Regulation of Plant Genes with Exogenous RNAs
Abstract
1. Introduction
| Target | RNA Treatment | RNA Amount | RNA Application | Accessory Carrier/Surfactant | Plant Host for Treatment | Effect | Effect Assessment | Reference |
|---|---|---|---|---|---|---|---|---|
| YFP transgene | In vitro-synthesized short siRNA (21 bp) in a complex with a carrier peptide | 100 µL of the RNA-peptide complex (20 pmol siRNA) | Infiltration | Carrier peptide (KH)9-Bp100 | YFP-transgenic Arabidopsis thaliana and poplar Populus tremula × tremuloides (fully expanded leaves) | - Suppression of YFP protein level and fluorescence | Assessed 1, 3, 6, 9, 12, 24, and 36 hpt | Numata et al. (2014) [48] |
| GFP transgene | In vitro-synthesized siRNAs (21, 22, and 24 nt) | 100 µL of aqueous siRNA solutions (10 µM) | High-pressure spraying and simple spraying | With or without Silwet L-77 surfactant | GFP-transgenic tobacco Nicotiana benthamiana (leaves and buds) | - Local and systemic GFP fluorescence suppression - Systemic silencing after spraying of 22 bp siRNAs | Assessed 2 and 20 dpt | Dalakouras et al. (2016) [49] |
| GUS transgene | Total RNA from dsRNA-expressing Escherichia coli HT115 (~504 bp) | 100 µg of dsRNA with or without LDH | Spraying | LDH clay nanosheets or BioClay | GUS-transgenic A. thaliana (5-day-old seedlings) | Reduction in GUS activity | Assessed 7 dpt | Mitter et al. (2017) [50] |
| EGFP and NPTII transgenes | In vitro-synthesized dsRNAs (EGFP 720 bp; NPTII 599 bp) | 0.35 µg/µL (100 µL | Spreading with brushes | - | EGFP- and NPTII-transgenic A. thaliana (4-week-old rosettes) | - Sequence-specific suppression of EGFP and NPTII mRNA; - Suppression of EGFP protein and fluorescence levels - Induction of EGFP and NPTII DNA methylation | Assessed 1, 7, and 14 dpt | Dubrovina et al. (2019); Kiselev et al. (2022) [44,51] |
| EGFP transgene | In vitro-synthesized siRNAs (EGFP 21 bp) linked to DNA nanostructures | 100 μL of siRNA (100 nM) | Infiltration | siRNA-linked DNA nanostructures (3D tetrahedron, 1D monomer, 1D nanostring) | mGFP5-transgenic tobacco N. benthamiana (4-week-old plants) | - siRNA-linked 3D DNA nanostructures show EGFP silencing at both the mRNA and protein - siRNA-linked to 1D DNA nanostructures shows gene silencing at the protein level, but increased mRNA levels | Assessed 12 and 36 hpt or 3 and 7 dpt | Zhang et al. (2019) [52] |
| GFP transgene | In vitro-synthesized GFP-dsRNAs (322 and 139 bp) | 200 μL of dsRNA (10, 20, 200, and 240 ng/μL) | High-pressure spraying | - | GFP-transgenic Nb-16C N. benthamiana (10–12 cm tall plants) | - No effect on GFP fluorescence; - Exogenous dsRNAs were not processed into specific siRNAs. | Assessed for 21 dpt Assessed 5 dpt | Uslu et al. (2020) [53] |
| GFP transgene | Synthetic siRNAs (22 bp) | 200 μL of aqueous siRNA solution at 1 μM concentration | High-pressure spraying | - | 25-day-old GFP-transgenic N. benthamiana Nb-16C plants | - Lowered GFP fluorescence - siRNA targeting the 5′ GFP and middle regions were more efficient when compared with the siRNAs targeting the 3′ GFP region | Assessed 6, 18, and 35 dpt | Uslu et al. (2022) [54] |
| NPTII transgene | In vitro-synthesized NPTII-siRNAs (NPTII 21 bp) methylated and non-methylated at 3′ ends Heterogeneous NPTII-siRNA mix (digestion of the NPTII-dsRNA) | 50 pmol/μL (100 µL per plant) | Soft brushes | - | NPTII-transgenic A. thaliana (4-week-old rosettes) | - Suppression of NPTII mRNA levels; - A higher effect was observed for NPTII-siRNAs methylated at 3′ ends - Induction of NPTII DNA methylation | Assessed 1 and 7 dpt | Dubrovina et al. (2020) [45] |
| EGFP transgene | In vitro-synthesized dsRNAs (EGFP 500 bp) | 1, 2, 4, or 8 μg of dsRNA per 1-week-old plant (5, 10, 20, or 40 ng/μL in 2 mL of water) | Spraying and dipping | - | EGFP-transgenic A. thaliana DR5-EGFP line (1-week-old seedlings) | Suppression of EGFP-induced fluorescence as well as EGFP mRNA levels | - Assessed 1, 2, 4, and 6 dpt | Park et al. (2022) [55] |
| GFP transgene | Synthetic siRNAs (22 bp) in a complex with MSNs | 100 μL of the MSN-siRNA (siRNAs 10 μg mL−1) | Spraying Infiltration | MSNs and 0.03% Tween 20 | GFP-transgenic N. benthamiana 16 C line (4–6-week-old plants) | - Reduction in GFP mRNA expression levels - Lowered GFP protein level and fluorescence - Reduction in GFP mRNA expression levels | - Assessed 1, 3, and 5 dpt - Long-term effect assessed 4, 7, 11, and 13 dpt | Cai et al. (2024) [56] |
| GFP transgene | GFP-RNA nanoparticles: triangle (474 nt), square (630 nt), pentagon (786 nt), and hexagon (942 nt) extracted from E. coli HT115 GFP-dsRNA | 100 ng µL−1 | Spraying | - | GFP-transgenic A. thaliana (2-week-old plants) | - Suppression of GFP fluorescence and mRNA - RNA squares had the highest RNAi efficiency, followed by RNA triangles | Assessed 1, 4, and 7 dpt | Zhao et al. (2024) [57] |
| GFP transgene | GFP-dsRNA (185 bp) with CPP | 500 ng of gfp-dsRNA and 5000 ng of CPP6 | Infiltration | Cationic poly-aspartic acid-derived polymer (CPP6) | GFP-transgenic A. thaliana (3-week-old plants) | - Suppression of GFP mRNA, GFP protein, and fluorescence levels | Assessed 1, 24, and 48 hpt | Pal et al. (2024) [58] |
| Plant Gene Target | RNA Treatment | RNA Amount | RNA Application | RNA Carrier | Plant Material for Treatment | Effect Assessment | Effect Assessment | Reference |
|---|---|---|---|---|---|---|---|---|
| EPSPS gene of 5-enolpyruvylshikimate-3-phosphate synthase in Palmer amaranth | In vitro-synthesized short dsRNAs (24 bp); long dsRNAs (200–250 bp) | 10 µL of dsRNA on each of four leaves per plant (0.024–0.8 nM) | Leaves pre-treatment by carborundum solution or surfactant solution | - | Palmer amaranth (glyphosate-tolerant) | - Suppressed EPSPS transcript and protein levels - Improved glyphosate efficacy | At least for 48–72 hpt | Sammons et al. (2011) [59] |
