCM/MRE11 | SLCO2A1/NLRP1 | |
| Pt8 | PLCG2 | POLD1 | |
| ATP7B/CXCR1/CD36 | |||
| Pt9 | POLE/DOCK8/MAP3K21/SIGLEC1/DNM2 | HS3ST6 | |
| Pt10 | SPINK5/CXCR1/DOCK1/PLCL2 | POLR3A | |
| HYOU1/POLE/RANBP2 | |||
| Pt11 | EP300/ORAI1 | PSMB11 | |
| CHD7/PIK3C3/RAD50 | |||
| Pt12 | MYO9B/NBAS/PDGFRB/XRCC2 | NLRC4 | |
| ORAI1/PIK3C3/CTC1/PEPD | CBLB | ||
| Pt13 | DOCK1/EPX/FANCM/HYOU1 | TICAM1 | |
| Pt14 | ADAMTS13/PRKCH/ZNFX1 | HAX1 | CTNNB1 |
| EP300 | |||
| Pt15 | CASP5/DOCK8/PIEZO1 | GATA2 | |
| PRKCH | |||
| Pt16 | ARHGAP21/PRKCH | TET2 | PSMG2/SP110 |
| Pt17 | BRCA1/MRE11/RIF1/ADAMTS13 | RFXAP | |
| PEPD/RAD50 | |||
| Pt18 | ABCB6/IRF7/MST1R/PLCG2 | AEN/ATG4D/RSF1 | |
| ALPI/LYST/PEPD/RAD50/RASAL3 | |||
| Pt19 | BRCA1/MAP3K21/RIF1 | ARPC1B/C6 | |
| VPS13B/RASAL3/CTC1/MYOF | |||
| Pt20 | CASP8/DOCK8/SIGLEC1 | GJB4/OVCA2/ZYX | MEFV/PLG |
| Pt21 | ATP7B/N4BP2L2/PEPD/RAD50 | DCLRE1C | |
| MST1R | RTEL1 | ||
| Pt22 | EP300/MSH4 | ACTN1/G6PC3 | FANCC |
| PLCH2/ALPI | TCF3/PGD | ||
| N4BP2L2 | STEAP3 | ||
| Pt23 | PIK3C2G/MYOF | ITGB3/IGSF6/MSRA/SERPINB3 | EXTL3/POLA1 |
| Pt24 | PDGFRB/ARHGAP21 | LTB | NFAT5/TLN1 |
| MSH4/LYST | |||
| Pt25 | BCLAF1/PIEZO1 | SAMD9 | CARD11 |
| CHD7/PEPD | CXCR4/RAD23A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grossi, A.; Tsaknakis, G.; Rosamilia, F.; Rusmini, M.; Uva, P.; Ceccherini, I.; Giarratana, M.C.; Vozzi, D.; Mavroudi, I.; Dufour, C.; et al. Genetic Landscape of Non-Remitting Neutropenia in Children and Chronic Idiopathic Neutropenia in Adults. Int. J. Mol. Sci. 2025, 26, 6929. https://doi.org/10.3390/ijms26146929
Grossi A, Tsaknakis G, Rosamilia F, Rusmini M, Uva P, Ceccherini I, Giarratana MC, Vozzi D, Mavroudi I, Dufour C, et al. Genetic Landscape of Non-Remitting Neutropenia in Children and Chronic Idiopathic Neutropenia in Adults. International Journal of Molecular Sciences. 2025; 26(14):6929. https://doi.org/10.3390/ijms26146929
Chicago/Turabian StyleGrossi, Alice, Grigorios Tsaknakis, Francesca Rosamilia, Marta Rusmini, Paolo Uva, Isabella Ceccherini, Maria Carla Giarratana, Diego Vozzi, Irene Mavroudi, Carlo Dufour, and et al. 2025. "Genetic Landscape of Non-Remitting Neutropenia in Children and Chronic Idiopathic Neutropenia in Adults" International Journal of Molecular Sciences 26, no. 14: 6929. https://doi.org/10.3390/ijms26146929
APA StyleGrossi, A., Tsaknakis, G., Rosamilia, F., Rusmini, M., Uva, P., Ceccherini, I., Giarratana, M. C., Vozzi, D., Mavroudi, I., Dufour, C., Papadaki, H. A., & Fioredda, F. (2025). Genetic Landscape of Non-Remitting Neutropenia in Children and Chronic Idiopathic Neutropenia in Adults. International Journal of Molecular Sciences, 26(14), 6929. https://doi.org/10.3390/ijms26146929








