Salinity Stress in Rice: Multilayered Approaches for Sustainable Tolerance
Abstract
1. Introduction
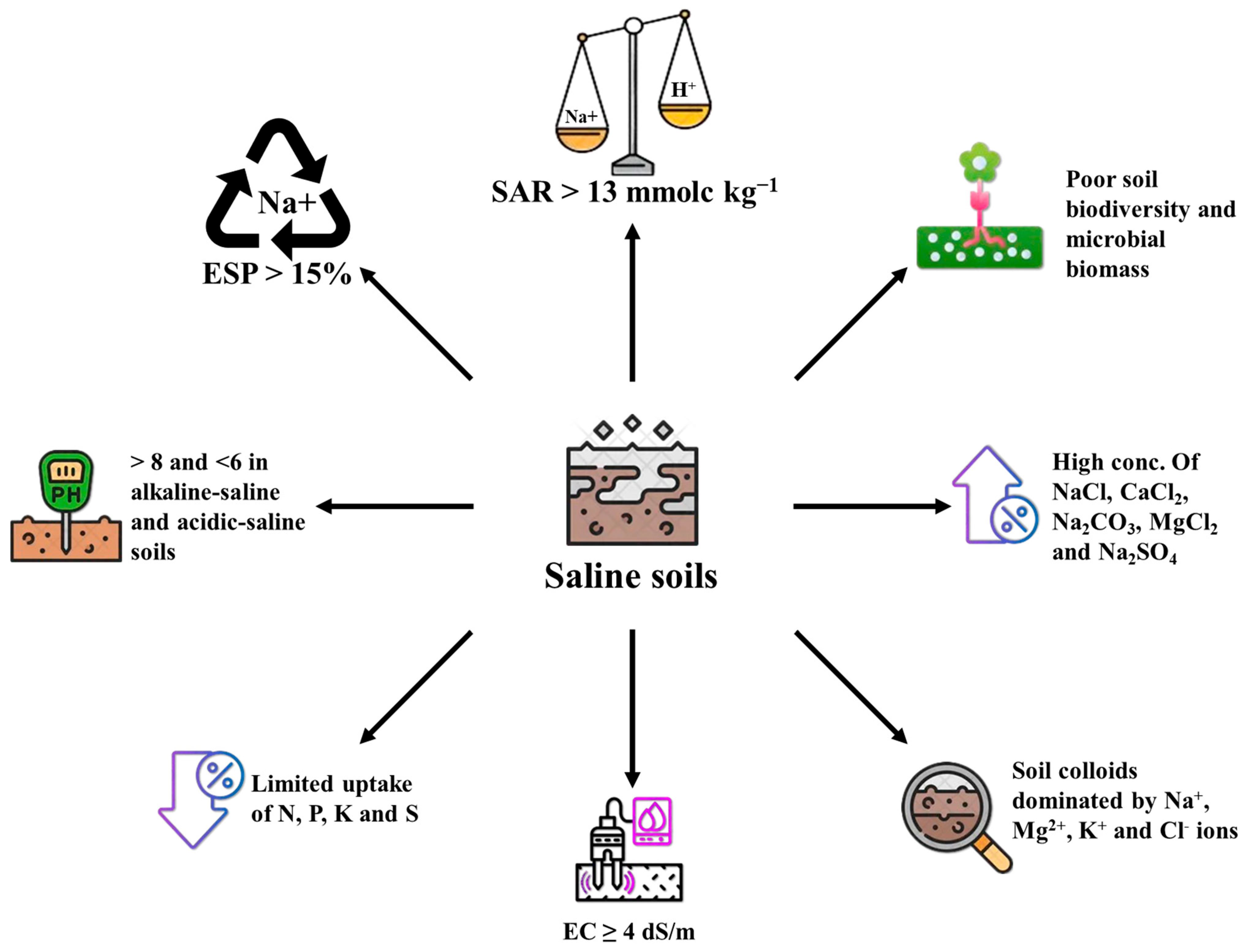
2. Impact of Salinity on Soil Characteristics and Productivity
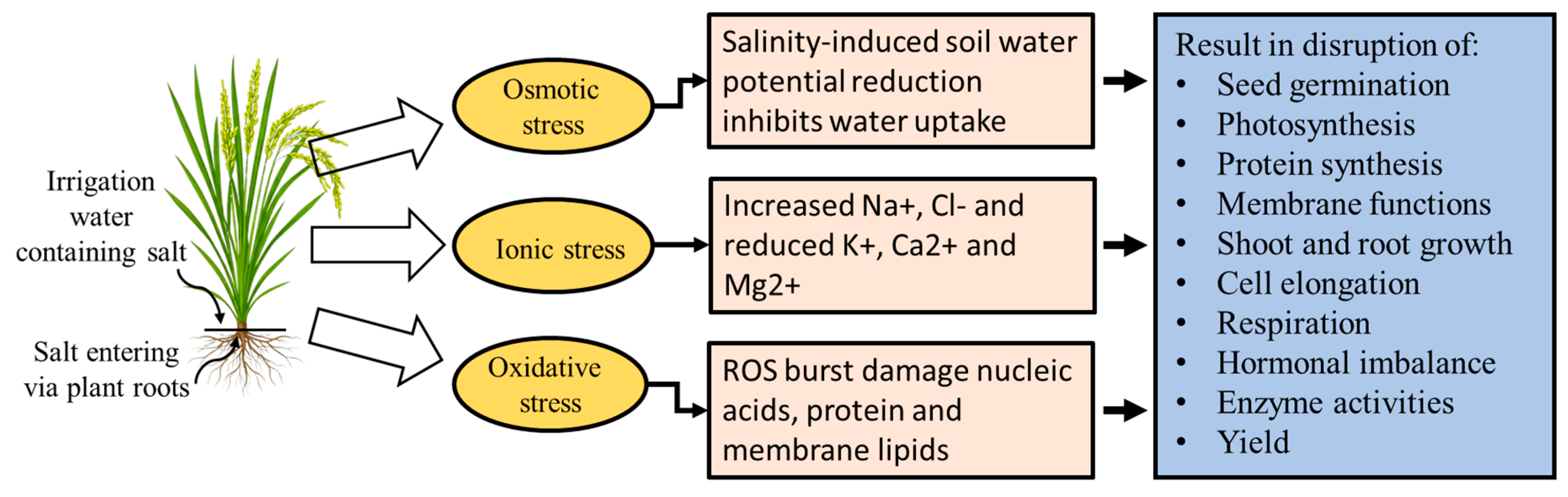
3. Effects of Salt Stress on Rice
3.1. Osmotic Stress Mechanism in Rice Under Salinity
3.2. Mechanism of Ionic Imbalance (Ion Toxicity) in Rice Under Salt Stress
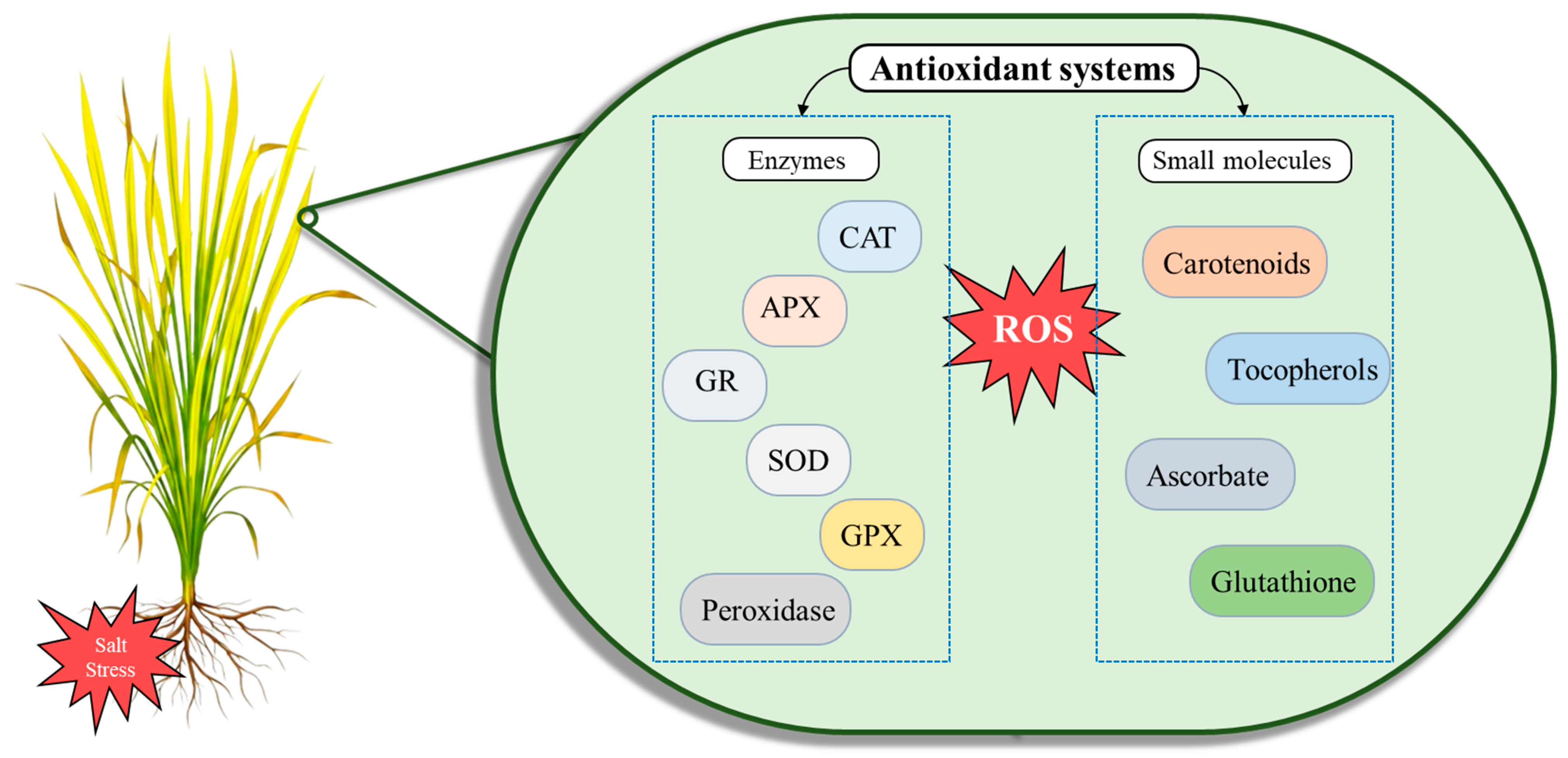
3.3. Mechanism of Oxidative Damage Under Salt Stress in Rice
3.4. Impact of Salt Stress on Rice Quality
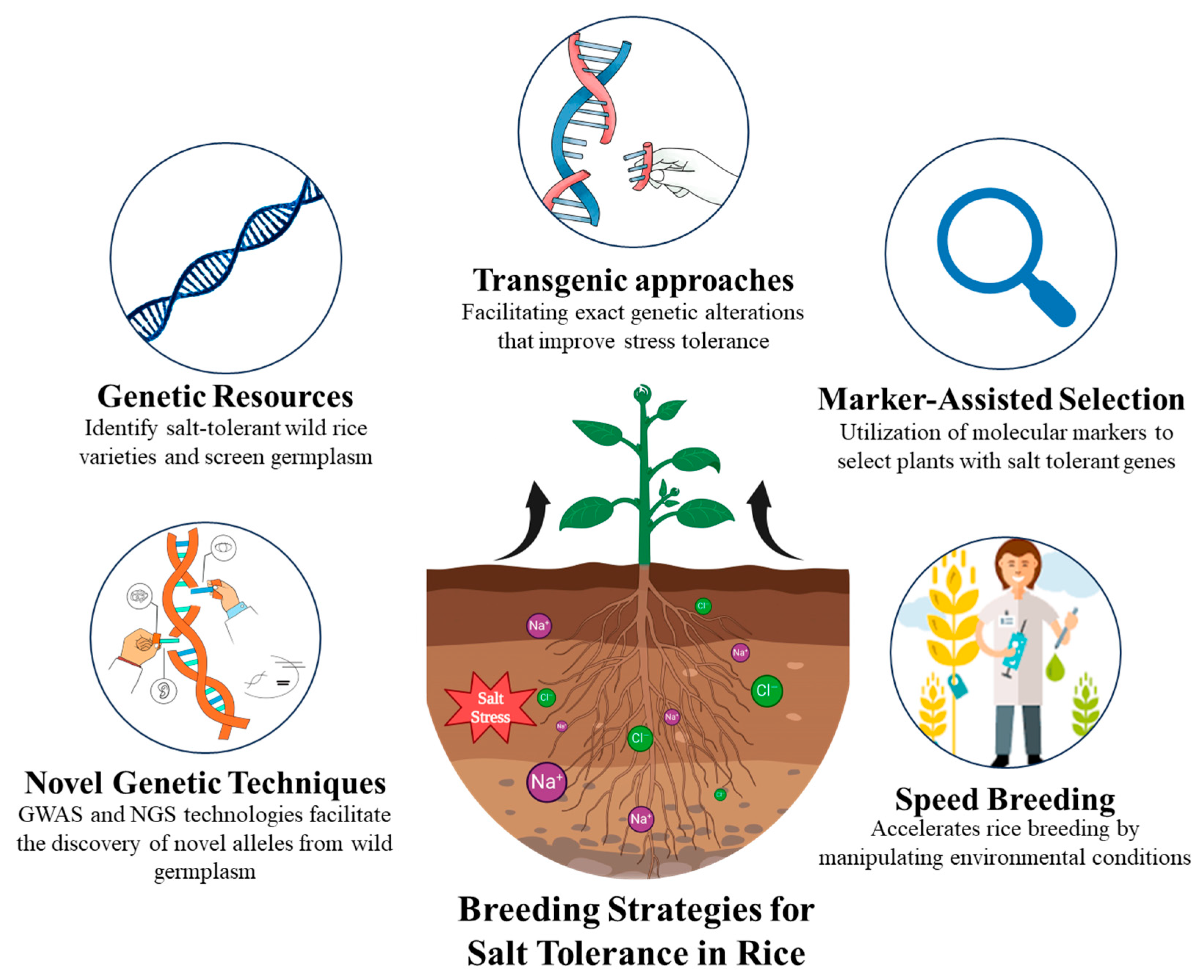
4. Key Sustainable Strategies for Rice Health in Saline Conditions
4.1. Genetic Resources and Breeding Strategies
4.1.1. Utilization of Wild Relatives and Novel Genetic Resources
4.1.2. Marker-Assisted Selection and Speed-Breeding
4.1.3. Transgenic Approaches
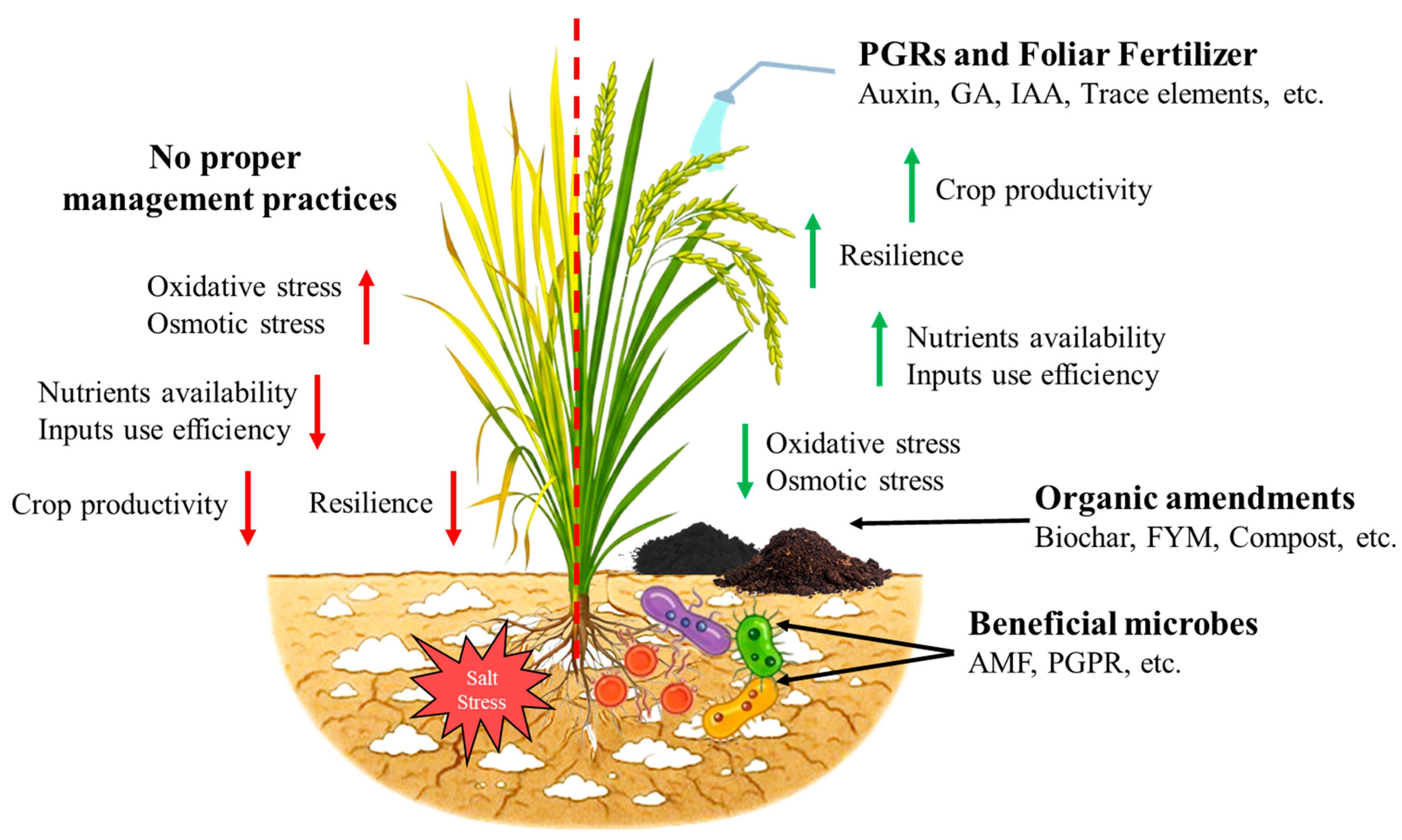
4.2. Management Strategies for Salt Tolerance in Rice
4.2.1. Agronomic Approaches
4.2.2. Organic Amendments and Fertilization
4.2.3. Symbiotic Microbes Enhancing Salt Tolerance in Rice
4.2.4. Plant Growth Regulators (PGRs)
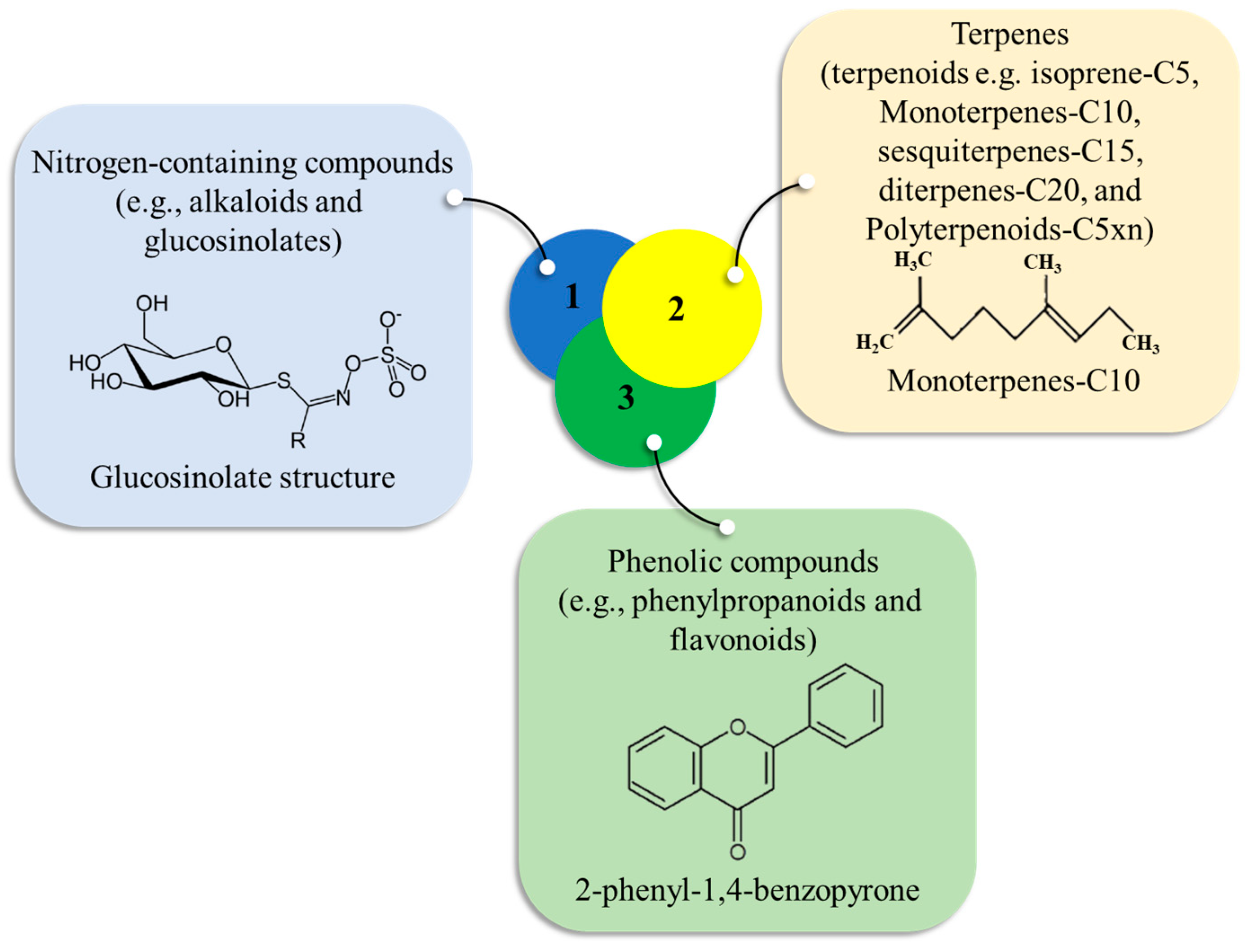
4.3. Primary and Secondary Metabolites in Salt-Stressed Rice
4.4. Modification of Plant Antioxidant Pathways in Salt-Stressed Rice
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef]
- Kivi, S.T.; Bailey, R.T. Modeling sulfur cycling and sulfate reactive transport in an agricultural groundwater system. Agric. Water Manag. 2017, 185, 78–92. [Google Scholar] [CrossRef]
- Bello, S.K. An overview of the morphological, genetic and metabolic mechanisms regulating phosphorus efficiency via root traits in soybean. J. Soil Sci. Plant Nutr. 2021, 21, 1013–1029. [Google Scholar] [CrossRef]
- Wani, S.H.; Kumar, V.; Khare, T.; Guddimalli, R.; Parveda, M.; Solymosi, K.; Suprasanna, P.; Kavi Kishor, P. Engineering salinity tolerance in plants: Progress and prospects. Planta 2020, 251, 76. [Google Scholar]
- Elmeknassi, M.; Elghali, A.; de Carvalho, H.W.P.; Laamrani, A.; Benzaazoua, M. A review of organic and inorganic amendments to treat saline-sodic soils: Emphasis on waste valorization for a circular economy approach. Sci. Total Environ. 2024, 921, 171087. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Bello, S.K.; Alayafi, A.H.; Al-Solaimani, S.G.; Abo-Elyousr, K.A. Mitigating soil salinity stress with gypsum and bio-organic amendments: A review. Agronomy 2021, 11, 1735. [Google Scholar] [CrossRef]
- Oelviani, R.; Adiyoga, W.; Suhendrata, T.; Bakti, I.G.M.Y.; Sutanto, H.A.; Fahmi, D.A.; Chanifah, C.; Jatuningtyas, R.K.; Samijan, S.; Malik, A. Effects of soil salinity on rice production and technical efficiency: Evidence from the northern coastal region of Central Java, Indonesia. Case Stud. Chem. Environ. Eng. 2024, 10, 101010. [Google Scholar] [CrossRef]
- Ahmed, M.F.; Haider, M.Z. Impact of salinity on rice production in the south-west region of Bangladesh. Environ. Sci. 2014, 9, 135–141. [Google Scholar]
- Bayoumy, M.; Khalifa, T.; Aboelsoud, H. Impact of some organic and inorganic amendments on some soil properties and wheat production under saline-sodic soil. J. Soil Sci. Agric. Eng. 2019, 10, 307–313. [Google Scholar] [CrossRef]
- Tang, S.; She, D.; Wang, H. Effect of salinity on soil structure and soil hydraulic characteristics. Can. J. Soil Sci. 2020, 101, 62–73. [Google Scholar] [CrossRef]
- Osman, K.T. Management of Soil Problems; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Brady, N. The Nature and Properties of Soils; Prentice Hall: Upper Saddle River, NJ, USA, 1984. [Google Scholar]