| Chalcone synthase CHS gene | In vitro-synthesized short dsRNA (21 bp) in a complex with a carrier peptide | 100 µL of protein carrier in a complex with the siRNA (6 pmol) | Infiltration | Carrier peptide (KH)9-Bp100 | Arabidopsis thaliana | - Local loss of anthocyanin pigmentation | Assessed 2 dpt | Numata et al. (2014) [48] |
| SHOOT MERISTEMLESS (STM) and WEREWOLF (WER) transcription factor genes | A mixture of cationic fluorescent nanoparticles G2 and in vitro-synthesized dsRNA (STM 450 bp; WER 550 bp) | G2 nanoparticles/dsRNA complexes 2: 1 (1 mg dsRNA once per 24 h) for 3–5 days | By pipette | Cationic fluorescent nanoparticles G2 | The root tip of a 10- day-old seedling of wild-type A. thaliana | - Suppressed STM and WER transcripts - Retarded growth and reduced meristem size; - Fluorescence observed throughout the root system (24 hpt) | At least for 5–7 dpt | Jiang et al. (2014) [60] |
| DhMYB1 transcription factor gene of Dendrobium hybrida | Crude extract of E. coli HT115 containing DhMYB1 dsRNA (430 bp) | 50 µL of crude bacterial extract (2 µg/µL at 5-day intervals) | Mechanical inoculation onto a flower bud | - | Flower buds of hybrid orchid, Dendrobium hybrida (D. bobby messina × D. chao phraya) | - Suppressed DhMYB1expression - Changed phenotype of floral cells (22, 25, and 29 dpt) | At least for 29 dpt | Lau et al. (2015) [34] |
| Mob1A and WRKY23 transcription factor genes in A. thaliana, Actin gene in rice | In vitro-synthesized dsRNAs (Mob1A 554 bp; WRKY23 562 bp) | Arabidopsis and rice seeds or seedlings soaked in 0.2 or 1 mL dsRNA (1.0 mg/mL) | Root soaking | - | Arabidopsis, rice | - Suppression of Mob1A and WRKY23 - Repressed root growth and seed germination - Plants could not bolt or flower - Suppression of Actin - Repressed root growth | Assessed 1 dpt and 5 dpt | Li et al. (2015) [61] |
| STP1 and STP2 sugar transporter genes in tomato | In vitro-synthesized dsRNAs STP1 and STP2 dsRNA | 300 µL of 10 ng/µL dsRNA solution per 10 germinated seeds | Seed soaking | - | Tomato Solanum lycopersicum seeds on the first day post-radicle emergence | - Downregulation of tomato STP1 and STP2 genes; - Reductions of glucose and fructose, but not xylose, in root exudate | Assessed 1 dpt | Warnock et al. (2016) [62] |
| The S-gene LBDIf7 transcription factor gene in grapevine | In vitro-synthesized dsRNAs (VviLBDIf7 412 bp) | 100 μg/plant of dsRNA in 1 mL of water | Spraying | 6-year-old Vitis vinifera cv. Pinot noir | - Decreased VviLBDIf7 gene expression - Reduced Plasmopara viticola infection and sporulation | At least for 7 dpt | Marcianò et al. (2021) [63] | |
| CHS, MYBL2, and ANAC032 transcription factor genes in A. thaliana | - In vitro-synthesized dsRNAs (736 bp for CHS; 588 bp for MYBL; 762 bp for ANAC032) - In vitro-synthesized siRNA (21 bp for CHS) | 0.35 µg/µL (100 μL per plant) 50 pmol/µL (100 μL per plant) | Individual soft brushes (natural pony hair) | Four-week-old rosettes of A. thaliana | - Decreased anthocyanin levels after AtCHS-dsRNA and AtCHS-siRNA application - Increased anthocyanin levels and AtCHS expression after AtMYBL-dsRNA and AtANAC032-dsRNA | Assessed on day 2 and 7 dpt | Kiselev et al. (2021) [37] | |
| The E2 conjugase PHO2 gene in A. thaliana SPL9 transcription factor gene in A. thaliana | - Total RNA, extracted from wild-type (WT) plants or from plants overexpressing either miR399 or miR156 Synthetic ds-miR399 and ds-miR156 | 0.01 and 1 μg in 2 mL of nutrient medium - 0.2 μM synthetic ds-miR156 | Seedling soaking (50 seedlings per well) | - | A. thaliana (6-day-old or 8-day-old seedlings) | - Silencing of target genes PHO2 and SPL9 - Inhibition of primary root development - Exogenous miRNAs are translocated by the xylematic route - Exogenous miRNA-triggered RNAi requires AGO1 and RDR6 | Assessed after 24 h of incubation Assessed 5 dpt | Betti et al. (2021) [38] |
| Phytoene desaturase PDS gene in citrus | In vitro-synthesized PDS-dsRNA (391 bp) | 500 ng µL−1, 20 µL per leaf | Foliar application to laser-treated leaves | Laser light for leaf microperforation | Citrus macrophylla (leaves of 12-month-old plants) | Decreased expression of the PDS gene; Leaf photobleaching phenotype | Assessed 3 dpt | Killiny et al. (2021) [64] |
| IAA9 and AGL6 transcription factor genes in tomato | In vitro-synthesized dsRNAs (SlIAA9 717 bp; SlAGL6 702 bp) coupled with LDH nanoparticles | 50 µL of dsRNA-LDHs (5 µg:1 mg; 1:200 w/w) or dsRNA alone | Pedicel injection | LDHs nanoparticles | Tomato S. lycopersicum cv UC82 | - Decreased expression of SlIAA9 and SlAGL6 - Increase in ovary weight - RNAi was induced by the processing of injected dsRNA to 21–24 siRNAs | Assessed 5 dpt Assessed 15 dpt Assessed 5 dpt | Molesini et al. (2022) [47] |
| A putative glutathione S-transferase GST40 gene in grapevine | In vitro-synthesized dsRNAs (VvGST40 688 bp) | 50 µg of dsRNA per plant | High-pressure spraying (10 bar) 7 and 4 days before the drought | - | 1-year-old V. vinifera cv Chardonnay | - Decreased VvGST40 gene expression - Increased resilience to severe drought | Assessed 18 dpt | Nerva et al. (2022) [35] |
| Isoamylase genes ISA1, ISA2, and ISA3 in potato | In vitro-synthesized dsRNAs (250 bp) | No data | Spraying (every 2 weeks, for a total of 6 sprays over a 15-week growth period) | lmPEI nanoparticles | Leaves of potato Solanum tuberosum L. cv. ’Desiree’ | - Decreased expression of ISA1 and ISA2 genes in leaves and ISA3 gene in tubers; - Reduced starch granule size and increased sucrose content - Early sprouting phenotype | Assessed 2, 4, 6, 8, and 10 weeks after treatment) Assessed during 120 days of cultivation | Simon et al. (2023) [65] |
