Selected Pathway Analyses to Gain Mechanistic Insights into the Pathogenesis of Feline Hypertrophic Cardiomyopathy
Abstract
1. Introduction
2. Results
2.1. Study Population and Histological Examination
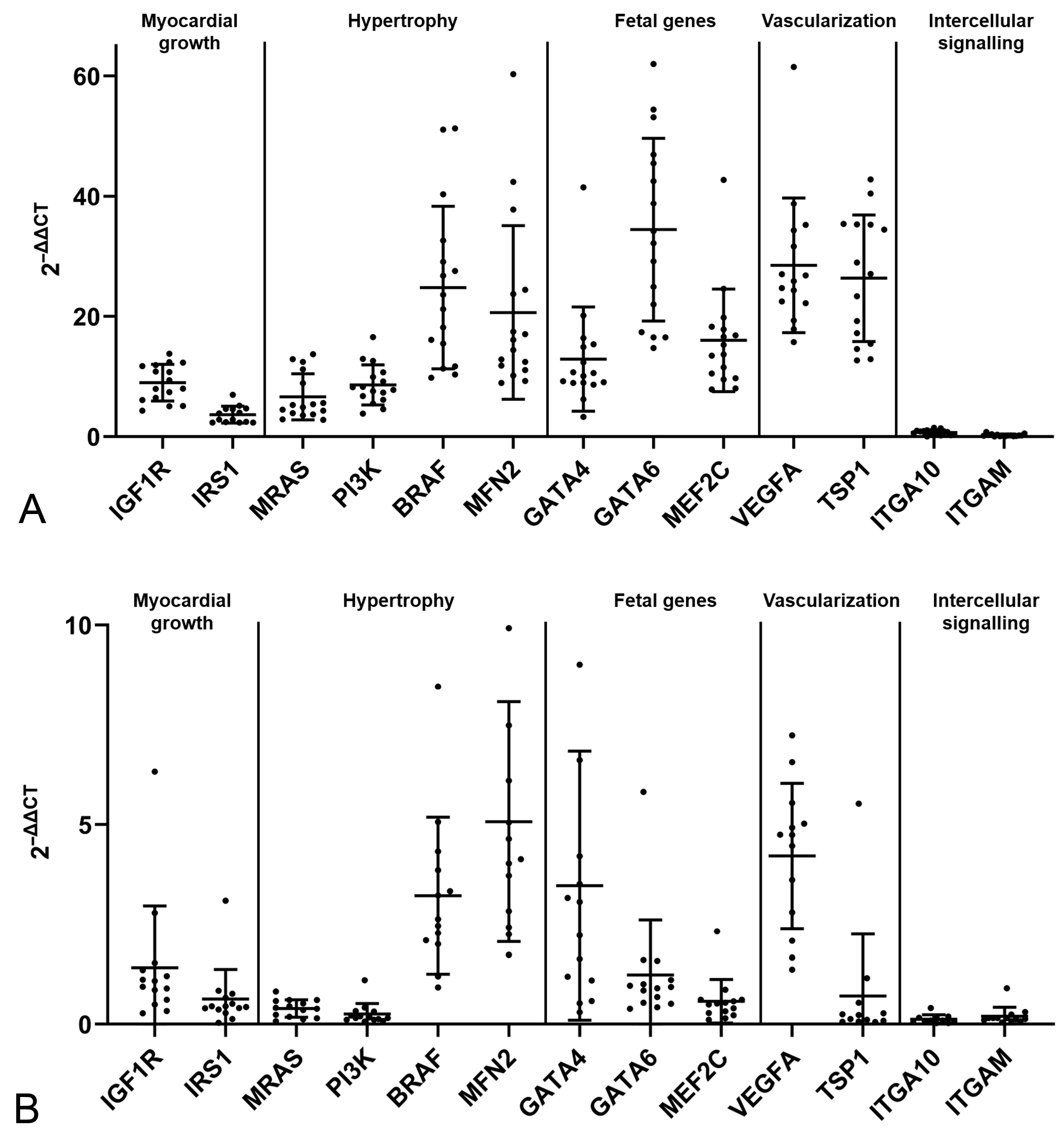
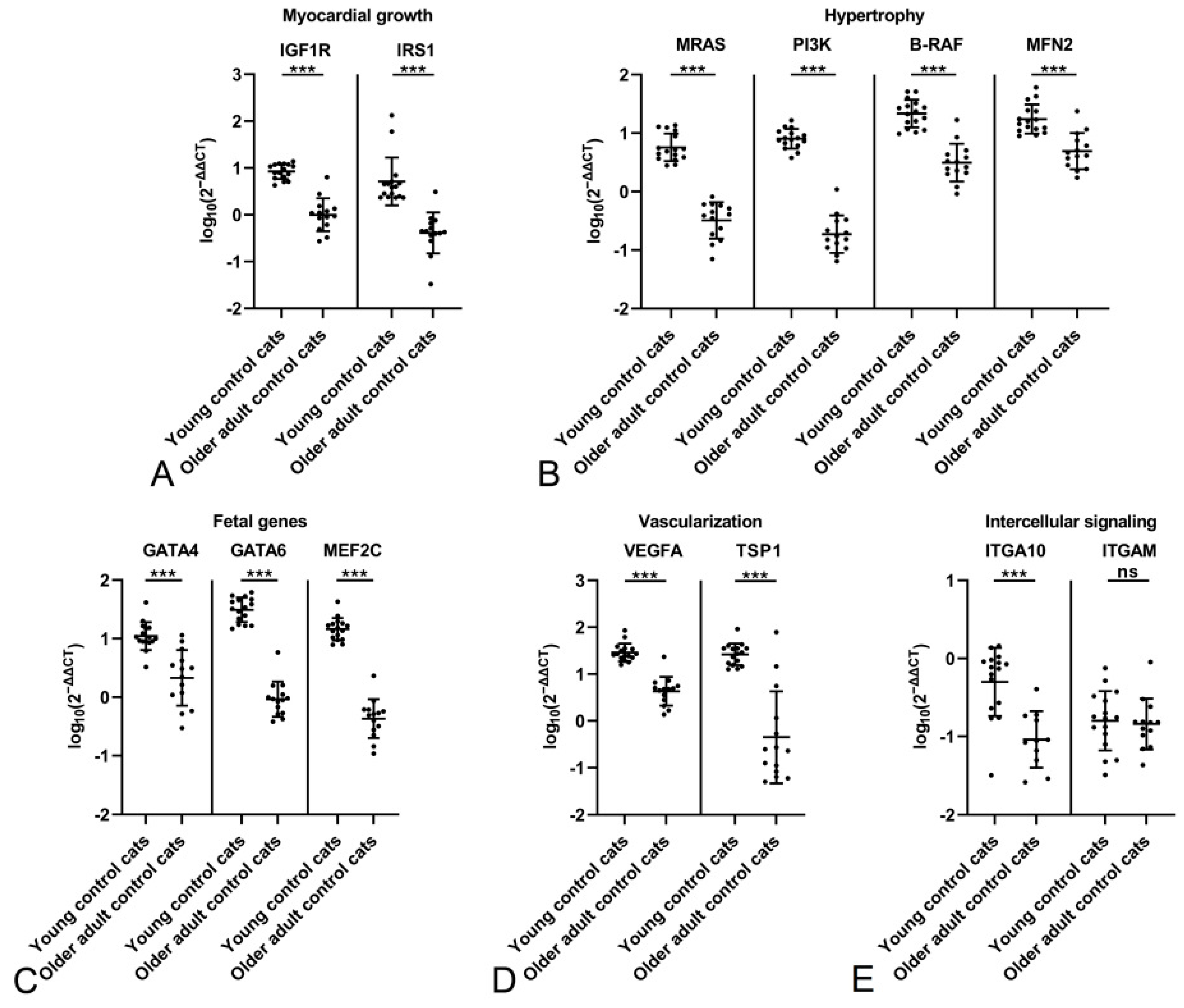
2.2. The Feline Myocardium of Both Sexes Constitutively Transcribes Markers Involved in Growth and Hypertrophy, Fetal Genes, and Markers of Vascularization and Intercellular Signaling, with Higher Expression in Young Cats
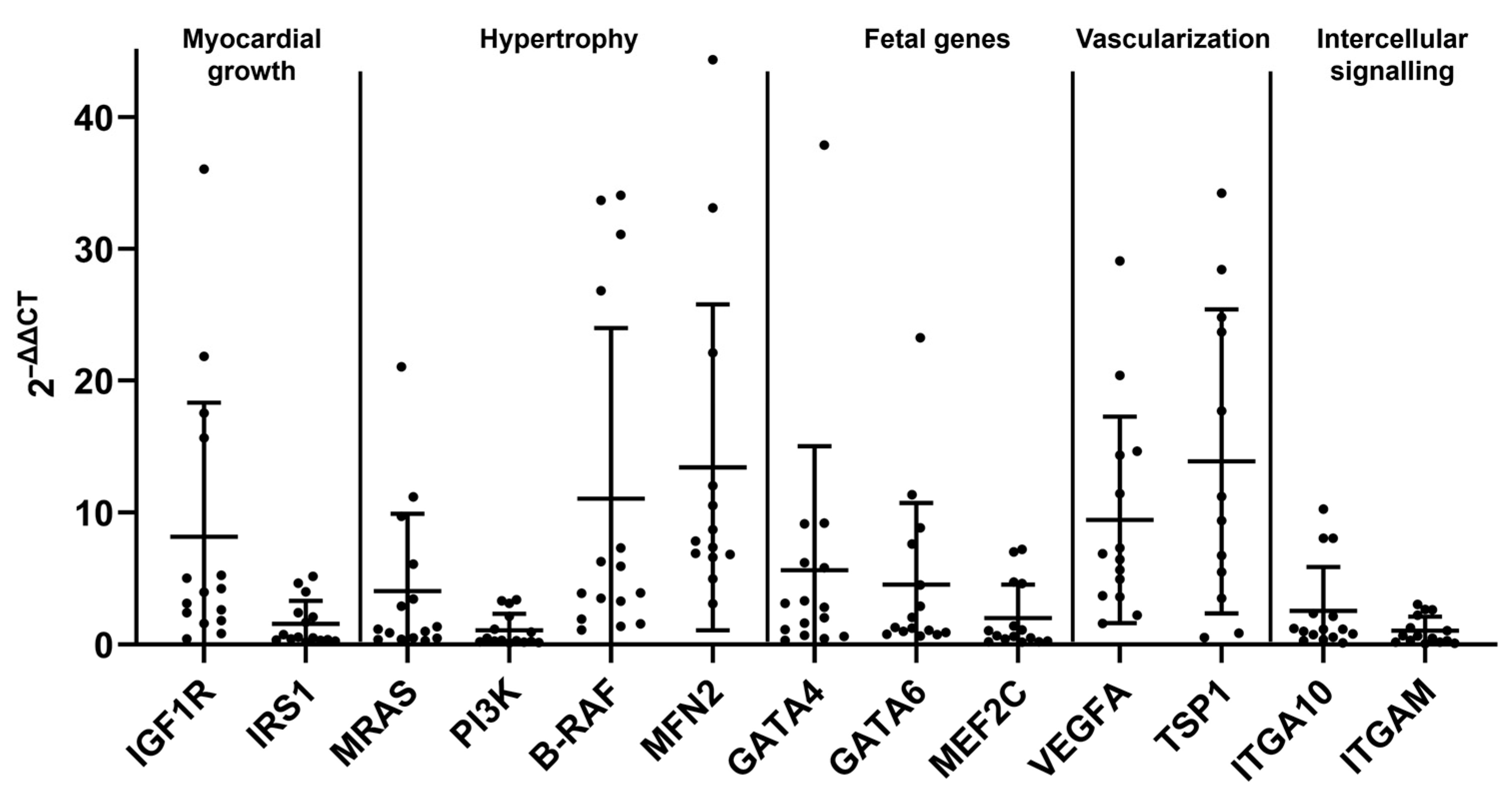
2.3. The HCM Myocardium Is Markedly More Dynamic than the Myocardium of Healthy Adult Cats
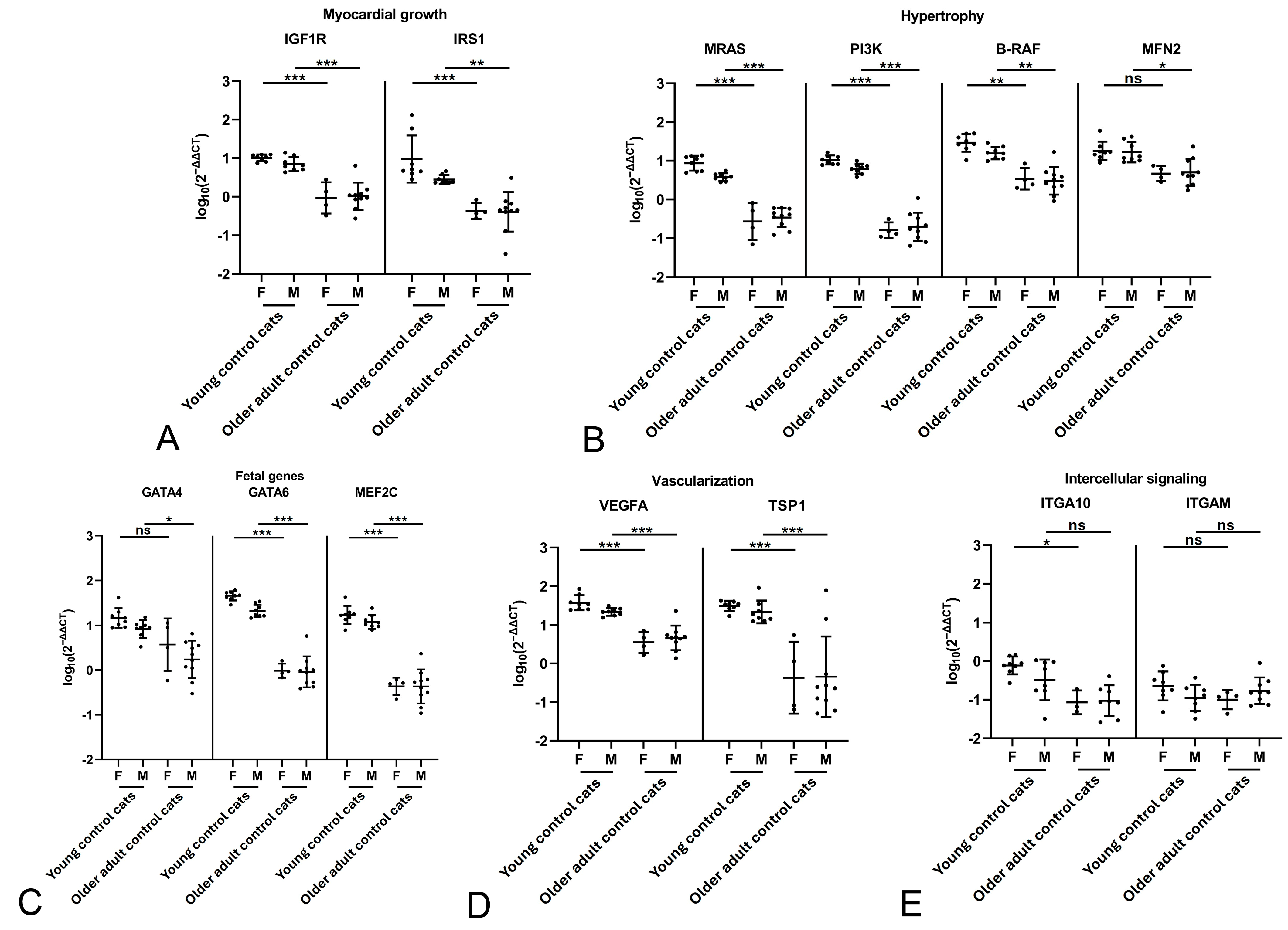
2.4. Male Cats with HCM Exhibit a More Dynamic Myocardium than Female Cats with HCM
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Tissue Sampling
4.3. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.3.1. Primer and Probe Design
4.3.2. RNA Extraction
4.3.3. DNase Treatment
4.3.4. cDNA Synthesis
4.3.5. Conventional Polymerase Chain Reaction
4.3.6. Gel Electrophoresis
4.3.7. Quantitative Polymerase Chain Reaction
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of Variance |
| B-RAF | Serine-threonine-protein kinase B-raf |
| cDNA | complementary deoxyribonucleic acid |
| DNA | Deoxyribonucleic acid |
| ERK1/2 | Extracellular signaling-regulated kinase 1/2 |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| GATA4 | GATA binding protein 4 |
| GATA6 | GATA binding protein 6 |
| IGF1R | Insulin-like growth factor 1 receptor |
| INSR | Insulin receptor |
| IRS1 | Insulin receptor substrate 1 |
| ITGA10 | Integrin β1 subunit α10 |
| ITGAM | Integrin β2 subunit M (synonym CD11b) |
| MAPK | Mitogen-activated protein kinase |
| MEF2C | Myocyte enhancer factor 2C |
| MFN2 | Mitofusin 2 |
| MMP | Matrix metalloproteinase |
| MRAS | RAS-related protein M-Ras isoform |
| NCBI BLAST | National Library of Medicine Basic Local Alignment Search Tool |
| PCR | Polymerase chain reaction |
| PI3K | Phosphatidylinositol 3-kinase |
| RNA | Ribonucleic acid |
| RT | Reverse transcription |
| RT-qPCR | Real-time quantitative polymerase chain reaction |
| TGF-β | Transforming growth factor beta |
| TIMP | Tissue inhibitor of metalloproteinase |
| TSP1 | Thrombospondin-1 |
| VEGFA | Vascular Endothelial Growth Factor A |
References
- Payne, J.R.; Brodbelt, D.C.; Luis Fuentes, V. Cardiomyopathy prevalence in 780 apparently healthy cats in rehoming centres (the CatScan study). J. Vet. Cardiol. 2015, 17, S244–S257. Available online: https://www.sciencedirect.com/science/article/pii/S1760273415000685?via%3Dihub (accessed on 13 September 2023). [CrossRef]
- Ferasin, L.; Sturgess, C.P.; Cannon, M.J.; Caney, S.; Gruffydd-Jones, T.J.; Wotton, P.R. Feline idiopathic cardiomyopathy: A retrospective study of 106 cats (1994–2001). J. Feline Med. Surg. 2003, 5, 151–159. [Google Scholar] [CrossRef]
- Paige, C.F.; Abbott, J.A.; Elvinger, F.; Pyle, R.L. Prevalence of cardiomyopathy in apparently healthy cats. J. Am. Vet. Med. Assoc. 2009, 234, 1398–1403. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.R.; Borgeat, K.; Connolly, D.J.; Boswood, A.; Dennis, S.; Wagner, T.; Menaut, P.; Maerz, I.; Evans, D.; Simons, V.E.; et al. Prognostic indicators in cats with hypertrophic cardiomyopathy. J. Vet. Intern. Med. 2013, 27, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Rush, J.E.; Freeman, L.M.; Fenollosa, N.K.; Brown, D.J. Population and survival characteristics of cats with hypertrophic cardiomyopathy: 260 cases (1990–1999). J. Am. Vet. Med. Assoc. 2002, 220, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Riesen, S.C.; Kovaceciv, A.; Lombard, C.W.; Amberger, C. Echocardiographic screening of purebred cats: An overview from 2002 to 2005. Schweiz. Arch. Tierheilkd. 2007, 149, 73–76. [Google Scholar] [CrossRef]
- Payne, J.R.; Borgeat, K.; Brodbelt, D.C.; Connolly, D.J.; Luis Fuentes, V. Risk factors associated with sudden death vs. congestive heart failure or arterial thromboembolism in cats with hypertrophic cardiomyopathy. J. Vet. Cardiol. 2015, 17, S318–S328. [Google Scholar] [CrossRef]