- El-Ramady, H.; Prokisch, J.; Mansour, H.; Bayoumi, Y.A.; Shalaby, T.A.; Veres, S.; Brevik, E.C. Review of Crop Response to Soil Salinity Stress: Possible Approaches from Leaching to Nano-Management. Soil Syst. 2024. [Google Scholar]
- Hardie, M.; Doyle, R. Measuring soil salinity. In Plant Salt Tolerance: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2012; pp. 415–425. [Google Scholar]
- Mishra, A.K.; Das, R.; George Kerry, R.; Biswal, B.; Sinha, T.; Sharma, S.; Arora, P.; Kumar, M. Promising management strategies to improve crop sustainability and to amend soil salinity. Front. Environ. Sci. 2023, 10, 962581. [Google Scholar] [CrossRef]
- Maksimovic, I.; Ilin, Z. Effects of salinity on vegetable growth and nutrients uptake. Irrig. Syst. Pract. Challenging Environ. 2012, 9, 169–190. [Google Scholar]
- Tan, S.; Su, X.; Jiang, X.; Yao, W.; Chen, S.; Yang, Q.; Ning, S. Irrigation Salinity Affects Water Infiltration and Hydraulic Parameters of Red Soil. Agronomy 2023, 13, 2627. [Google Scholar] [CrossRef]
- Yan, S.; Zhang, T.; Zhang, B.; Zhang, T.; Cheng, Y.; Wang, C.; Luo, M.; Feng, H.; Siddique, K. The higher relative concentration of K+ to Na+ in saline water improves soil hydraulic conductivity, salt-leaching efficiency and structural stability. Soil 2023, 9, 339–349. [Google Scholar] [CrossRef]
- Libohova, Z.; Seybold, C.; Wysocki, D.; Wills, S.; Schoeneberger, P.; Williams, C.; Lindbo, D.; Stott, D.; Owens, P. Reevaluating the effects of soil organic matter and other properties on available water-holding capacity using the National Cooperative Soil Survey Characterization Database. J. Soil Water Conserv. 2018, 73, 411–421. [Google Scholar] [CrossRef]
- Gonçalo Filho, F.; da Silva Dias, N.; Suddarth, S.R.P.; Ferreira, J.F.; Anderson, R.G.; dos Santos Fernandes, C.; de Lira, R.B.; Neto, M.F.; Cosme, C.R. Reclaiming tropical saline-sodic soils with gypsum and cow manure. Water 2019, 12, 57. [Google Scholar] [CrossRef]
- Xie, Y.; Ning, H.; Zhang, X.; Zhou, W.; Xu, P.; Song, Y.; Li, N.; Wang, X.; Liu, H. Reducing the Sodium Adsorption Ratio Improves the Soil Aggregates and Organic Matter in Brackish-Water-Irrigated Cotton Fields. Agronomy 2024, 9, 2169. [Google Scholar] [CrossRef]
- Haj-Amor, Z.; Araya, T.; Kim, D.G.; Bouri, S.; Lee, J.; Ghiloufi, W.; Yang, Y.; Kang, H.; Jhariya, M.K.; Banerjee, A.; et al. Soil salinity and its associated effects on soil microorganisms, greenhouse gas emissions, crop yield, biodiversity and desertification: A review. Sci. Total Environ. 2022, 843, 156946. [Google Scholar] [CrossRef]
- Zhang, W.-W.; Chong, W.; Rui, X.; Wang, L.-j. Effects of salinity on the soil microbial community and soil fertility. J. Integr. Agric. 2019, 18, 1360–1368. [Google Scholar] [CrossRef]
- Sharma, S.; Gupta, N.; Chakkal, A.S.; Sharma, N.; Alamri, S.; Siddiqui, M.H.; Haider, F.U. Changes in Enzyme Activities in Salt-Affected Soils during Incubation Study of Diverse Particle Sizes of Rice Straw. Agriculture 2023, 13, 1694. [Google Scholar] [CrossRef]
- Wichern, F.; Islam, M.R.; Hemkemeyer, M.; Watson, C.; Joergensen, R.G. Organic amendments alleviate salinity effects on soil microorganisms and mineralisation processes in aerobic and anaerobic paddy rice soils. Front. Sustain. Food Syst. 2020, 4, 30. [Google Scholar] [CrossRef]
- Abhayawickrama, B.; Gimhani, D.; Kottearachchi, N.; Herath, V.; Liyanage, D.; Senadheera, P. In silico identification of QTL-based polymorphic genes as salt-responsive potential candidates through mapping with two reference genomes in rice. Plants 2020, 9, 233. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chu, L.; Lu, H.; Qi, W.; Chen, X.; Liu, J.; Kuang, S.; Tang, B.; Wong, V. Towards sustainable agriculture for the salt-affected soil. Land Degrad. Dev. 2019, 30, 574–579. [Google Scholar] [CrossRef]