| - The polyphenol oxidase PPO and the phenylalanine ammonia-lyase PAL2 potato genes - MYB12 transcription factor gene in potato | dsRNA-PPO and dsRNA-PALX dsRNA-MYB12 (500 bp all dsRNAs) | 20 μL of dsRNA (0.1 g/L) | Spraying | - | Fresh-cut potato slices of S. tuberosum | - Decreased expression of StPPO, StPAL2, and StMYB12 genes - reduced activities of PPO and PAL - Decrease in fresh-cut potato browning | Assessed 12, 24, 48, 72, and 120 h dpt | Chen et al. (2023) [66] |
| SlMYBATV1, SlMYB32, SlMYB76, and SlTRY transcription factor genes in tomato | In vitro-synthesized dsRNAs (599 bp for SlMYBATV1; 500 bp for SlMYB32; 386 bp for SlMYB76; 285 bp for SlTRY) | 70 µg of the dsRNA diluted in 400 µL of water | Spraying | - | Four-week-old tomato S. lycopersicum | - Downregulated mRNAs of the SlMYBATV1, SlMYB32, SlMYB76, and SlTRY genes - Upregulated expression of anthocyanin biosynthesis genes - Enhanced anthocyanin content in leaves | Assessed at 7 dpt | Suprun et al. (2023) [67] |
| - Flowering locus FT and phytochrome interacting factor 4 PIF4 genes in A. thaliana Phytoene desaturase PDS gene in rice; ZIP23 transcription factor gene in rice The RING-finger containing E3 ligase SDIR1 gene in A. thaliana and rice Sugar transporter SWEET14 gene in rice | ft-dsRNA-CPP6 (219 bp); pif4-dsRNA-CPP (210 bp) pds-dsRNA-CPP6 (481 bp); zip23-dsRNA-CPP6 sdir1-dsRNA-CPP6 (133 bp) for A. thaliana; sdir1-dsRNA-CPP6 (179 bp) for rice; sweet14-dsRNA-CPP6 (189 bp) for rice | 125 ng/plant mixed with CPP6 in a 1:10 ratio 2 μg/seedling of pds-dsRNA-CPP6; 150 ng/seedling of zip23-dsRNA-CPP6 250 ng of dsRNAs per leaf 250 ng of dsRNAs per leaf | Foliar spray Root uptake (3-day-old seedlings transferred to tubes with dsRNA solution) Spraying Spraying | Cationic poly-aspartic acid-derived polymer (CPP6) | Three-week-old A. thaliana plants Three-day-old seedlings of O. sativa Three-week-old A. thaliana plants 45-days-old O. sativa plants 45-days-old O. sativa plants | - Decreased expression of FT and PIF4 genes - Delayed flowering - Enhanced biomass - Decreased seedling height associated with dwarf and albino plant phenotypes - PDS transcript levels did not show any reduction compared to control plants. - Decreased expression of the AtSDIR1 gene; - Improved resistance against Pseudomonas syringae pv. tomato - Decreased expression of the OsSDIR1 gene; - Improved resistance against Xanthomonas oryzae pv. oryzae - AGO and DICER expression increased - Decreased expression of OsSWEET14 gene; - Improved resistance against Xanthomonas oryzae pv. oryzae - Prolonged survival of bacteria | Assessed 48 hpt (the number of bolts); 10 dpt (bolting length and the number of leaves); 2, 4, 6, 8, and 10 dpt (leaves collected); 10 dpt (flowers collected) Assessed 48 hpt (gene expression) and 10 dpt (seedling height) Assessed 48 hpt (gene expression, shoot, and root length) Assessed 3 dpt Assessed at 2, 4, 6, and 10 dpt Assessed 10 dpt Assessed at 2, 4, 6, and 10 dpt Assessed 10 dpt Assessed until 60 dpt | Pal et al. (2024) [58] |
| The tobacco genes of magnesium chelatase ChlH, phytoene desaturase PDS, the chloroplast protein HHL1, and thylakoid membrane-bound protease FtsH2. A disease-resistant R protein, ROQ, and an SOS protein gene | Synthetic siRNAs (19–25 bp) in a complex with mesoporous silica nanoparticles (MSNs) | 100 μL of the MSN-siRNA solution with 0.03% Tween 20 (siRNAs 10 μg mL−1) | Infiltration spraying | MSNs and 0.03% Tween 20 | 4–6-week-old N. benthamiana Plants | - Reduction in PDS, ChlH, HHL1 and FtsH2 mRNA levels - Photobleaching phenotype (white and yellow leaf spots) - Reduction in ROQ and a SOS mRNA ex- pression levels | Assessed 1, 3, and 5 dpt | Cai et al. (2024) [56] |
| CTR4sv3 protein kinase gene in tomato | Synthetic CTR4sv3-siRNA (21 bp) designed based on the interaction site between miR1917 and CTR4sv3 | 10 pmol/µL and 100 pmol/µL of a siRNA Fruit injection—5 µL of a solution with 2000 pmol (400 pmol/µL) of each siRNA | - Seed soaking - Fruit injection (into green-mature tomato fruits) | - | S. lycopersicum cv Micro-Tom (7-d-old seedlings and mature-green fruits) | - Reduction in CTR4sv3 mRNA; - Triple response to ethylene phenotype - Increase in the ethylene biosynthesis gene ACO1 - Reduction in CTR4sv3 mRNA levels - Increase in ACO1 - No noticeable changes in the fruit phenotype | Assessed 7 dpt (seedlings) Assessed 72 h after injection (fruits) | Cedillo-Jimenez et al. (2024) [36] |
| MYB2 transcription factor gene from ginseng GUT gene from oil-seed camellia | RNA nanoparticles of square shape based on MYB2-siRNAs and GUT-siRNAs. The RNA NPs were synthesized and extracted from E. coli. | 100 ng µL−1 of the RNA Nanoparticles | Spraying | - | Individual leaves of Panax notoginsen, Camellia oleifera | - Inhibition of PnMYB2 and CoGUT expression; - RNA squares had the highest RNAi efficiency, followed by RNA triangles | Assessed at 5 and 10 dpt | Zhao et al. (2024) [57] |
| CPC, MYBL2, and ANAC032 transcription factor genes; CBP60g calmodulin-binding protein gene; and AtBAN, anthocyanidin reductase gene in A. thaliana | In vitro-synthesized dsRNAs applied individually or in mixtures (218 bp for CPC; 588 bp for MYBL2; 762 bp for ANAC032; 724 bp for dsCBP60g; 486 bp for dsBAN) | 0.35 µg of a dsRNA in 100 μL of water per plant (0.35 µg/µL) Five dsRNAs in mixtures (50 µg, 100 µg, or 150 µg of total dsRNAs equally mixed together | Individual soft brushes (natural pony hair) | - | Four-week-old rosettes of A. thaliana | - Significant downregulation of all five target genes - Enhanced expression of CHS gene - Increased anthocyanin content; - Application of the five dsRNAs in mixtures was more efficient than individual dsRNAs | Assessed 2 and 7 dpt | Kiselev et al. (2024) [68] |