- Trehiou-Sechi, E.; Tissier, R.; Gouni, V.; Misbach, C.; Petit, A.; Balouka, D.; Sampedrano, C.C.; Castaignet, M.; Pouchelon, J.; Chetboul, V. Comparative echocardiographic and clinical features of hypertrophic cardiomyopathy in 5 breeds of cats: A retrospective analysis of 344 cases (2001–2011). J. Vet. Intern. Med. 2012, 26, 532–541. [Google Scholar] [CrossRef]
- Ironside, V.A.; Tricklebank, P.R.; Boswood, A. Risk indictors in cats with preclinical hypertrophic cardiomyopathy: A prospective cohort study. J. Feline Med. Surg. 2021, 23, 149–159. [Google Scholar] [CrossRef]
- März, I.; Wilkie, L.J.; Harrington, N.; Payne, J.R.; Muzzi, R.A.L.; Häggström, J.; Smith, K.; Fuentes, V.L. Familial cardiomyopathy in norwegian forest cats. J. Feline Med. Surg. 2015, 17, 681–691. [Google Scholar] [CrossRef]
- Maron, B.J.; Fox, P.R. Hypertrophic cardiomyopathy in man and cats. J. Vet. Cardiol. 2015, 17, S6–S9. [Google Scholar] [CrossRef]
- Schipper, T.; Van Poucke, M.; Sonck, L.; Smets, P.; Ducatelle, R.; Broeckx, B.J.G.; Peelman, L.J. A feline orthologue of the human MYH7 c.5647G>A (p.(Glu1883Lys)) variant causes hypertrophic cardiomyopathy in a Domestic Shorthair cat. Eur. J. Human. Genet. 2019, 27, 1724–1730. [Google Scholar] [CrossRef] [PubMed]
- Meurs, K.M.; Williams, B.G.; DeProspero, D.; Friedenberg, S.G.; Malarkey, D.E.; Ezzell, J.A.; Keene, B.W.; Adin, D.B.; DeFrancesco, T.C.; Tou, S. A deleterious mutation in the ALMS1 gene in a naturally occurring model of hypertrophic cardiomyopathy in the Sphynx cat. Orphanet. J. Rare Dis. 2021, 16, 108. [Google Scholar] [CrossRef]
- Gil-Ortuño, C.; Sebastián-Marcos, P.; Sabater-Molina, M.; Nicolas-Rocamora, E.; Gimeno-Blanes, J.R.; Fernández del Palacio, M.J. Genetics of feline hypertrophic cardiomyopathy. Clin. Genet. 2020, 98, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Fox, P.R.; Keene, B.W.; Lamb, K.; Schober, K.A.; Chetboul, V.; Fuentes, V.L.; Wess, G.; Payne, J.R.; Hogan, D.F.; Motsinger-Reif, A.; et al. International collaborative study to assess cardiovascular risk and evaluate long-term health in cats with preclinical hypertrophic cardiomyopathy and apparently healthy cats: The REVEAL Study. J. Vet. Intern. Med. 2018, 32, 930–943. [Google Scholar] [CrossRef] [PubMed]
- Novo Matos, J.; Payne, J.R.; Seo, J.; Luis Fuentes, V. Natural history of hypertrophic cardiomyopathy in cats from rehoming centers: The CatScan II study. J. Vet. Intern. Med. 2022, 36, 1900–1912. [Google Scholar] [CrossRef]
- Uentes, V.L.; Abbott, J.; Chetboul, V.; Côté, E.; Fox, P.R.; Häggström, J.; Kittleson, M.D.; Schober, K.; Stern, J.A. ACVIM consensus statement guidelines for the classification, diagnosis, and management of cardiomyopathies in cats. J. Vet. Intern. Med. 2020, 34, 1062–1077. [Google Scholar]
- Luis Fuentes, V.; Wilkie, L.J. Asymptomatic Hypertrophic Cardiomyopathy: Diagnosis and Therapy. Vet. Clin. North Am.—Small Anim. Pract. 2017, 47, 1041–1054. [Google Scholar] [CrossRef]
- Fox, P.R.; Liu, S.K.; Maron, B.J. Echocardiographic assessment of spontaneously occurring feline hypertrophic cardiomyopathy: An animal model of human disease. Circulation. 1995, 92, 2645–2651. [Google Scholar] [CrossRef]
- Fox, P.R. Hypertrophic Cardiomyopathy. Clinical and Pathologic Correlates. J. Vet. Cardiol. 2003, 5, 39–45. [Google Scholar] [CrossRef]
- Robinson, W.F.; Robinson, N.A. Myocardial disease: Cardiomyopathies. In Jubb, Kennedy and Palmer’s Pathology of Domestic Animals, 6th ed.; Maxie, M.G., Ed.; Elsevier Inc.: St. Louis, MO, USA, 2016; pp. 44–50. [Google Scholar]
- Biasato, I.; Francescone, L.; La Rosa, G.; Tursi, M. Anatomopathological staging of feline hypertrophic cardiomyopathy through quantitative evaluation based on morphometric and histopathological data. Res. Vet. Sci. 2015, 102, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Khor, K.H.; Campbell, F.E.; Owen, H.; Shiels, I.A.; Mills, P.C. Myocardial collagen deposition and inflammatory cell infiltration in cats with pre-clinical hypertrophic cardiomyopathy. Vet. J. 2015, 203, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.K.; Maron, B.J.; Tilley, L.P. Feline Hypertrophic Cardiomyopathy Gross Anatomic and Quantitative Histologic Features. Am. J. Pathol. 1981, 102, 388–395. [Google Scholar]
- Rodríguez Monné, J.M. Feline Hypertrophic Cardiomyopathy: An Out-of-Hand Cardiac Remodelling Scenario? Ph.D. Thesis, Vetsuisse Faculty of the University of Zurich, Zurich, Switzerland, 2021. [Google Scholar]