- Rengasamy, P. Soil processes affecting crop production in salt-affected soils. Funct. Plant Biol. 2010, 37, 613–620. [Google Scholar] [CrossRef]
- Cheeseman, J.M. The evolution of halophytes, glycophytes and crops, and its implications for food security under saline conditions. New Phytol. 2015, 206, 557–570. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef]
- Razzaq, A.; Ali, A.; Safdar, L.B.; Zafar, M.M.; Rui, Y.; Shakeel, A.; Shaukat, A.; Ashraf, M.; Gong, W.; Yuan, Y. Salt stress induces physiochemical alterations in rice grain composition and quality. J. Food Sci. 2020, 85, 14–20. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Rahman, A.; Alam, M.; Hossain, M.; Mahmud, J.; Nahar, K.; Fujita, M.; Hasanuzzaman, M. Exogenous Gallic Acid Confers Salt Tolerance in Rice Seedlings: Modulation of Ion Homeostasis, Osmoregulation, Antioxidant Defense, and Methylglyoxal Detoxification Systems. Agronomy 2022, 13, 16. [Google Scholar] [CrossRef]
- Qin, H.; Huang, R. The phytohormonal regulation of Na+/K+ and reactive oxygen species homeostasis in rice salt response. Mol. Breed. 2020, 40, 47. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.-K.; Shabala, S. Mechanisms of Plant Responses and Adaptation to Soil Salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef] [PubMed]
- Hakim, M.; Juraimi, A.; Hanafi, M.; Ismail, M.; Rafii, M.; Islam, M.; Selamat, A. The effect of salinity on growth, ion accumulation and yield of rice varieties. J. Anim. Plant Sci. 2014, 24, 874–885. [Google Scholar]
- Byrt, C.S.; Munns, R.; Burton, R.A.; Gilliham, M.; Wege, S. Root cell wall solutions for crop plants in saline soils. Plant Sci. 2018, 269, 47–55. [Google Scholar] [CrossRef]
- Xu, Y.; Bu, W.; Xu, Y.; Fei, H.; Zhu, Y.; Ahmad, I.; Nimir, N.E.A.; Zhou, G.; Zhu, G. Effects of Salt Stress on Physiological and Agronomic Traits of Rice Genotypes with Contrasting Salt Tolerance. Plants 2024, 13, 1157. [Google Scholar] [CrossRef]
- Koc, Y.E.; Aycan, M.; Mitsui, T. Self-Defense Mechanism in Rice to Salinity: Proline. J 2024, 7, 103–115. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, P.; Yu, X.; Xu, J.; Liu, G. Physiological and Molecular Mechanisms of Rice Tolerance to Salt and Drought Stress: Advances and Future Directions. Int. J. Mol. Sci. 2024, 25, 9404. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, X.; Giraldo, J.P.; Shabala, S. It is not all about sodium: Revealing tissue specificity and signalling roles of potassium in plant responses to salt stress. Plant Soil 2018, 431, 1–17. [Google Scholar] [CrossRef]
- Demidchik, V. Mechanisms and physiological roles of K+ efflux from root cells. J. Plant Physiol. 2014, 171, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Bazihizina, N.; Colmer, T.D.; Cuin, T.A.; Mancuso, S.; Shabala, S. Friend or foe? Chloride patterning in halophytes. Trends Plant Sci. 2019, 24, 142–151. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef] [PubMed]
- Hongqiao, L.; Suyama, A.; Mitani-Ueno, N.; Hell, R.; Maruyama-Nakashita, A. A Low Level of NaCl Stimulates Plant Growth by Improving Carbon and Sulfur Assimilation in Arabidopsis thaliana. Plants 2021, 10, 2138. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Tomar, N.S.; Tittal, M.; Argal, S.; Agarwal, R. Plant growth under water/salt stress: ROS production; antioxidants and significance of added potassium under such conditions. Physiol. Mol. Biol. Plants 2017, 23, 731–744. [Google Scholar] [CrossRef]