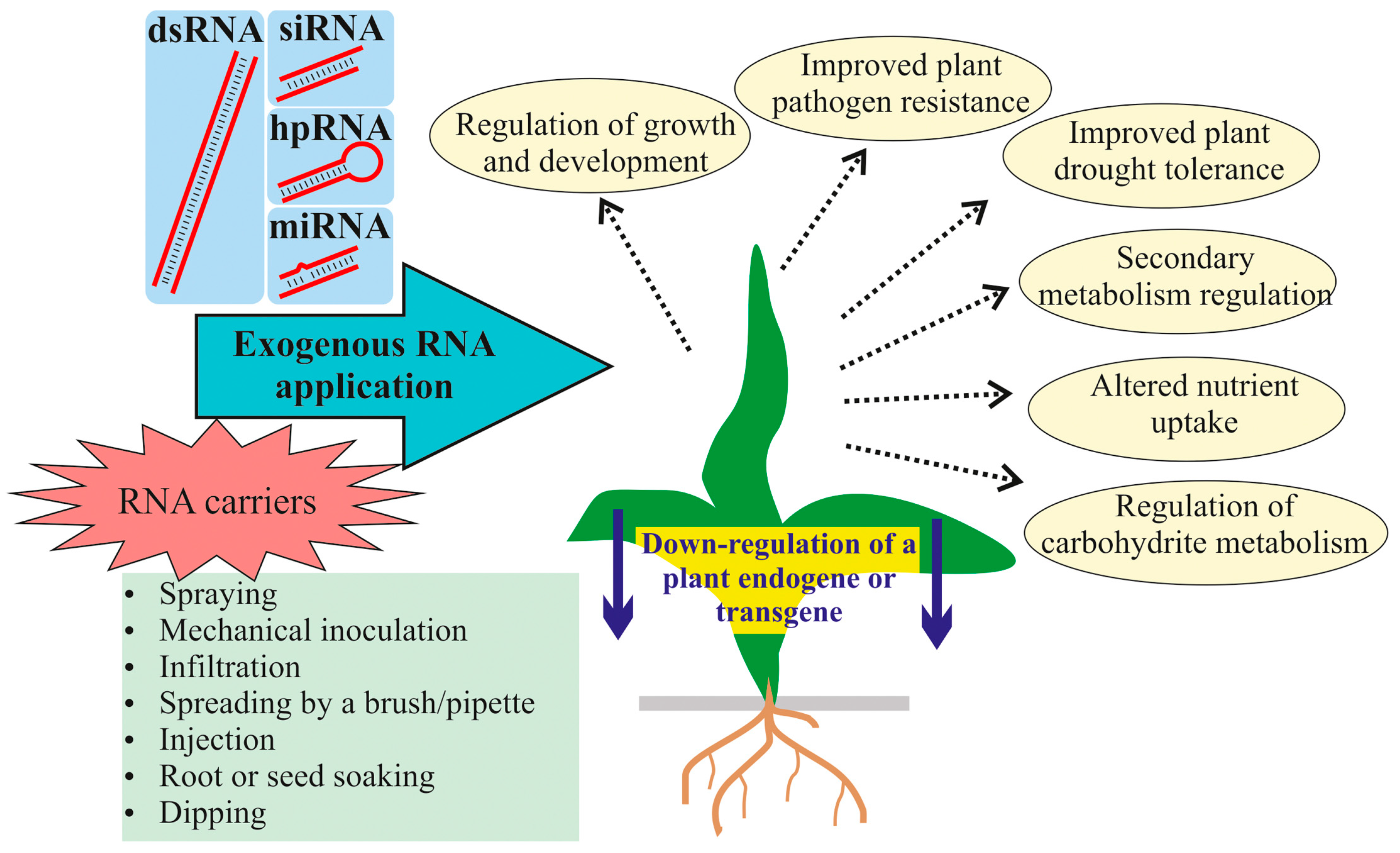
2. Silencing Transgenes in Plants Through the Application of Exogenous RNAs
3. Silencing Plant Endogenes by the Application of Exogenous RNAs
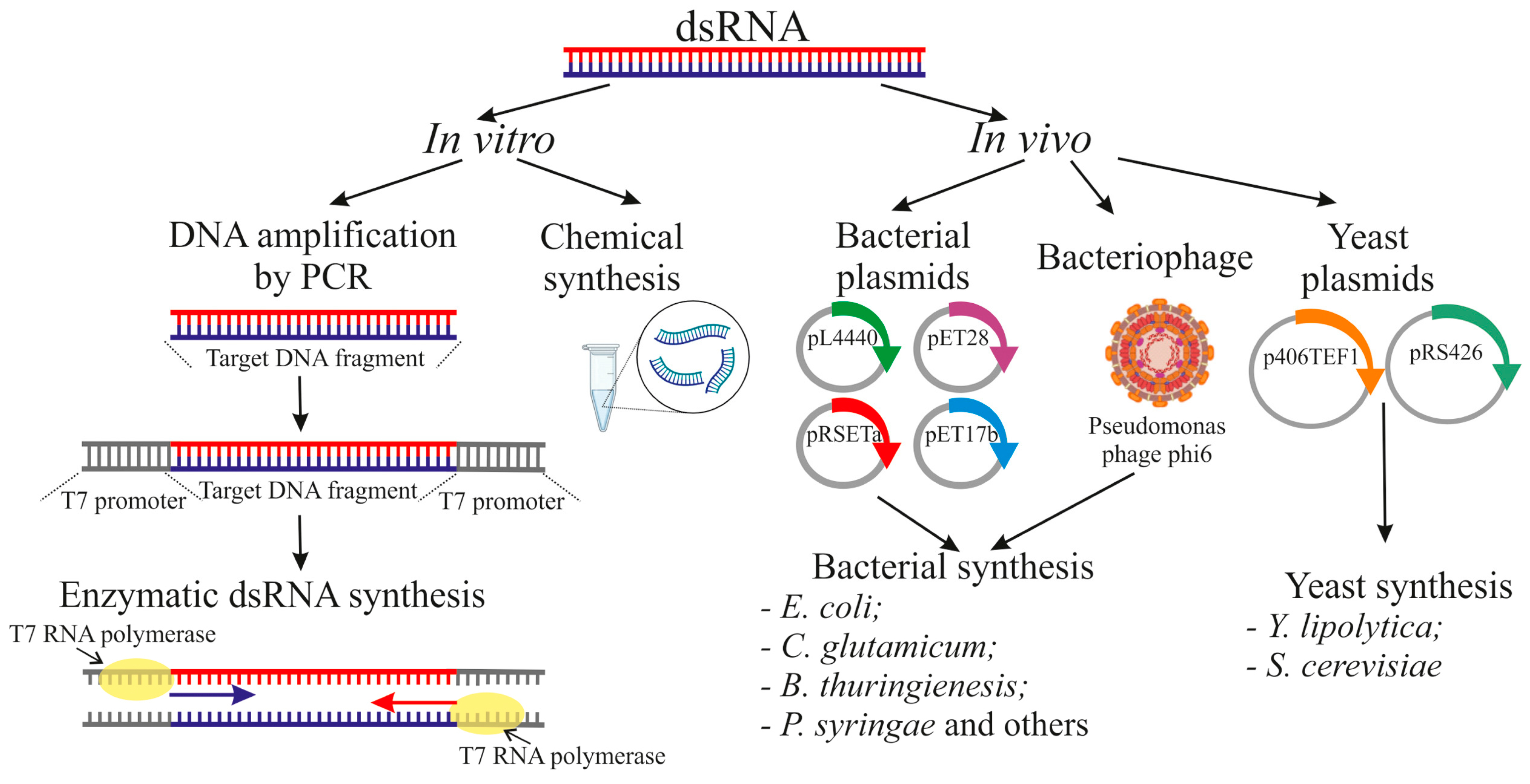
4. RNA Production for Plant Exogenous Treatments
5. RNA Delivery Methods for Exogenous Plant Treatments
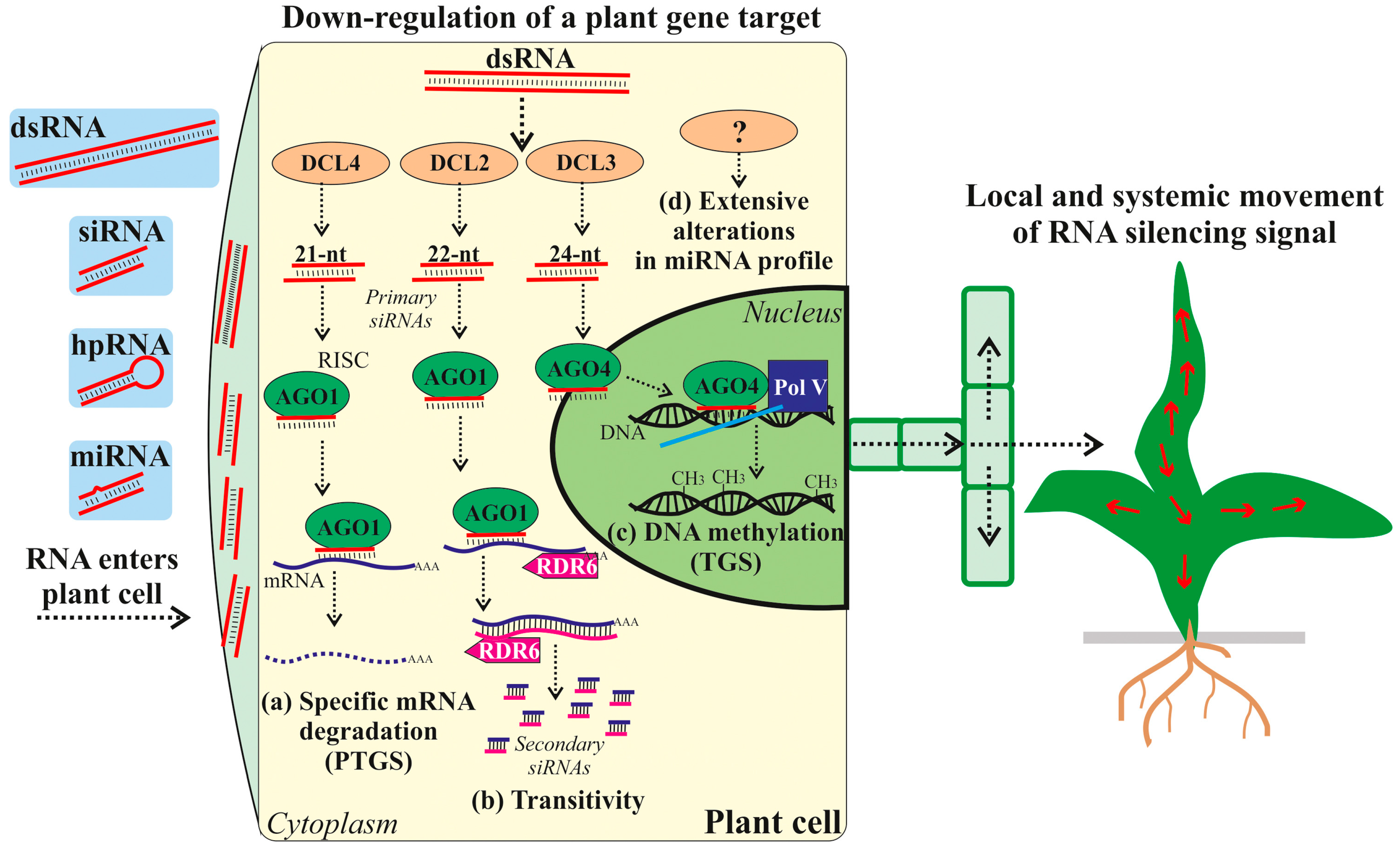
6. Exogenous RNA Recognition, Uptake, and Transport in Plants
7. Processing of Exogenous dsRNA into siRNA in Plants
8. Impact of Exogenous RNAs on Plant Epigenetics
9. Mechanism of Exogenously Induced RNAi
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
Abbreviations
| CHS | Chalcone synthase |
| CPP | Cell-penetrating peptides |
| dsRNA | double-stranded RNA |
| DCL | Dicer-like proteins |
| dpt | Days post-treatment |
| EGFP | Enhanced green fluorescent protein |
| exoRNAi | Exogenous RNA interference |
| GFP | Green fluorescent protein |
| GUS | β-glucuronidase |
| hpRNA | Hairpin RNA |
| hpt | Hours post-treatment |
| LDH | Layered double hydroxide clay nanosheets |
| lmPEI | Lipid-modified PEI nanoparticles |
| miRNA | microRNA |
| MSN | Mesoporous silica nanoparticles |
| NPTII | Neomycin phosphotransferase II |
| RISC | RNA-induced silencing complex |
| RDP | RNA-dependent RNA-polymerase |
| RdDM | RNA-directed DNA methylation |
| RNAi | RNA interference or gene silencing |
| SIGS | Spray-induced gene silencing |
| siRNA | Small interfering RNA |
| sRNA | Small RNA |
| sRNA-seq | Small RNA sequencing |
| YFP | Yellow fluorescent protein |
References
- Morozov, S.Y.; Solovyev, A.G.; Kalinina, N.O.; Taliansky, M.E. Double-stranded RNAs in plant protection against pathogenic organisms and viruses in agriculture. Acta Naturae 2019, 11, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.J.; Mammarella, F.; Ariel, F. Exogenous RNAs: Promising tools for the second green revolution. J. Exp. Bot. 2023, 74, 2323–2337. [Google Scholar] [CrossRef] [PubMed]
- Vatanparast, M.; Merkel, L.; Amari, K. Exogenous application of dsRNA in plant protection: Efficiency, safety concerns and risk assessment. Int. J. Mol. Sci. 2024, 25, 6530. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jin, H. Spray-induced gene silencing: A powerful innovative strategy for crop protection. Trends Microbiol. 2017, 25, 4–6. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V. Exogenous RNAs for gene regulation and plant resistance. Int. J. Mol. Sci. 2019, 20, 2282. [Google Scholar] [CrossRef]
- Dalakouras, A.; Wassenegger, M.; Dadami, E.; Ganopoulos, I.; Pappas, M.L.; Papadopoulou, K. Genetically modified organism-free RNA interference: Exogenous application of RNA molecules in plants. Plant Physiol. 2020, 182, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Ito, H. Small RNAs and regulation of transposons in plants. Genes. Genet. Syst. 2013, 88, 3–7. [Google Scholar] [CrossRef]
- Ding, S.W. Transgene Silencing, RNA Interference, and the Antiviral Defense Mechanism Directed by Small Interfering RNAs. Phytopathology 2023, 113, 616–625. [Google Scholar] [CrossRef]
- Singh, A.; Gautam, V.; Singh, S.; Sarkar Das, S.; Verma, S.; Mishra, V.; Mukherjee, S.; Sarkar, A.K. Plant small RNAs: Advancement in the understanding of biogenesis and role in plant development. Planta 2018, 248, 545–558. [Google Scholar] [CrossRef]
- Guleria, P.; Mahajan, M.; Bhardwaj, J.; Yadav, S.K. Plant small RNAs: Biogenesis, mode of action and their roles in abiotic stresses. Genom. Proteom. Bioinform. 2011, 9, 183–199. [Google Scholar] [CrossRef]
- Kryovrysanaki, N.; James, A.; Tselika, M.; Bardani, E.; Kalantidis, K. RNA silencing pathways in plant development and defense. Int. J. Dev. Biol. 2022, 66, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Martinez, G. Enjoy the silence: Canonical and non-canonical RNA silencing activity during plant sexual reproduction. Curr. Opin. Plant Biol. 2024, 82, 102654. [Google Scholar] [CrossRef] [PubMed]
- Malakondaiah, S.; Julius, A.; Ponnambalam, D.; Gunthoti, S.S.; Ashok, J.; Krishana, P.S.; Rebecca, J. Gene silencing by RNA interference: A review. Genome Instab. Dis. 2024, 5, 225–241. [Google Scholar] [CrossRef]
- Krzyszton, M.; Kufel, J.; Zakrzewska-Placzek, M. RNA interference and turnover in plants -a complex partnership. Front. Plant Sci. 2025, 16, 1608888. [Google Scholar] [CrossRef]
- El-Sappah, A.H.; Yan, K.; Huang, Q.; Islam, M.M.; Li, Q.; Wang, Y.; Khan, M.S.; Zhao, X.; Mir, R.R.; Li, J.; et al. Comprehensive mechanism of gene silencing and its role in plant growth and development. Front. Plant Sci. 2021, 12, 705249. [Google Scholar] [CrossRef]
- Lopez-Gomollon, S.; Baulcombe, D.C. Roles of RNA silencing in viral and non-viral plant immunity and in the crosstalk between disease resistance systems. Nat. Rev. Mol. Cell Biol. 2022, 23, 645–662. [Google Scholar] [CrossRef]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef]
- Ipsaro, J.J.; Joshua-Tor, L. From guide to target: Molecular insights into eukaryotic RNA-interference machinery. Nat. Struct. Mol. Biol. 2015, 22, 20–28. [Google Scholar] [CrossRef]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef]
- Ali, S.; Tang, Y. Noncoding RNA-mediated regulation of DNA methylation: Insights into plant epigenetic mechanisms. J. Plant Growth Regul. 2024, 44, 373–388. [Google Scholar] [CrossRef]
- Zhan, J.; Meyers, B.C. Plant small RNAs: Their biogenesis, regulatory roles, and functions. Annu. Rev. Plant Biol. 2023, 74, 21–51. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Zapletal, D.; Kubicek, K.; Svoboda, P.; Stefl, R. Dicer structure and function: Conserved and evolving features. EMBO Rep. 2023, 24, e57215. [Google Scholar] [CrossRef]