- Kitz, S.; Fonfara, S.; Hahn, S.; Hetzel, U.; Kipar, A. Feline hypertrophic cardiomyopathy: The consequence of cardiomyocyte-initiated and macrophage-driven remodeling processes? Vet. Pathol. 2019, 56, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Kittleson, M.D.; Meurs, K.M.; Munro, M.J.; Kittleson, J.A.; Liu, S.-K.; Pion, P.D.; Towbin, J.A. Familial hypertrophic cardiomyopathy in Maine Coon cats. Circulation 1999, 99, 3172–3180. [Google Scholar] [CrossRef]
- Kershaw, O.; Heblinski, N.; Lotz, F.; Dirsch, O.; Gruber, A.D. Diagnostic value of morphometry in feline hypertrophic cardiomyopathy. J. Comp. Pathol. 2012, 147, 73–83. [Google Scholar] [CrossRef]
- Colpitts, M.E.; Caswell, J.L.; Monteith, G.; Joshua, J.; O’SUllivan, M.L.; Raheb, S.; Fonfara, S. Cardiac gene activation varies between young and adult cats and in the presence of hypertrophic cardiomyopathy. Res. Vet. Sci. 2022, 152, 38–47. [Google Scholar] [CrossRef]
- Fonfara, S.; Hetzel, U.; Hahn, S.; Kipar, A. Age- and gender-dependent myocardial transcription patterns of cytokines and extracellular matrix remodelling enzymes in cats with non-cardiac diseases. Exp. Gerontol. 2015, 72, 117–123. [Google Scholar] [CrossRef]
- Fonfara, S.; Kitz, S.; Monteith, G.; Hahn, S.; Kipar, A. Myocardial transcription of inflammatory and remodeling markers in cats with hypertrophic cardiomyopathy and systemic diseases associated with an inflammatory phenotype. Res. Vet. Sci. 2022, 136, 484–494. Available online: https://pubmed.ncbi.nlm.nih.gov/33848803/ (accessed on 31 March 2022). [CrossRef]
- Joshua, J.; Caswell, J.; Lynne OSullivan, M.; Wood, G.; Fonfara, S. Feline myocardial transcriptome in health and in hypertrophic cardiomyopathy-A translational animal model for human disease. PLoS ONE 2023, 18, e0283244. [Google Scholar] [CrossRef]
- Liu, T.J.; Lai, H.C.; Wu, W.; Chinn, S.; Wang, P.H. Developing a strategy to define the effects of insulin-like growth factor-1 on gene expression profile in cardiomyocytes. Circ. Res. 2001, 88, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Samson, W.K.; Sowers, J.R. Insulin-like Growth Factor I as a cardiac hormone: Physiological and pathophysiological implications in heart disease. J. Mol. Cell Cardiol. 1999, 31, 2049–2061. [Google Scholar] [CrossRef] [PubMed]
- Joseph D’ercole, A. Insulin-like growth factors and their receptors in growth. Endocrinol. Metab. Clin. North Am. 1996, 25, 573–590. [Google Scholar] [CrossRef]
- Liu, F.; Yang, X.; Geng, M.; Huang, M. Targeting ERK, an Achilles’ Heel of the MAPK pathway, in cancer therapy. Acta Pharm. Sin. B 2018, 8, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Muniyappa, R.; Montagnani, M.; Koh, K.K.; Quon, M.J. Cardiovascular actions of insulin. Endocr. Rev. 2007, 28, 463–491. [Google Scholar] [CrossRef]
- Belke, D.D.; Betuing, S.; Tuttle, M.J.; Graveleau, C.; Young, M.E.; Pham, M.; Zhang, D.; Cooksey, R.C.; McClain, D.A.; Litwin, S.E.; et al. Insulin signaling coordinately regulates cardiac size, metabolism, and contractile protein isoform expression. J. Clin. Investig. 2002, 109, 629–639. [Google Scholar] [CrossRef]
- DeBosch, B.J.; Muslin, A.J. Insulin signaling pathways and cardiac growth. J. Mol. Cell Cardiol. 2008, 44, 855–864. [Google Scholar] [CrossRef]
- Olson, E.N.; Schneider, M.D. Sizing up the heart: Development redux in disease. Genes Dev. 2003, 17, 1937–1956. [Google Scholar] [CrossRef]
- Sala, V.; Gallo, S.; Leo, C.; Gatti, S.; Gelb, B.D.; Crepaldi, T. Signaling to cardiac hypertrophy: Insights from human and mouse RASopathies. Mol. Med. 2012, 18, 938–947. [Google Scholar] [CrossRef]
- Molkentin, J.D.; Dorn, G.W. Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu. Rev. Physiol. 2001, 63, 391–426. [Google Scholar] [CrossRef]
- McMullen, J.R.; Shioi, T.; Zhang, L.; Tarnavski, O.; Sherwood, M.C.; Kang, P.M.; Izumo, S. Phosphoinositide 3-kinase(p110α) plays a critical role for the induction of physiological, but not pathological, cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 2003, 100, 12355–12360. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, T.; Matsui, T. Phosphoinositide-3 kinase signaling in cardiac hypertrophy and heart failure. Curr. Pharm. Des. 2011, 17, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Shioi, T.; Kang, P.M.; Douglas, P.S.; Hampe, J.; Yballe, C.M.; Lawitts, J.; Cantley, L.C.; Izumo, S. The conserved phosphoinositide 3-kinase pathway determines heart size in mice. EMBO J. 2000, 19, 2537–2548. [Google Scholar] [CrossRef]