- Khare, T.; Seth, C.S.; Kumar, V. Sodium stress-induced oxidative damage and antioxidant responses during grain filling in Indica rice. Plant Cell Rep. 2024, 43, 239. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Raihan, M.R.H.; Masud, A.A.C.; Rahman, K.; Nowroz, F.; Rahman, M.; Nahar, K.; Fujita, M. Regulation of Reactive Oxygen Species and Antioxidant Defense in Plants under Salinity. Int. J. Mol. Sci. 2021, 22, 9326. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, L.M.; Feng, N.; Zheng, D.; Shen, X.F.; Zhou, H.; Jiang, W.; Du, Y.; Zhao, H.; Lu, X.; et al. Plant growth regulators mitigate oxidative damage to rice seedling roots by NaCl stress. PeerJ 2024, 12, e17068. [Google Scholar] [CrossRef]
- Fang, X.; Mo, J.; Zhou, H.; Shen, X.; Xie, Y.; Xu, J.; Yang, S. Comparative transcriptome analysis of gene responses of salt-tolerant and salt-sensitive rice cultivars to salt stress. Sci. Rep. 2023, 13, 19065. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Niu, S.; Yan, Y.; Zhou, G.; Peng, Y.; He, Y.; Zhou, J.; Li, Y.; Xie, X. Moderate Salinity Stress Affects Rice Quality by Influencing Expression of Amylose- and Protein-Content-Associated Genes. Int. J. Mol. Sci. 2024, 25, 4042. [Google Scholar] [CrossRef] [PubMed]
- Cui, R.; Zhou, T.; Shu, C.; Zhu, K.; Ye, M.; Zhang, W.; Zhang, H.; Liu, L.; Wang, Z.; Gu, J.; et al. Effects of Salt Stress on Grain Quality and Starch Properties of High-Quality Rice Cultivars. Agronomy 2024, 14, 444. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Hussain, S.; Yang, S.; Li, R.; Liu, S.; Chen, Y.; Wei, H.; Dai, Q.; Hou, H. Study on the Effect of Salt Stress on Yield and Grain Quality Among Different Rice Varieties. Front. Plant Sci. 2022, 13, 918460. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, T.; Zhu, K.; Wang, W.; Zhang, W.; Zhang, H.; Liu, L.; Zhang, Z.; Wang, Z.; Wang, B.; et al. Effects of Salt Stress on Grain Yield and Quality Parameters in Rice Cultivars with Differing Salt Tolerance. Plants 2023, 12, 3243. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Khan, A.L.; Muneer, S.; Kim, Y.-H.; Al-Rawahi, A.; Al-Harrasi, A. Silicon and salinity: Crosstalk in crop-mediated stress tolerance mechanisms. Front. Plant Sci. 2019, 10, 1429. [Google Scholar]
- Safdar, H.; Amin, A.; Shafiq, Y.; Ali, A.; Yasin, R.; Shoukat, A.; Hussan, M.U.; Sarwar, M.I. A review: Impact of salinity on plant growth. Nat. Sci 2019, 17, 34–40. [Google Scholar]
- Padmavathi, G.; Bangale, U.; Rao, K.N.; Balakrishnan, D.; Arun, M.N.; Singh, R.K.; Sundaram, R.M. Progress and prospects in harnessing wild relatives for genetic enhancement of salt tolerance in rice. Front. Plant Sci. 2024, 14, 1253726. [Google Scholar] [CrossRef]
- Quan, R.; Wang, J.; Hui, J.; Bai, H.; Lyu, X.; Zhu, Y.; Zhang, H.; Zhang, Z.; Li, S.; Huang, R. Improvement of Salt Tolerance Using Wild Rice Genes. Front. Plant Sci. 2018, 8, 2269. [Google Scholar] [CrossRef]
- Hellwig, T.; Abbo, S.; Sherman, A.; Ophir, R. Prospects for the natural distribution of crop wild-relatives with limited adaptability: The case of the wild pea Pisum fulvum. Plant Sci. 2021, 310, 110957. [Google Scholar] [CrossRef]
- Nurbekova, Z.; Satkanov, M.; Beisekova, M.; Akbassova, A.; Ualiyeva, R.; Cui, J.; Chen, Y.; Wang, Z.; Zhangazin, S. Strategies for Achieving High and Sustainable Plant Productivity in Saline Soil Conditions. Horticulturae 2024, 10, 878. [Google Scholar] [CrossRef]