- Hung, Y.H.; Slotkin, R.K. The initiation of RNA interference (RNAi) in plants. Curr. Opin. Plant Biol. 2021, 61, 102014. [Google Scholar] [CrossRef]
- Zulfiqar, S.; Farooq, M.A.; Zhao, T.; Wang, P.; Tabusam, J.; Wang, Y.; Xuan, S.; Zhao, J.; Chen, X.; Shen, S.; et al. Virus-induced gene silencing (VIGS): A powerful tool for crop improvement and its advancement towards epigenetics. Int. J. Mol. Sci. 2023, 24, 5608. [Google Scholar] [CrossRef]
- Zand, K.H.; Innes, R.W. Molecular mechanisms underlying host-induced gene silencing. Plant Cell 2022, 34, 3183–3199. [Google Scholar] [CrossRef]
- Koch, A.; Wassenegger, M. Host-induced gene silencing-mechanisms and applications. New Phytol. 2021, 231, 54–59. [Google Scholar] [CrossRef]
- Kamthan, A.; Chaudhuri, A.; Kamthan, M.; Datta, A. Small RNAs in plants: Recent development and application for crop improvement. Front. Plant Sci. 2015, 6, 208. [Google Scholar] [CrossRef]
- Taliansky, M.; Samarskaya, V.; Zavriev, S.K.; Fesenko, I.; Kalinina, N.O.; Love, A.J. RNA-based technologies for engineering plant virus resistance. Plants 2021, 10, 82. [Google Scholar] [CrossRef]
- Gebremichael, D.E.; Haile, Z.M.; Negrini, F.; Sabbadini, S.; Capriotti, L.; Mezzetti, B.; Baraldi, E. RNA interference strategies for future management of plant pathogenic fungi: Prospects and challenges. Plants 2021, 10, 650. [Google Scholar] [CrossRef] [PubMed]
- Voloudakis, A.E.; Kaldis, A.; Patil, B.L. RNA-based vaccination of plants for control of viruses. Annu. Rev. Virol. 2022, 9, 521–548. [Google Scholar] [CrossRef] [PubMed]
- Singewar, K.; Fladung, M. Double-stranded RNA (dsRNA) technology to control forest insect pests and fungal pathogens: Challenges and opportunities. Funct. Integr. Genom. 2023, 23, 185. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.E.; Schwarzacher, T.; Othman, R.Y.; Harikrishna, J.A. dsRNA silencing of an R2R3-MYB transcription factor affects flower cell shape in a Dendrobium hybrid. BMC Plant Biol. 2015, 15, 194. [Google Scholar] [CrossRef]
- Nerva, L.; Guaschino, M.; Pagliarani, C.; De Rosso, M.; Lovisolo, C.; Chitarra, W. Spray-induced gene silencing targeting a glutathione S-transferase gene improves resilience to drought in grapevine. Plant Cell Environ. 2022, 45, 347–361. [Google Scholar] [CrossRef]
- Cedillo-Jimenez, C.A.; Guevara-Gonzalez, R.G.; Cruz-Hernandez, A. Exogenous dsRNA sequence based on miR1917 downregulates its target gene related to ethylene signaling in tomato seedlings and fruit. Sci. Hortic. 2024, 331, 113090. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Kalachev, A.V.; Dubrovina, A.S. External dsRNA downregulates anthocyanin biosynthesis-related genes and affects anthocyanin accumulation in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 6749. [Google Scholar] [CrossRef]
- Betti, F.; Ladera-Carmona, M.J.; Weits, D.A.; Ferri, G.; Iacopino, S.; Novi, G.; Svezia, B.; Kunkowska, A.B.; Santaniello, A.; Piaggesi, A.; et al. Exogenous miRNAs induce post-transcriptional gene silencing in plants. Nat. Plants 2021, 7, 1379–1388. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef]
- Faustinelli, P.C.; Power, I.L.; Arias, R.S. Detection of exogenous double-stranded RNA movement in in vitro peanut plants. Plant Biol. 2018, 20, 444–449. [Google Scholar] [CrossRef]
- Biedenkopf, D.; Will, T.; Knauer, T.; Jelonek, L.; Furch, A.C.U.; Busche, T.; Koch, A. Systemic spreading of exogenous applied RNA biopesticides in the crop plant Hordeum vulgare. ExRNA 2020, 2, 12. [Google Scholar] [CrossRef]
- Pampolini, F.; Rieske, L.K. Root uptake, translocation and persistence of EAB-specific dsRNA in ash seedlings. Sci. Rep. 2025, 15, 6378. [Google Scholar] [CrossRef] [PubMed]
- Samarskaya, V.O.; Spechenkova, N.; Ilina, I.; Suprunova, T.P.; Kalinina, N.O.; Love, A.J.; Taliansky, M.E. A non-canonical pathway induced by externally applied virus-specific dsRNA in potato plants. Int. J. Mol. Sci. 2023, 24, 15769. [Google Scholar] [CrossRef] [PubMed]
- Dubrovina, A.S.; Aleynova, O.A.; Kalachev, A.V.; Suprun, A.R.; Ogneva, Z.V.; Kiselev, K.V. Induction of transgene suppression in plants via external application of synthetic dsRNA. Int. J. Mol. Sci. 2019, 20, 1585. [Google Scholar] [CrossRef] [PubMed]
- Dubrovina, A.S.; Aleynova, O.A.; Suprun, A.R.; Ogneva, Z.V.; Kiselev, K.V. Transgene suppression in plants by foliar application of in vitro-synthesized small interfering RNAs. Appl. Microbiol. Biotechnol. 2020, 104, 2125–2135. [Google Scholar] [CrossRef]
- Nityagovsky, N.N.; Kiselev, K.V.; Suprun, A.R.; Dubrovina, A.S. Exogenous dsRNA induces RNA interference of a chalcone synthase gene in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 5325. [Google Scholar] [CrossRef]
- Molesini, B.; Pennisi, F.; Cressoni, C.; Vitulo, N.; Dusi, V.; Speghini, A.; Pandolfini, T. Nanovector-mediated exogenous delivery of dsRNA induces silencing of target genes in very young tomato flower buds. Nanoscale Adv. 2022, 4, 4542–4553. [Google Scholar] [CrossRef]
- Numata, K.; Ohtani, M.; Yoshizumi, T.; Demura, T.; Kodama, Y. Local gene silencing in plants via synthetic dsRNA and carrier peptide. Plant Biotechnol. J. 2014, 12, 1027–1034. [Google Scholar] [CrossRef]