- Alharbi, H.O.; Hardyman, M.A.; Cull, J.J.; Markou, T.; Cooper, S.T.; Glennon, P.E.; Fuller, S.J.; Sugden, P.H.; Clerk, A. Cardiomyocyte BRAF is a key signalling intermediate in cardiac hypertrophy in mice. Clin. Sci. 2022, 136, 1661–1681. [Google Scholar] [CrossRef]
- Gilbert, C.J.; Longenecker, J.Z.; Accornero, F. Erk1/2: An integrator of signals that alters cardiac homeostasis and growth. Biology 2021, 10, 346. [Google Scholar] [CrossRef]
- Chen, L.; Liu, B.; Qin, Y.; Li, A.; Gao, M.; Liu, H.; Gong, G. Mitochondrial Fusion Protein Mfn2 and Its Role in Heart Failure. Front. Mol. Biosci. 2021, 8, 681237. [Google Scholar] [CrossRef]
- Bach, D.; Pich, S.; Soriano, F.X.; Vega, N.; Baumgartner, B.; Oriola, J.; Daugaard, J.R.; Lloberas, J.; Camps, M.; Zierath, J.R.; et al. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. J. Biol. Chem. 2003, 278, 17190–17197. [Google Scholar] [CrossRef] [PubMed]
- Dorn, G.W.; Kitsis, R.N. The mitochondrial dynamism-mitophagy-cell death interactome: Multiple roles performed by members of a mitochondrial molecular ensemble. Circ. Res. 2015, 116, 167–182. [Google Scholar] [CrossRef]
- Chen, H.; Detmer, S.A.; Ewald, A.J.; Griffin, E.E.; Fraser, S.E.; Chan, D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003, 160, 189–200. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Dorn, G.W. Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ. Res. 2011, 109, 1327–1331. [Google Scholar] [CrossRef]
- Papanicolaou, K.N.; Kikuchi, R.; Ngoh, G.A.; Coughlan, K.A.; Dominguez, I.; Stanley, W.C.; Walsh, K. Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ. Res. 2012, 111, 1012–1026. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Li, A.; Liu, B.; Jiang, W.; Gao, M.; Tian, X.; Gong, G. Mitochondrial fusion mediated by fusion promotion and fission inhibition directs adult mouse heart function toward a different direction. FASEB J. 2020, 34, 663–675. [Google Scholar] [CrossRef]
- McKinsey, T.A.; Zhang, C.L.; Lu, J.; Olson, E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 2000, 408, 106–111. [Google Scholar] [CrossRef]
- Xu, J.; Gong, N.L.; Bodi, I.; Aronow, B.J.; Backx, P.H.; Molkentin, J.D. Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J. Biol. Chem. 2006, 281, 9152–9162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.L.; Mckinsey, T.A.; Chang, S.; Antos, C.L.; Hill, J.A.; Olson, E.N. Class II Histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 2002, 110, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Pikkarainen, S.; Tokola, H.; Kerkelä, R.; Ruskoaho, H. GATA transcription factors in the developing and adult heart. Cardiovasc. Res. 2004, 63, 196–207. [Google Scholar] [CrossRef]
- Välimäki, M.J.; Ruskoaho, H.J. Targeting GATA4 for cardiac repair. IUBMB Life 2020, 72, 68–79. [Google Scholar] [CrossRef]
- van Berlo, J.H.; Elrod, J.W.; Hoogenhof, M.M.v.D.; York, A.J.; Aronow, B.J.; Duncan, S.A.; Molkentin, J.D. The transcription factor GATA-6 regulates pathological cardiac hypertrophy. Circ. Res. 2010, 107, 1032–1040. [Google Scholar] [CrossRef]
- Narita, N.; Heikinheimo, M.; Bielinska, M.; White, R.A.; Wilson, D.B. The gene for transcription factor GATA-6 resides on mouse chromosome 18 and is expressed in myocardium and vascular smooth muscle. Genomics 1996, 36, 345–348. [Google Scholar] [CrossRef]
- Zhuang, T.; Liu, J.; Chen, X.; Pi, J.; Kuang, Y.; Wang, Y.; Tomlinson, B.; Chan, P.; Zhang, Q.; Li, Y.; et al. Cell-specific effects of GATA (GATA zinc finger transcription tactor family)-6 in vascular smooth muscle and endothelial cells on vascular injury neointimal formation. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 888–901. [Google Scholar] [CrossRef]
- Tang, J.; Wang, J.; Kong, X.; Yang, J.; Guo, L.; Zheng, F.; Zhang, L.; Huang, Y.; Wan, Y. Vascular endothelial growth factor promotes cardiac stem cell migration via the PI3K/Akt pathway. Exp. Cell Res. 2009, 315, 3521–3531. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. Matricellular proteins in cardiac adaptation and disease. Physiol. Rev. 2012, 92, 635–688. [Google Scholar] [CrossRef]