- Reddy, I.N.B.L.; Kim, B.-K.; Yoon, I.-S.; Kim, K.-H.; Kwon, T.-R. Salt Tolerance in Rice: Focus on Mechanisms and Approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Rana, M.; Takamatsu, T.; Baslam, M.; Kaneko, K.; Itoh, K.; Harada, N.; Sugiyama, T.; Ohnishi, T.; Kinoshita, T.; Takagi, H.; et al. Salt Tolerance Improvement in Rice through Efficient SNP Marker-Assisted Selection Coupled with Speed-Breeding. Int. J. Mol. Sci. 2019, 20, 2585. [Google Scholar] [CrossRef]
- Saminadane, T.; Geddam, S.; Krishnaswamy, P.; Jothiganapathy, K.; Tamilselvan, A.; Ramadoss, B.R.; Sri Hari Reddy, P.; Singh, U.S.; Singh, R.K.; Platten, J.D.; et al. Development of early maturing salt-tolerant rice variety KKL(R) 3 using a combination of conventional and molecular breeding approaches. Front. Genet. 2024, 14, 1332691. [Google Scholar] [CrossRef] [PubMed]
- Ćeran, M.; Miladinović, D.; Đorđević, V.; Trkulja, D.; Radanović, A.; Glogovac, S.; Kondić-Špika, A. Genomics-assisted speed breeding for crop improvement: Present and future. Front. Sustain. Food Syst. 2024, 8, 1383302. [Google Scholar] [CrossRef]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Fujii-Kashino, M.; Horiuchi, T. OsHKT1; 5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef]
- Biradar, H. Overexpression of a Plasma Membrane Protein Gene, SaPMP3, from Spartina alterniflora L. Enhances Salinity Tolerance in Rice (Oryza sativa L.); Louisiana State University and Agricultural & Mechanical College: Baton Rouge, LA, USA, 2012. [Google Scholar]
- Biradar, H.; Karan, R.; Subudhi, P.K. Transgene pyramiding of salt responsive protein 3-1 (SaSRP3-1) and SaVHAc1 from Spartina alterniflora L. enhances salt tolerance in rice. Front. Plant Sci. 2018, 9, 1304. [Google Scholar] [CrossRef]
- Gahlaut, V.; Jaiswal, V.; Kumar, S. Role of small RNA and RNAi technology toward improvement of abiotic stress tolerance in plants. In CRISPR and RNAi Systems; Elsevier: Amsterdam, The Netherlands, 2021; pp. 491–507. [Google Scholar]
- Junedi, M.A.; Mukhopadhyay, R.; Manjari, K.S. Alleviating salinity stress in crop plants using new engineered nanoparticles (ENPs). Plant Stress 2023, 9, 100184. [Google Scholar] [CrossRef]
- Yin, W.; Xiao, Y.; Niu, M.; Meng, W.; Li, L.; Zhang, X.; Liu, D.; Zhang, G.; Qian, Y.; Sun, Z. ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell 2020, 32, 2292–2306. [Google Scholar] [CrossRef]
- Santosh Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Hu, J.; Dong, L.; Zeng, D.; Guo, L.; Zhang, G.; Zhu, L.; Qian, Q. The tolerance of salinity in rice requires the presence of a functional copy of FLN2. Biomolecules 2019, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, D.; Shan, T.; Xu, S.; Qin, R.; Li, H.; Negm, M.; Wu, D.; Li, J. The trihelix transcription factor OsGTγ-2 is involved adaption to salt stress in rice. Plant Mol. Biol. 2020, 103, 545–560. [Google Scholar] [CrossRef]
- Mo, W.; Tang, W.; Du, Y.; Jing, Y.; Bu, Q.; Lin, R. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 interaction controls seedling growth under salt stress. Plant Physiol. 2020, 184, 506–517. [Google Scholar] [CrossRef]
- Alfatih, A.; Wu, J.; Jan, S.U.; Zhang, Z.S.; Xia, J.Q.; Xiang, C.B. Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant Cell Environ. 2020, 43, 2743–2754. [Google Scholar] [CrossRef]
- Wang, W.-C.; Lin, T.-C.; Kieber, J.; Tsai, Y.-C. Response Regulators 9 and 10 Negatively Regulate Salinity Tolerance in Rice. Plant Cell Physiol. 2019, 60, 2549–2563. [Google Scholar] [CrossRef]