- Dalakouras, A.; Wassenegger, M.; McMillan, J.N.; Cardoza, V.; Maegele, I.; Dadami, E.; Runne, M.; Krczal, G.; Wassenegger, M. Induction of silencing in plants by high-pressure spraying of in vitro-synthesized small RNAs. Front. Plant Sci. 2016, 7, 1327. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Kostetsky, E.Y.; Dubrovina, A.S. The specificity of transgene suppression in plants by exogenous dsRNA. Plants 2022, 11, 715. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Demirer, G.S.; Zhang, H.; Ye, T.; Goh, N.S.; Aditham, A.J.; Cunningham, F.J.; Fan, C.; Landry, M.P. DNA nanostructures coordinate gene silencing in mature plants. Proc. Natl. Acad. Sci. USA 2019, 116, 7543–7548. [Google Scholar] [CrossRef] [PubMed]
- Uslu, V.V.; Bassler, A.; Krczal, G.; Wassenegger, M. High-pressure-sprayed double stranded RNA does not induce RNA interference of a reporter gene. Front. Plant Sci. 2020, 11, 534391. [Google Scholar] [CrossRef]
- Uslu, V.V.; Dalakouras, A.; Steffens, V.A.; Krczal, G.; Wassenegger, M. High-pressure sprayed siRNAs influence the efficiency but not the profile of transitive silencing. Plant J. 2022, 109, 1199–1212. [Google Scholar] [CrossRef]
- Park, M.; Um, T.Y.; Jang, G.; Choi, Y.D.; Shin, C. Targeted gene suppression through double-stranded RNA application using easy-to-use methods in Arabidopsis thaliana. Appl. Biol. Chem. 2022, 65, 4. [Google Scholar] [CrossRef]
- Cai, Y.; Liu, Z.; Wang, H.; Meng, H.; Cao, Y. Mesoporous silica nanoparticles mediate SiRNA delivery for long-term multi-gene silencing in intact plants. Adv. Sci. 2024, 11, e2301358. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Z.; Liu, Y.; Lu, M.; Xu, J.; Wu, F.; Jin, W. Development and application of an RNA nanostructure to induce transient RNAi in difficult transgenic plants. Biotechnol. J. 2024, 19, e2400024. [Google Scholar] [CrossRef]
- Pal, G.; Ingole, K.D.; Yavvari, P.S.; Verma, P.; Kumari, A.; Chauhan, C.; Chaudhary, D.; Srivastava, A.; Bajaj, A.; Vemanna, R.S. Exogenous application of nanocarrier-mediated double-stranded RNA manipulates physiological traits and defence response against bacterial diseases. Mol. Plant Pathol. 2024, 25, e13417. [Google Scholar] [CrossRef] [PubMed]
- Sammons, R.; Ivashuta, S.; Liu, H.; Wang, D.; Feng, P.; Kouranov, A.; Andersen, S. Polynucleotide Molecules for Gene Regulation in Plants. U.S. Patent 20110296556 A1, 8 March 2011. [Google Scholar]
- Jiang, L.; Ding, L.; He, B.; Shen, J.; Xu, Z.; Yin, M.; Zhang, X. Systemic gene silencing in plants triggered by fluorescent nanoparticle-delivered double-stranded RNA. Nanoscale 2014, 6, 9965–9969. [Google Scholar] [CrossRef]
- Li, H.; Guan, R.; Guo, H.; Miao, X. New insights into an RNAi approach for plant defence against piercing-sucking and stem-borer insect pests. Plant Cell Environ. 2015, 38, 2277–2285. [Google Scholar] [CrossRef]
- Warnock, N.D.; Wilson, L.; Canet-Perez, J.V.; Fleming, T.; Fleming, C.C.; Maule, A.G.; Dalzell, J.J. Exogenous RNA interference exposes contrasting roles for sugar exudation in host-finding by plant pathogens. Int. J. Parasitol. 2016, 46, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Marcianò, D.; Ricciardi, V.; Marone Fassolo, E.; Passera, A.; Bianco, P.A.; Failla, O.; Casati, P.; Maddalena, G.; De Lorenzis, G.; Toffolatti, S.L. RNAi of a putative grapevine susceptibility gene as a possible downy mildew control strategy. Front. Plant Sci. 2021, 12, 667319. [Google Scholar] [CrossRef]
- Killiny, N.; Gonzalez-Blanco, P.; Gowda, S.; Martini, X.; Etxeberria, E. Plant functional genomics in a few days: Laser-assisted delivery of double-stranded RNA to higher plants. Plants 2021, 10, 93. [Google Scholar] [CrossRef] [PubMed]
- Simon, I.; Persky, Z.; Avital, A.; Harat, H.; Schroeder, A.; Shoseyov, O. Foliar application of dsRNA targeting endogenous potato (Solanum tuberosum) isoamylase genes ISA1, ISA2, and ISA3 confers transgenic phenotype. Int. J. Mol. Sci. 2023, 24, 190. [Google Scholar] [CrossRef]
- Chen, N.; Dai, X.; Hu, Q.; Tan, H.; Qiao, L.; Lu, L. Sprayable double-stranded RNA mediated RNA interference reduced enzymatic browning of fresh-cut potatoes. Posthar. Biol. Technol. 2023, 206, 112563. [Google Scholar] [CrossRef]
- Suprun, A.R.; Kiselev, K.V.; Dubrovina, A.S. Exogenously induced silencing of four MYB transcription repressor genes and activation of anthocyanin accumulation in Solanum lycopersicum. Int. J. Mol. Sci. 2023, 24, 9344. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Dubrovina, A.S. Simultaneous application of several exogenous dsRNAs for the regulation of anthocyanin biosynthesis in Arabidopsis thaliana. Plants 2024, 13, 541. [Google Scholar] [CrossRef] [PubMed]
- Vermeersch, L.; De Winne, N.; Nolf, J.; Bleys, A.; Kovařík, A.; Depicker, A. Transitive RNA silencing signals induce cytosine methylation of a transgenic but not an endogenous target. Plant J. 2013, 74, 867–879. [Google Scholar] [CrossRef]
- Dadami, E.; Moser, M.; Zwiebel, M.; Krczal, G.; Wassenegger, M.; Dalakouras, A. An endogene-resembling transgene delays the onset of silencing and limits siRNA accumulation. FEBS Lett. 2013, 18, 706–710. [Google Scholar] [CrossRef]
- Dadami, E.; Dalakouras, A.; Zwiebel, M.; Krczal, G.; Wassenegger, M. An endogene-resembling transgene is resistant to DNA methylation and systemic silencing. RNA Biol. 2014, 11, 934–941. [Google Scholar] [CrossRef]
- Dalakouras, A.; Jarausch, W.; Buchholz, G.; Bassler, A.; Braun, M.; Manthey, T.; Krczal, G.; Wassenegger, M. Delivery of hairpin RNAs and small RNAs into woody and herbaceous plants by trunk injection and petiole absorption. Front. Plant Sci. 2018, 9, 1253. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Dubrovina, A.S. Physiological conditions and dsRNA application approaches for exogenously induced RNA interference in Arabidopsis thaliana. Plants 2021, 10, 264. [Google Scholar] [CrossRef]