- Xia, Y.; Dobaczewski, M.; Gonzalez-Quesada, C.; Chen, W.; Biernacka, A.; Li, N.; Lee, D.-W.; Frangogiannis, N.G. Endogenous thrombospondin 1 protects the pressure-overloaded myocardium by modulating fibroblast phenotype and matrix metabolism. Hypertension 2011, 58, 902–911. [Google Scholar] [CrossRef]
- Zhang, K.; Li, M.; Yin, L.; Fu, G.; Liu, Z. Role of thrombospondin–1 and thrombospondin–2 in cardiovascular diseases. Int. J. Mol. Med. 2020, 45, 1275–1293. [Google Scholar] [CrossRef]
- Cai, H.; Yuan, Z.; Fei, Q.; Zhao, J. Investigation of thrombospondin-1 and transforming growth factor-β expression in the heart of aging mice. Exp. Ther. Med. 2012, 3, 433–436. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, Q.; Kong, M.; Mao, L.; Yang, Y.; Xu, Y. Myocardin-related transcription factor A regulates integrin beta 2 transcription to promote macrophage infiltration and cardiac hypertrophy in mice. Cardiovasc. Res. 2022, 118, 844–858. [Google Scholar] [CrossRef]
- Li, R.; Frangogiannis, N.G. Integrins in cardiac fibrosis. J. Mol. Cell Cardiol. 2022, 172, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, R.; Ross, R.S.; Manso, A.M. Integrins and integrin-related proteins in cardiac fibrosis. J. Mol. Cell Cardiol. 2016, 93, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Shai, S.-Y.; Harpf, A.E.; Babbitt, C.J.; Jordan, M.C.; Fishbein, M.C.; Chen, J.; Omura, M.; Leil, T.A.; Becker, K.D.; Jiang, M.; et al. Cardiac myocyte-specific excision of the β1 integrin gene results in myocardial fibrosis and cardiac failure. Circ. Res. 2002, 90, 458–464. [Google Scholar] [CrossRef]
- Payne, J.; Luis Fuentes, V.; Boswood, A.; Connolly, D.; Koffas, H.; Brodbelt, D. Population characteristics and survival in 127 referred cats with hypertrophic cardiomyopathy (1997 to 2005). J. Small Anim. Pract. 2010, 51, 540–547. [Google Scholar] [CrossRef]
- McKenzie, B.A. Comparative veterinary geroscience: Mechanism of molecular, cellular, and tissue aging in humans, laboratory animal models, and companion dogs and cats. Am. J. Vet. Res. 2022, 83, 1–16. [Google Scholar] [CrossRef]
- Meschiari, C.A.; Ero, O.K.; Pan, H.; Finkel, T.; Lindsey, M.L. The impact of aging on cardiac extracellular matrix. Geroscience 2017, 39, 7–18. [Google Scholar] [CrossRef]
- Walker, C.J.; Schroeder, M.E.; Aguado, B.A.; Anseth, K.S.; Leinwand, L.A. Matters of the heart: Cellular sex differences. J. Mol. Cell Cardiol. 2021, 160, 42–55. [Google Scholar] [CrossRef]
- Abdellatif, M.; Trummer-Herbst, V.; Heberle, A.M.; Humnig, A.; Pendl, T.; Durand, S.; Cerrato, G.; Hofer, S.J.; Islam, M.; Voglhuber, J.; et al. Fine-tuning cardiac Insulin-Like Growth Factor 1 Receptor signaling to promote health and longevity. Circulation 2022, 145, 1853–1866. [Google Scholar] [CrossRef] [PubMed]
- Schellings, M.W.M.; Van Almen, G.C.; Sage, E.H.; Heymans, S. Thrombospondins in the heart: Potential functions in cardiac remodeling. J. Cell Commun. Signal. 2009, 3, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Mondragón, R.; Galindo, C.A.; Avila, G. Role of TGF-β on cardiac structural and electrical remodeling. Vasc. Health Risk Manag. 2008, 4, 1289–1300. [Google Scholar] [PubMed]
- Joshua, J.; Caswell, J.L.; Rodriguez, J.M.M.; Kipar, A.; O’SUllivan, M.L.; Wood, G.; Fonfara, S. MicroRNA profiling of the feline left heart identifies chamber-specific expression signatures in health and in advanced hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. Plus 2023, 4, 100037. [Google Scholar] [CrossRef]
- Lakdawala, N.K.; Olivotto, I.; Day, S.M.; Han, L.; Ashley, E.A.; Michels, M.; Ingles, J.; Semsarian, C.; Jacoby, D.; Jefferies, J.L.; et al. Associations between female sex, sarcomere variants, and clinical outcomes in hypertrophic cardiomyopathy. Circ. Genom. Precis. Med. 2021, 14, E003062. [Google Scholar] [CrossRef]
- Thomas, W.P.; Gaber, C.E.; Jacobs, G.J.; Kaplan, P.M.; Lombard, C.W.; Vet, M.; Moise, N.S.; Moses, B.L. Recommendations for Standards in Transthoracic Two-Dimensional Echocardiography in the Dog and Cat. J. Vet. Intern. Med. 1993, 7, 247–252. [Google Scholar] [CrossRef]
- Clustal Omega. Available online: https://www.ebi.ac.uk/jdispatcher/msa/clustalo (accessed on 11 November 2024).