- Qin, H.; Wang, J.; Chen, X.; Wang, F.; Peng, P.; Zhou, Y.; Miao, Y.; Zhang, Y.; Gao, Y.; Qi, Y. Rice Os DOF 15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 2019, 223, 798–813. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Zheng, Y.; Su, Z.; Yu, S.; Song, H.; Zheng, X.; Lin, G.; Wu, W. OsSPL10, a SBP-Box Gene, Plays a Dual Role in Salt Tolerance and Trichome Formation in Rice (Oryza sativa L.). G3 Genes|Genomes|Genet 2019, 9, 4107–4114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Srivastava, A.K.; Sadanandom, A. Targeted mutagenesis of the SUMO protease, Overly Tolerant to Salt1 in rice through CRISPR/Cas9-mediated genome editing reveals a major role of this SUMO protease in salt tolerance. BioRxiv 2019, 555706. [Google Scholar] [CrossRef]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cui, L.; Xie, Z.; Zhang, Z.; Liu, E.; Peng, X. Two NCA1 isoforms interact with catalase in a mutually exclusive manner to redundantly regulate its activity in rice. BMC Plant Biol. 2019, 19, 105. [Google Scholar] [CrossRef] [PubMed]
- Bo, W.; Zhaohui, Z.; Huanhuan, Z.; Xia, W.; Binglin, L.; Lijia, Y.; Xiangyan, H.; Deshui, Y.; Xuelian, Z.; Chunguo, W.; et al. Targeted Mutagenesis of NAC Transcription Factor Gene, OsNAC041, Leading to Salt Sensitivity in Rice. Rice Sci. 2019, 26, 98–108. [Google Scholar] [CrossRef]
- Zeng, D.-D.; Yang, C.-C.; Qin, R.; Alamin, M.; Yue, E.-K.; Jin, X.-L.; Shi, C.-H. A guanine insert in OsBBS1 leads to early leaf senescence and salt stress sensitivity in rice (Oryza sativa L.). Plant Cell Rep. 2018, 37, 933–946. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, K.; Cheng, Y.; Zhong, Z.; Tian, L.; Tang, X.; Tang, A.; Zheng, X.; Zhang, T.; Qi, Y. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front. Plant Sci. 2017, 8, 1598. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.-B.; Li, J.; Qin, R.-Y.; Xu, R.-F.; Li, H.; Yang, Y.-C.; Ma, H.; Li, L.; Wei, P.-C.; Yang, J.-B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Ju, C.; Ma, X.; Han, B.; Zhang, W.; Zhao, Z.; Geng, L.; Cui, D.; Han, L. Candidate gene discovery for salt tolerance in rice (Oryza sativa L.) at the germination stage based on genome-wide association study. Front. Plant Sci. 2022, 13, 1010654. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, T.; Wen, Y.; Zheng, X.; Liu, H.; Chen, X.; Diao, Y.; Hu, Z.; Feng, W.; Chu, Z. Genetic Variation and Assessment of Seven Salt-Tolerance Genes in an Indica/Xian Rice Population. Agronomy 2025, 15, 570. [Google Scholar] [CrossRef]
- Yuan, H.; Cheng, M.; Wang, R.; Wang, Z.; Fan, F.; Wang, W.; Si, F.; Gao, F.; Li, S. miR396b/GRF6 module contributes to salt tolerance in rice. Plant Biotechnol. J. 2024, 22, 2079–2092. [Google Scholar] [CrossRef]
- Gupta, S.; Groen, S.C.; Zaidem, M.L.; Sajise, A.G.C.; Calic, I.; Natividad, M.A.; McNally, K.L.; Vergara, G.V.; Satija, R.; Franks, S.J.; et al. Systems Genomics of Salinity Stress Response in Rice; eLife 2025, 13, RP99352. 13. [CrossRef]
- Alam, M.S.; Kong, J.; Tao, R.; Ahmed, T.; Alamin, M.; Alotaibi, S.S.; Abdelsalam, N.R.; Xu, J.-H. CRISPR/Cas9 Mediated Knockout of the OsbHLH024 Transcription Factor Improves Salt Stress Resistance in Rice (Oryza sativa L.). Plants 2022, 11, 1184. [Google Scholar] [CrossRef]
- Sheng, X.; Ai, Z.; Tan, Y.; Hu, Y.; Guo, X.; Liu, X.; Sun, Z.; Yu, D.; Chen, J.; Tang, N.; et al. Novel Salinity-Tolerant Third-Generation Hybrid Rice Developed via CRISPR/Cas9-Mediated Gene Editing. Int. J. Mol. Sci. 2023, 24, 8025. [Google Scholar] [CrossRef] [PubMed]
- Lian, T.; Huang, Y.; Xie, X.; Huo, X.; Shahid, M.Q.; Tian, L.; Lan, T.; Jin, J. Rice SST Variation Shapes the Rhizosphere Bacterial Community, Conferring Tolerance to Salt Stress through Regulating Soil Metabolites. mSystems 2020, 5, e00721-20. [Google Scholar] [CrossRef] [PubMed]