- Suprun, A.R.; Manyakhin, A.Y.; Trubetskaya, E.V.; Kiselev, K.V.; Dubrovina, A.S. Regulation of anthocyanin accumulation in tomato Solanum lycopersicum L. by exogenous synthetic dsRNA targeting different regions of SlTRY gene. Plants 2024, 13, 2489. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.M.; Chia, L.S.; Goh, N.K.; Chia, T.F.; Brouillard, R. Analysis and biological activities of anthocyanins. Phytochemistry 2003, 64, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef]
- Takiff, H.E.; Chen, S.M.; Court, D.L. Genetic analysis of the Rnc operon of Escherichia coli. J. Bacteriol. 1989, 171, 2581–2590. [Google Scholar] [CrossRef]
- Zotti, M.; Dos Santos, E.A.; Cagliari, D.; Christiaens, O.; Taning, C.N.T.; Smagghe, G. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag. Sci. 2018, 74, 1239–1250. [Google Scholar] [CrossRef]
- Höfle, L.; Biedenkopf, D.; Werner, B.T.; Shrestha, A.; Jelonek, L.; Koch, A. Study on the efficiency of dsRNAs with increasing length in RNA-based silencing of the Fusarium CYP51 genes. RNA Biol. 2020, 17, 463–473. [Google Scholar] [CrossRef]
- Chen, X.; Shi, T.; Tang, T.; Chen, C.; Liang, Y.; Zuo, S. Nanosheet-facilitated spray delivery of dsRNAs represents a potential tool to control Rhizoctonia solani infection. Int. J. Mol. Sci. 2022, 23, 12922. [Google Scholar] [CrossRef]
- Hu, D.; Chen, Z.-Y.; Zhang, C.; Ganiger, M. Reduction of Phakopsora pachyrhizi infection on soybean through host- and spray-induced gene silencing. Mol. Plant Pathol. 2020, 21, 794–807. [Google Scholar] [CrossRef]
- Sarkar, A.; Roy-Barman, S. Spray-induced silencing of pathogenicity gene MoDES1 via exogenous double-stranded RNA can confer partial resistance against fungal blast in rice. Front. Plant Sci. 2021, 12, 733129. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Greenwald, E.; Ahmad, S.; Hur, S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018, 46, 5239–5249. [Google Scholar] [CrossRef]
- Timmons, L.; Court, D.L.; Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 2001, 263, 103–112. [Google Scholar] [CrossRef]
- Hashiro, S.; Mitsuhashi, M.; Chikami, Y.; Kawaguchi, H.; Niimi, T.; Yasueda, H. Construction of Corynebacterium glutamicum cells as containers encapsulating dsRNA overexpressed for agricultural pest control. Appl. Microbiol. Biotechnol. 2019, 103, 8485–8496. [Google Scholar] [CrossRef]
- Jiang, Y.-X.; Chen, J.-Z.; Li, M.-W.; Zha, B.-H.; Huang, P.-R.; Chu, X.-M.; Chen, J.; Yang, G. The combination of Bacillus thuringiensis and its engineered strain expressing dsRNA increases the toxicity against Plutella xylostella. Int. J. Mol. Sci. 2022, 23, 444. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Soininen, M.; Poranen, M.M.; Heinlein, M. Synthetic biology approach for plant protection using dsRNA. Plant Biotechnol. J. 2018, 16, 1679–1687. [Google Scholar] [CrossRef]
- Murphy, K.A.; Tabuloc, C.A.; Cervantes, K.R.; Chiu, J.C. Ingestion of genetically modified yeast symbiont rduces fitness of an insect pest via RNA interference. Sci. Rep. 2016, 6, 22587. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhou, Y.; Mo, Q.; Huang, Y.; Tang, X. Spray-induced gene silencing in phytopathogen: Mechanisms, applications, and progress. Adv. Agrochem. 2024, 3, 289–297. [Google Scholar] [CrossRef]
- Dvorak, P.; Chrast, L.; Nikel, P.I.; Fedr, R.; Soucek, K.; Sedlackova, M.; Chaloupkova, R.; de Lorenzo, V.; Prokop, Z.; Damborsky, J. Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21(DE3) carrying a synthetic metabolic pathway. Microb. Cell Fact. 2015, 14, 201. [Google Scholar] [CrossRef]
- Ma, Z.-Z.; Zhou, H.; Wei, Y.-L.; Yan, S.; Shen, J. A novel plasmid-Escherichia coli system produces large batch dsRNAs for insect gene silencing. Pest Manag. Sci. 2020, 76, 2505–2512. [Google Scholar] [CrossRef]
- Guan, R.; Chu, D.; Han, X.; Miao, X.; Li, H. Advances in the development of microbial double-stranded RNA production systems for application of RNA interference in agricultural pest control. Front. Bioeng. Biotechnol. 2021, 9, 753790. [Google Scholar] [CrossRef] [PubMed]
- Fadeev, R.R.; Kudryavtseva, Y.S.; Bayazyt, K.-D.K.; Shuhalova, A.G.; Dolgikh, V.V. The optimized method to isolate heterologous dsRNA expressed in Escherichia coli HT115(DE3). Agricult. Biol. 2024, 59, 460–472. [Google Scholar] [CrossRef]
- Zhong, C.; Smith, N.A.; Zhang, D.; Goodfellow, S.; Zhang, R.; Shan, W.; Wang, M.-B. Full-length hairpin RNA accumulates at high levels in yeast but not in bacteria and plants. Genes 2019, 10, 458. [Google Scholar] [CrossRef]
- Riet, J.; Costa-Filho, J.; Dall’Agno, L.; Medeiros, L.; Azevedo, R.; Nogueira, L.F.; Maggioni, R.; Pedrosa, V.F.; Romano, L.A.; Altenbuchner, J.; et al. Bacillus subtilis expressing double-strand RNAs (dsRNAs) induces RNA interference mechanism (RNAi) and increases survival of WSSV-challenged Litopenaeus vannamei. Aquaculture 2021, 541, 736834. [Google Scholar] [CrossRef]
- Duman-Scheel, M. Saccharomyces cerevisiae (baker’s yeast) as an interfering RNA expression and delivery system. Curr. Drug Targets 2019, 20, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Bento, F.M.; Marques, R.N.; Campana, F.B.; Demétrio, C.G.; Leandro, R.A.; Parra, J.R.P.; Figueira, A. Gene silencing by RNAi via oral delivery of dsRNA by bacteria in the South American tomato pinworm, Tuta absoluta. Pest Manag. Sci. 2020, 76, 287–295. [Google Scholar] [CrossRef]
- Levanova, A.A.; Poranen, M.M. Utilization of Bacteriophage phi6 for the Production of High-Quality Double-Stranded RNA Molecules. Viruses 2024, 16, 166. [Google Scholar] [CrossRef]
- Aalto, A.P.; Sarin, L.P.; van Dijk, A.A.; Saarma, M.; Poranen, M.M.; Arumäe, U.; Bamford, D.H. Large-scale production of dsRNA and siRNA pools for RNA interference utilizing bacteriophage Φ6 RNA-dependent RNA polymerase. RNA 2007, 13, 422–429. [Google Scholar] [CrossRef]