- Primer Blast. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi (accessed on 11 November 2024).
- Applied Biosystem. Getting Started Guide; Applied Biosystem: Waltham, MA, USA, 2004. [Google Scholar]
- Tm Calculator. 2024. Available online: https://www.thermofisher.com/ch/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/tm-calculator.html. (accessed on 11 October 2024).
- Multiple Primer Analyzer. 2024. Available online: https://www.thermofisher.com/ch/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/multiple-primer-analyzer.html (accessed on 11 October 2024).
- Oligonucleotide Properties Calculator. 2024. Available online: https://www.biosyn.com/gizmo/tools/oligo/oligonucleotide%20properties%20calculator.htm (accessed on 3 July 2025).
- Thermo Fisher Scientific. Primer Express Software v. 3.0; Applied Biosystems, Thermo Fisher Scientific: Waltham, MA, USA, 2022. [Google Scholar]
- National Center for Biotechnology Information Web BLAST 2024; v. 1.30+. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 11 October 2024).
- Leutenegger, C.M.; Mislin, C.N.; Sigrist, B.; Ehrengruber, M.U.; Hofmann-Lehmann, R.; Lutz, H. Quantitative real-time PCR for the measurement of feline cytokine mRNA. Vet. Immunol. Immunopathol. 1999, 71, 291–305. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.r-project.org/ (accessed on 13 November 2024).
- R Studio Team. R Studio: Integrated Development Environment for R Posit Software; PBC: Boston, MA, USA, 2023; Available online: http://www.posit.co/ (accessed on 13 November 2024).
- Wickham, H.; Vaughan, D.; Girlich, M. Tidyr: Tidy Messy Data. 2024. Available online: https://tidyr.tidyverse.org (accessed on 13 November 2024).
- Wickham, H. Stringr: Simple, Consistent Wrappers for Common String Operations. 2023. Available online: https://stringr.tidyverse.org (accessed on 12 November 2024).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 13 November 2024).
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. Dplyr: A Grammar of Data Manipulation. 2023. Available online: https://dplyr.tidyverse.org (accessed on 13 November 2024).
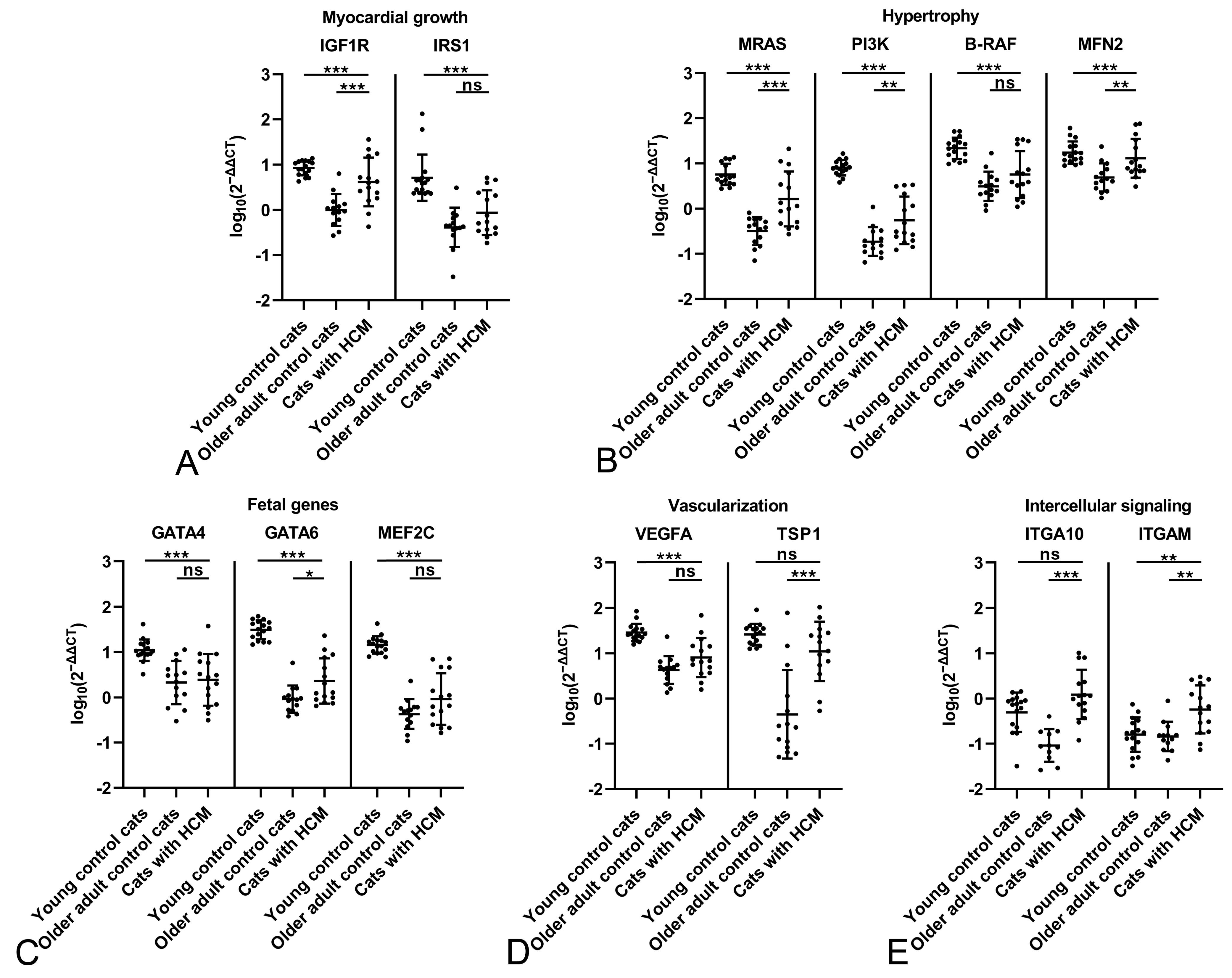

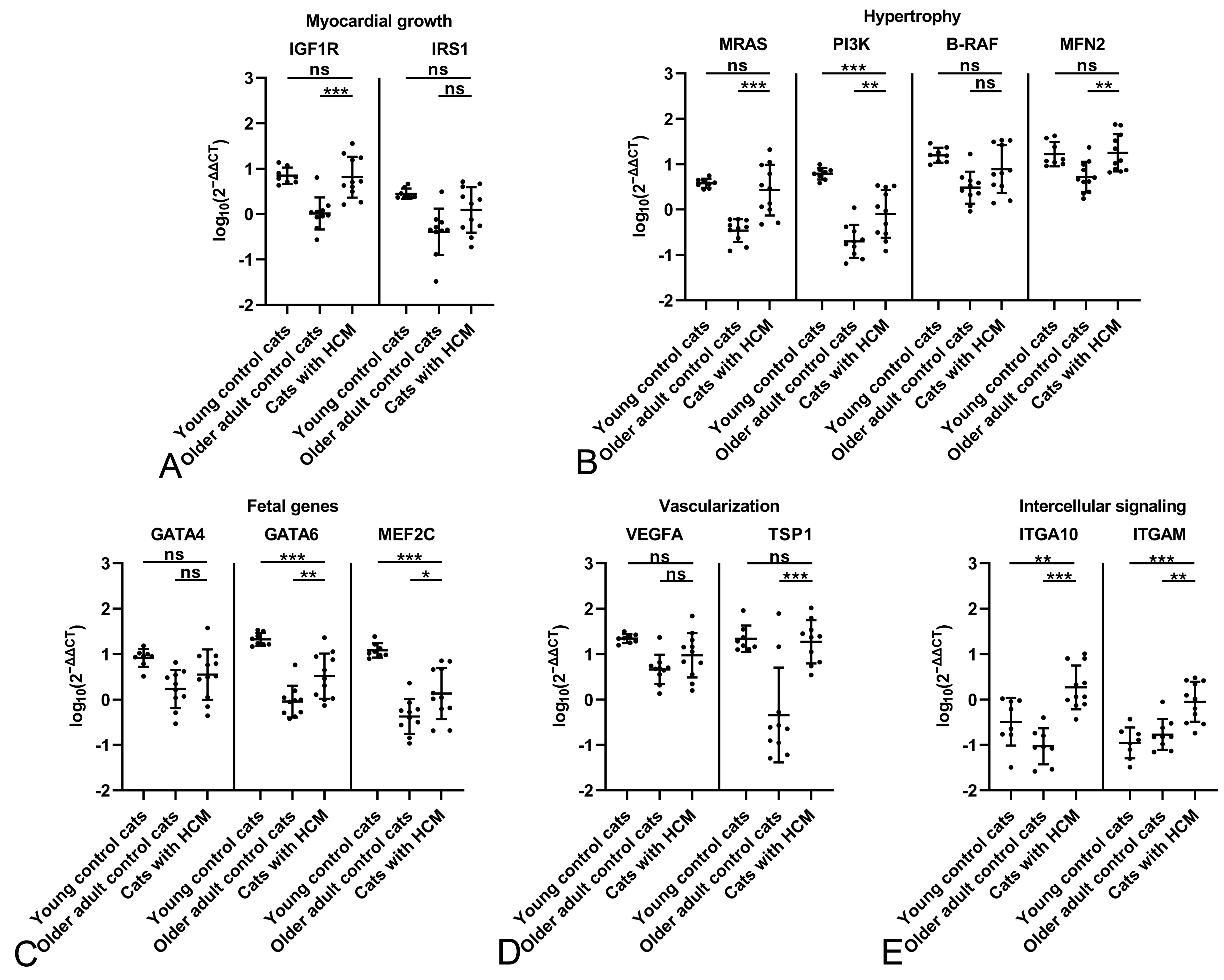
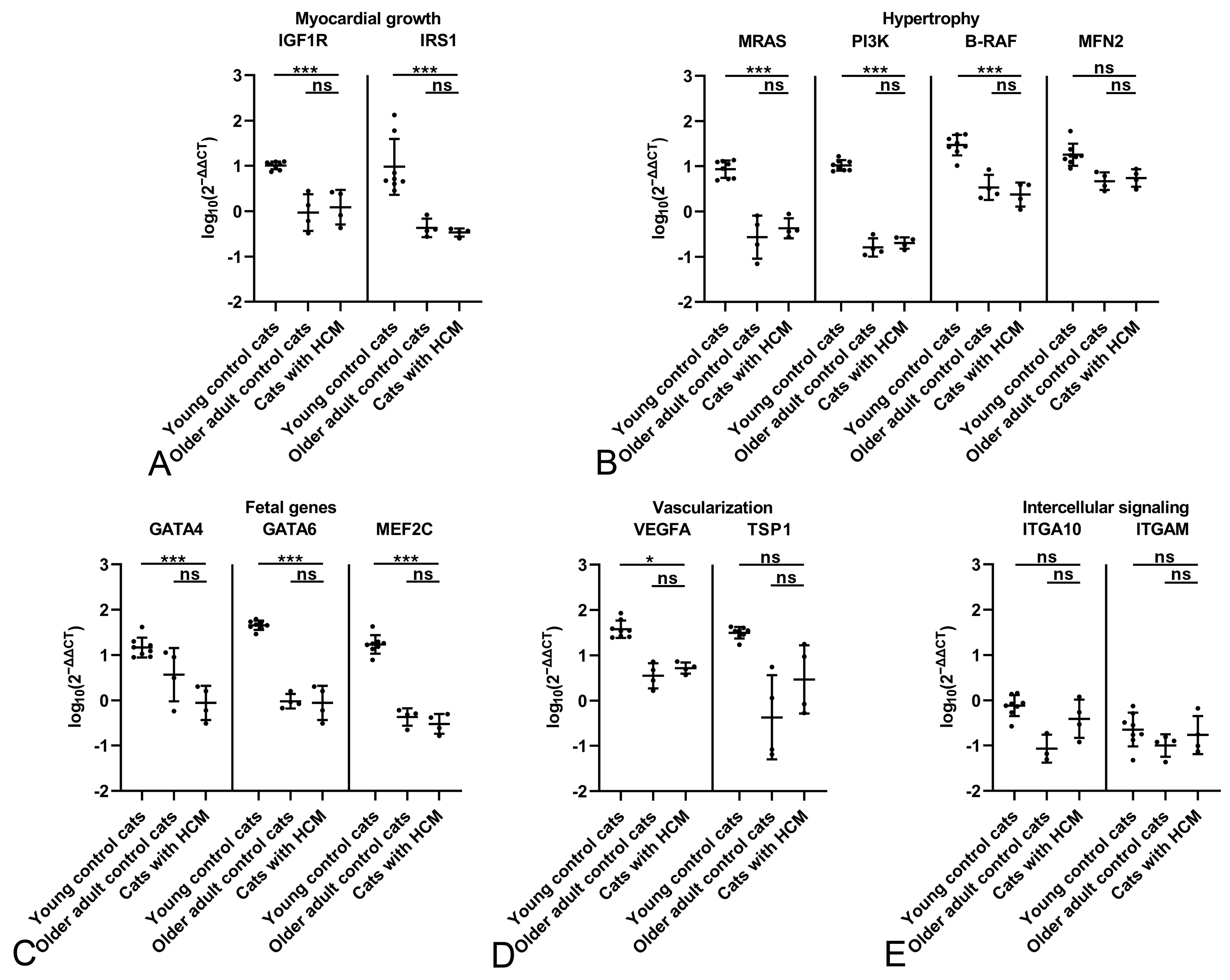
| Functional Group | Marker | Characteristics and Function |
|---|---|---|
| Myocardial growth | IGF1R | Transmembrane receptor. Activates PI3K pathway and MAPK pathway [33], thereby promoting cell proliferation, differentiation, survival and apoptosis [34,35,36]. |
| IRS1 | Signaling protein. Activated by INSR and IGF1R. In addition to effects of IGF1R, insulin signaling is involved in several cardiac regulatory processes, such as cardiomyocyte growth, contractility, apoptosis and increased myocardial perfusion through vasodilation [37,38,39]. | |
| Hypertrophy | MRAS | GTPase. Activates PI3K and the MAPK signaling pathways, thereby regulating a variety of cell functions (proliferation, differentiation, survival and apoptosis), including the activation of transcription factors involved in the hypertrophic gene program, such as MEF2c and GATA4, and leading to fetal cardiac gene expression [40,41,42]. |