- Ly, L.K.; Ho, T.M.; Bui, T.P.; Nguyen, L.T.; Phan, Q.; Le, N.T.; Khuat, L.T.M.; Le, L.H.; Chu, H.H.; Pham, N.B.; et al. CRISPR/Cas9 targeted mutations of OsDSG1 gene enhanced salt tolerance in rice. Funct. Integr. Genom. 2024, 24, 70. [Google Scholar] [CrossRef]
- Mishra, P.; Jain, A.; Takabe, T.; Tanaka, Y.; Negi, M.; Singh, N.; Jain, N.; Mishra, V.; Maniraj, R.; Krishnamurthy, S. Heterologous expression of serine hydroxymethyltransferase-3 from rice confers tolerance to salinity stress in E. coli and Arabidopsis. Front. Plant Sci. 2019, 10, 217. [Google Scholar] [CrossRef]
- Li, M.; Guo, L.; Guo, C.; Wang, L.; Chen, L. Over-expression of a DUF1644 protein gene, SIDP361, enhances tolerance to salt stress in transgenic rice. J. Plant Biol. 2016, 59, 62–73. [Google Scholar] [CrossRef]
- Chen, M.; Chen, Q.; Niu, X.; Zhang, R.; Lin, H.; Xu, C.; Wang, X.; Wang, G.; Chen, J. Expression of OsNHX1 gene in maize confers salt tolerance and promotes plant growth in the field. Plant Soil Environ. 2007, 53, 490. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef]
- Kurotani, K.-i.; Yamanaka, K.; Toda, Y.; Ogawa, D.; Tanaka, M.; Kozawa, H.; Nakamura, H.; Hakata, M.; Ichikawa, H.; Hattori, T. Stress tolerance profiling of a collection of extant salt-tolerant rice varieties and transgenic plants overexpressing abiotic stress tolerance genes. Plant Cell Physiol. 2015, 56, 1867–1876. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jing, W.; Xiao, L.; Jin, Y.; Shen, L.; Zhang, W. The rice high-affinity potassium transporter1; 1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef]
- Miyamoto, T.; Ochiai, K.; Nonoue, Y.; Matsubara, K.; Yano, M.; Matoh, T. Expression level of the sodium transporter gene OsHKT2; 1 determines sodium accumulation of rice cultivars under potassium-deficient conditions. Soil Sci. Plant Nutr. 2015, 61, 481–492. [Google Scholar] [CrossRef]
- Li, Q.-L.; Gao, X.-R.; Yu, X.-H.; Wang, X.-Z.; An, L.-J. Molecular cloning and characterization of betaine aldehyde dehydrogenase gene from Suaeda liaotungensis and its use in improved tolerance to salinity in transgenic tobacco. Biotechnol. Lett. 2003, 25, 1431–1436. [Google Scholar] [CrossRef]
- Yasmin, F.; Biswas, S.; Jewel, G.N.A.; Elias, S.M.; Seraj, Z.I. Constitutive overexpression of the plasma membrane Na+/H+ antiporter for conferring salinity tolerance in rice. Plant Tissue Cult. Biotechnol. 2015, 25, 257–272. [Google Scholar] [CrossRef]
- Diédhiou, C.J.; Popova, O.V.; Dietz, K.-J.; Golldack, D. The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 2008, 8, 49. [Google Scholar] [CrossRef]
- Ahmad, I.; Mian, A.; Maathuis, F.J. Overexpression of the rice AKT1 potassium channel affects potassium nutrition and rice drought tolerance. J. Exp. Bot. 2016, 67, 2689–2698. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Zhao, Y.; Luo, D.; Zhang, H. Isolating the promoter of a stress-induced gene encoding betaine aldehyde dehydrogenase from the halophyte Atriplex centralasiatica Iljin. Biochim. Biophys. Acta Gene Struct. Expr. 2002, 1577, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.-D.; Lin, K.-H.; Chen, C.-C.; Chiang, C.-M. Oryza sativa protein phosphatase 1a (OsPP1a) involved in salt stress tolerance in transgenic rice. Mol. Breed. 2016, 36, 22. [Google Scholar] [CrossRef]
- Wang, W.-S.; Zhao, X.-Q.; Li, M.; Huang, L.-Y.; Xu, J.-L.; Zhang, F.; Cui, Y.-R.; Fu, B.-Y.; Li, Z.-K. Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling. J. Exp. Bot. 2016, 67, 405–419. [Google Scholar] [CrossRef]