- Bennett, M.; Deikman, J.; Hendrix, B.; Iandolino, A. Barriers to efficient foliar uptake of dsRNA and molecular barriers to dsRNA activity in plant cells. Front. Plant Sci. 2020, 11, 816. [Google Scholar] [CrossRef]
- Hoang, B.T.L.; Fletcher, S.J.; Brosnan, C.A.; Ghodke, A.B.; Manzie, N.; Mitter, N. RNAi as a foliar spray: Efficiency and challenges to field applications. Int. J. Mol. Sci. 2022, 23, 6639. [Google Scholar] [CrossRef]
- Doškářová, A. Uptake of RNA by the root system of tomato. Biol. Plant 1966, 8, 110–116. [Google Scholar] [CrossRef]
- Bragg, Z.; Rieske, L.K. Feasibility of systemically applied dsRNAs for pest-specific RNAi-induced gene silencing in white oak. Front. Plant Sci. 2022, 13, 638. [Google Scholar] [CrossRef]
- Pampolini, F.; Rodrigues, T.B.; Leelesh, R.S.; Kawashima, T.; Rieske, L.K. Confocal microscopy provides visual evidence and confirms the feasibility of dsRNA delivery to emerald ash borer through plant tissues. J. Pest Sci. 2020, 93, 1143–1153. [Google Scholar] [CrossRef]
- Khizar, S.; Alrushaid, N.; Alam Khan, F.; Zine, N.; Jaffrezic-Renault, N.; Errachid, A.; Elaissari, A. Nanocarriers based novel and effective drug delivery system. Int. J. Pharm. 2023, 632, 122570. [Google Scholar] [CrossRef] [PubMed]
- Moazzam, M.; Zhang, M.; Hussain, A.; Yu, X.; Huang, J.; Huang, Y. The landscape of nanoparticle-based siRNA delivery and therapeutic development. Mol. Ther. 2024, 32, 284–312. [Google Scholar] [CrossRef] [PubMed]
- Marquez, A.R.; Madu, C.O.; Lu, Y. An overview of various carriers for siRNA delivery. Oncomedicine 2018, 3, 48–58. [Google Scholar] [CrossRef]
- Komarova, T.; Ilina, I.; Taliansky, M.; Ershova, N. Nanoplatforms for the delivery of nucleic acids into Plant Cells. Int. J. Mol. Sci. 2023, 24, 16665. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Luo, L.; Hao, Z.; Liu, D. DNA-based nanostructures for RNA delivery. Med. Rev. 2024, 4, 207–224. [Google Scholar] [CrossRef]
- Yip, T.; Qi, X.; Yan, H.; Chang, Y. Therapeutic applications of RNA nanostructures. RSC Adv. 2024, 14, 28807–28821. [Google Scholar] [CrossRef]
- Song, X.S.; Gu, K.X.; Duan, X.X.; Xiao, X.M.; Hou, Y.P.; Duan, Y.B.; Wang, J.X.; Yu, N.; Zhou, M.G. Secondary amplification of siRNA machinery limits the application of spray-induced gene silencing. Mol. Plant Pathol. 2018, 19, 2543–2560. [Google Scholar] [CrossRef]
- Winston, W.M.; Sutherlin, M.; Wright, A.J.; Feinberg, E.H.; Hunter, C.P. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA 2007, 104, 10565–10570. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.; Ryu, C.M. Plant perceptions of extracellular DNA and RNA. Mol. Plant 2016, 9, 956–958. [Google Scholar] [CrossRef]
- Lee, B.; Park, Y.S.; Lee, S.; Song, G.C.; Ryu, C.M. Bacterial RNAs activate innate immunity in Arabidopsis. New Phytol. 2016, 209, 785–797. [Google Scholar] [CrossRef]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef]
- Vega-Muñoz, I.; Feregrino-Pérez, A.A.; Torres-Pacheco, I.; Guevara-González, R.G. Exogenous fragmented DNA acts as a damage-associated molecular pattern (DAMP) inducing changes in CpG DNA methylation and defence-related responses in Lactuca sativa. Funct. Plant Biol. 2018, 45, 1065–1072. [Google Scholar] [CrossRef]
- Samarskaya, V.O.; Spechenkova, N.; Markin, N.; Suprunova, T.P.; Zavriev, S.K.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Impact of exogenous application of potato virus Y-Specific dsRNA on RNA interference, pattern-triggered immunity and Poly(ADP-ribose) metabolism. Int. J. Mol. Sci. 2022, 23, 7915. [Google Scholar] [CrossRef]
- Holeva, M.C.; Sklavounos, A.; Rajeswaran, R.; Pooggin, M.M.; Voloudakis, A.E. Topical application of double-stranded RNA targeting 2b and CP genes of Cucumber mosaic virus protects plants against local and systemic viral infection. Plants 2021, 10, 963. [Google Scholar] [CrossRef] [PubMed]
- Power, I.L.; Faustinelli, P.C.; Orner, V.A.; Sobolev, V.S.; Arias, R.S. Analysis of small RNA populations generated in peanut leaves after exogenous application of dsRNA and dsDNA targeting aflatoxin synthesis genes. Sci. Rep. 2020, 10, 13820. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Khan, S.; Wani, I.A.; Gupta, R.; Verma, S.; Alam, P.; Alaklabi, A. Unravelling the role of epigenetic modifications in development and reproduction of angiosperms: A critical appraisal. Front. Genet. 2022, 13, 819941. [Google Scholar] [CrossRef]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA methylation. PLoS Genet. 2020, 16, e1009034. [Google Scholar] [CrossRef]
- Dalakouras, A.; Ganopoulos, I. Induction of promoter DNA methylation upon high-pressure spraying of double-stranded RNA in plants. Agronomy 2021, 11, 789. [Google Scholar] [CrossRef]
- Nityagovsky, N.N.; Kiselev, K.V.; Suprun, A.R.; Dubrovina, A.S. Impact of Exogenous dsRNA on miRNA Composition in Arabidopsis thaliana. Plants 2024, 13, 2335. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubrovina, A.S.; Suprun, A.R.; Kiselev, K.V. Regulation of Plant Genes with Exogenous RNAs. Int. J. Mol. Sci. 2025, 26, 6773. https://doi.org/10.3390/ijms26146773
Dubrovina AS, Suprun AR, Kiselev KV. Regulation of Plant Genes with Exogenous RNAs. International Journal of Molecular Sciences. 2025; 26(14):6773. https://doi.org/10.3390/ijms26146773
Chicago/Turabian StyleDubrovina, Alexandra S., Andrey R. Suprun, and Konstantin V. Kiselev. 2025. "Regulation of Plant Genes with Exogenous RNAs" International Journal of Molecular Sciences 26, no. 14: 6773. https://doi.org/10.3390/ijms26146773
APA StyleDubrovina, A. S., Suprun, A. R., & Kiselev, K. V. (2025). Regulation of Plant Genes with Exogenous RNAs. International Journal of Molecular Sciences, 26(14), 6773. https://doi.org/10.3390/ijms26146773