| PI3K | Protein kinase. Intracellular signaling protein involved in the PI3K/AKT/mTOR signaling pathway, thus regulating cell growth, proliferation and metabolism [43,44]; did cause cardiac and myocyte hypertrophy in mice when overexpressed [44,45]. | |
| B-RAF | Serine/threonine-specific protein kinase. Activation of ERK1/2. Facilitates cardiac adaptation to wall stress and promotes compensatory hypertrophy of cardiomyocytes [46,47]. | |
| MFN2 | Transmembrane GTPase, located in the outer mitochondrial membrane. Contributes to mitochondrial dynamics (fusion, fission) [48,49,50]. Essential for embryonic and postnatal heart development and maintenance of cardiac function [51,52,53,54]. | |
| Fetal genes | MEF2C | Transcription factor. Signaling can result in cardiomyocyte hypertrophy accompanied by sarcomeric derangement and elongation [40,55,56,57]. |
| GATA4 | Transcription factor. Crucial for cardiac morphogenesis by regulating cardiac protein expression [58]. Overexpression preserves cardiac function after cardiac injury [59]. | |
| GATA6 | Transcription factor. Expressed in both the developing and adult heart, regulates the same hypertrophic response as GATA4 [58,60], while also playing an important role in migration, differentiation, and modulation of vascular smooth muscle cells (VSMCs) [61,62]. | |
| Vascularization | VEGFA | Signaling protein and angiogenic factor. Promotes cardiac stem cell migration via the PI3K/Akt signaling pathway [63]. |
| TSP1 | Matricellular protein. Antagonizes VEGFA and activates latent TGF-β which stimulates extracellular matrix deposition, angiogenesis and recruitment of inflammatory cells [64,65,66]. Increased expression in the myocardium of aging mice suggests contribution to age-related cardiac fibrosis [67]. | |
| Intracellular signaling | ITGAM | Cell surface receptor located on leukocytes. Potential contribution to the development of cardiac fibrosis through recruitment of fibroblast-activating macrophages [68,69]. |
| ITGA10 | Cell surface receptor on cardiomyocytes. Binds fibrillar and non-fibrillar collagens. Can trigger force-dependent cellular responses, including cell growth and myofibroblast differentiation. Critical for maintaining cardiomyocyte membrane integrity and subsequent protection against dysfunction, death, and fibrosis [70,71]. |
| Gene | NCBI Accession No | Location | Sequence (5′-3′) | PCR Product Length (bp) |
|---|---|---|---|---|
| GAPDH | XM_006933438.4 | F: 57 | F: GCCGTGGAATTTGCCGT | 82 |
| R: 138 | R: GCCATCAATGACCCCTTCAT | |||
| P: 77 [90] | P: CTCAACTACATGGTCTACATGTT-CCAGTATGATTCCA | |||
| IGF1R | XM_023254968 | F: 975 | F: GCGTCTCCGAAATTTACCGC | 111 |
| R: 1085 | R: CGTCACTTTCACAGGAGGCT | |||
| P: 1015 | P: TAAGGGGCGCCAGAGCAAAG | |||
| IRS1 | XM_003991219.4 | F: 4179 | F: GCGAGGATTTAAGCGCCTATG | 99 |
| R: 4277 | R: CATTCAGGTCTTCATTCTGCTGT | |||
| P: 4207 | P: CAGTTTCCAGAAGCAGCCAGA | |||
| MRAS | XM_019810658.3 | F: 409 | F: CGGCTTCCTCATCGTCTACTC | 104 |
| R: 512 | R: TCGGAAATGACTCTCTGTCCT | |||
| P: 470 | P: CACCAGCTCATCCTGCGCGT | |||
| PI3K | XM_019840086.3 | F: 3818 | F: AGACGACTTTGTGACCTGCG | 111 |
| R: 3928 | R: CATGCCAATAGCAAAACCAATTTCT | |||
| P: 3865 | P: TGAGCCAGTAGGCAACCGTGA | |||
| B-RAF | XM_019825910.3 | F: 1456 | F: TTGGATCCGGGTCATTTGGG | 147 |
| R: 1602 | R: TCACATGCCGAGTTTTCCTGA | |||
| P: 1530 | P: TGACAGCACCCACACCTCAGC | |||
| MFN2 | XM_019836178.3 | F: 1008 | R: AAGAAGATGCGGTCTCCAGC | 104 |
| R: 1111 | P: CTGGGCGTGGTGGACCGAGGC | |||
| P: 1068 | R: AAGAAGATGCGGTCTCCAGC | |||
| GATA4 | XM_011281564.4 | F: 1771 | F: CATGGCCCAGACGTTCTCA | 119 |
| R: 1890 | R: TGTGGTGACTGGCTGACAGAAG | |||
| P:1835 | P: TCGGCCCTGAAGCTCTCCCC | |||
| GATA6 | XM_023241809.2 | F: 1629 | F: AGTCAAAAGCTTGCTCTGGTAG | 119 |
| R: 1747 | R: TGAGGATGTCGGTTGTGTTGT | |||
| P: 1661 | P: TCTGTTCCTATGACTCCAACTTC | |||
| MEF2c | XM_019836585.2 | F: 501 | F: ATCTTCAACAGCACCAACAAGCT | 155 |
| R: 632 | R: TTCTCAACGTCTCCACGATGTC | |||
| P: 567 | P: TACACGGAGTACAACGAGCCGCAC | |||
| VEGFA | XM_045057004.1 | F: 1047 | F: CTTCAAGCCATCCTGCGTG | 113 |
| R: 1159 | R: TGATCCGCATAATCTGCATGG | |||
| P: 1102 | P: CGAGGGCCTGGAGTGTGTGC | |||
| TSP1 | XM_011283077.4 | F: 2124 | F: CGCCAACAAACAGGTATGCAA | 118 |
| R: 2241 | R: CTCACAGCGGTACATGGGG | |||
| P: 2147 | P: CCCGCAACCCCTGCACAGAT | |||
| ITGAM | XM_006942117.4 | F: 938 | F: TCGCTATGTCATTGGGGTGG | 102 |
| R: 1039 | R: ACCCGGAACACATAATCACGA | |||
| P: 993 | P: CTTAATACCATTGCATCAAAGCCT | |||
| ITGA10 | XM_019837750.3 | F: 3546 | F: AGCAACACTCGGTGTCAAGT | 122 |
| R: 3667 | R: GACTTGAATTTGGCCCTCCG | |||
| P: 3575 | P: CCATCCTTGGGCGGCTGGCAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schurna, L.; Joshua, J.; Monné Rodríguez, J.; Prisco, F.; Baron Toaldo, M.; De Neck, S.; Baggio, F.; Fonfara, S.; Kipar, A. Selected Pathway Analyses to Gain Mechanistic Insights into the Pathogenesis of Feline Hypertrophic Cardiomyopathy. Int. J. Mol. Sci. 2025, 26, 6497. https://doi.org/10.3390/ijms26136497
Schurna L, Joshua J, Monné Rodríguez J, Prisco F, Baron Toaldo M, De Neck S, Baggio F, Fonfara S, Kipar A. Selected Pathway Analyses to Gain Mechanistic Insights into the Pathogenesis of Feline Hypertrophic Cardiomyopathy. International Journal of Molecular Sciences. 2025; 26(13):6497. https://doi.org/10.3390/ijms26136497
Chicago/Turabian StyleSchurna, Lea, Jessica Joshua, Josep Monné Rodríguez, Francesco Prisco, Marco Baron Toaldo, Simon De Neck, Francesca Baggio, Sonja Fonfara, and Anja Kipar. 2025. "Selected Pathway Analyses to Gain Mechanistic Insights into the Pathogenesis of Feline Hypertrophic Cardiomyopathy" International Journal of Molecular Sciences 26, no. 13: 6497. https://doi.org/10.3390/ijms26136497
APA StyleSchurna, L., Joshua, J., Monné Rodríguez, J., Prisco, F., Baron Toaldo, M., De Neck, S., Baggio, F., Fonfara, S., & Kipar, A. (2025). Selected Pathway Analyses to Gain Mechanistic Insights into the Pathogenesis of Feline Hypertrophic Cardiomyopathy. International Journal of Molecular Sciences, 26(13), 6497. https://doi.org/10.3390/ijms26136497