- Sahoo, R.K.; Ansari, M.W.; Tuteja, R.; Tuteja, N. OsSUV3 transgenic rice maintains higher endogenous levels of plant hormones that mitigates adverse effects of salinity and sustains crop productivity. Rice 2014, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, L.-H.; Zhao, J.-F.; Song, Y.; Zhang, C.-J.; Guo, Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 2009, 149, 916–928. [Google Scholar] [CrossRef]
- Campo, S.; Baldrich, P.; Messeguer, J.; Lalanne, E.; Coca, M.; San Segundo, B. Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 2014, 165, 688–704. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Pandian, S.K.; Ramesh, M. Transgenic indica rice cv. ADT 43 expressing a Δ 1-pyrroline-5-carboxylate synthetase (P5CS) gene from Vigna aconitifolia demonstrates salt tolerance. Plant Cell Tissue Organ Cult. 2011, 107, 383–395. [Google Scholar] [CrossRef]
- Su, J.; Hirji, R.; Zhang, L.; He, C.; Selvaraj, G.; Wu, R. Evaluation of the stress-inducible production of choline oxidase in transgenic rice as a strategy for producing the stress-protectant glycine betaine. J. Exp. Bot. 2006, 57, 1129–1135. [Google Scholar] [CrossRef]
- Pandey, S.; Patel, M.K.; Mishra, A.; Jha, B. In planta transformed cumin (Cuminum cyminum L.) plants, overexpressing the SbNHX1 gene showed enhanced salt endurance. PLoS ONE 2016, 11, e0159349. [Google Scholar] [CrossRef] [PubMed]
- Majee, M.; Maitra, S.; Dastidar, K.G.; Pattnaik, S.; Chatterjee, A.; Hait, N.C.; Das, K.P.; Majumder, A.L. A novel salt-tolerant L-myo-inositol-1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice: Molecular cloning, bacterial overexpression, characterization, and functional introgression into tobacco-conferring salt tolerance phenotype. J. Biol. Chem. 2004, 279, 28539–28552. [Google Scholar] [PubMed]
- Sakurai, J.; Ishikawa, F.; Yamaguchi, T.; Uemura, M.; Maeshima, M. Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 2005, 46, 1568–1577. [Google Scholar] [CrossRef]
- Hanba, Y.T.; Shibasaka, M.; Hayashi, Y.; Hayakawa, T.; Kasamo, K.; Terashima, I.; Katsuhara, M. Overexpression of the barley aquaporin HvPIP2; 1 increases internal CO2 conductance and CO2 assimilation in the leaves of transgenic rice plants. Plant Cell Physiol. 2004, 45, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Mallikarjuna, G.; Mallikarjuna, K.; Reddy, M.; Kaul, T. Expression of OsDREB2A transcription factor confers enhanced dehydration and salt stress tolerance in rice (Oryza sativa L.). Biotechnol. Lett. 2011, 33, 1689–1697. [Google Scholar] [CrossRef]
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000, 23, 319–327. [Google Scholar] [CrossRef]
- Singh, Y.P.; Mishra, V.K.; Singh, S.; Sharma, D.K.; Singh, D.; Singh, U.S.; Singh, R.K.; Haefele, S.M.; Ismail, A.M. Productivity of sodic soils can be enhanced through the use of salt tolerant rice varieties and proper agronomic practices. Field Crops Res. 2016, 190, 82–90. [Google Scholar] [CrossRef]
- Sheoran, P.; Kumar, A.; Sharma, R.; Barman, A.; Parjapat, K.; Singh, R.K.; Kumar, S.; Sharma, P.C.; Ismail, A.M.; Singh, R.K. Managing sodic soils for better productivity and farmers’ income by integrating use of salt tolerant rice varieties and matching agronomic practices. Field Crops Res. 2021, 270, 108192. [Google Scholar] [CrossRef]
- Sackey, O.K.; Feng, N.; Mohammed, Y.Z.; Dzou, C.F.; Zheng, D.; Zhao, L.; Shen, X. A comprehensive review on rice responses and tolerance to salt stress. Front. Plant Sci. 2025, 16, 1561280. [Google Scholar] [CrossRef]
- Khan, S.; Irshad, S.; Mehmood, K.; Hasnain, Z.; Nawaz, M.; Rais, A.; Gul, S.; Wahid, M.A.; Hashem, A.; Abd_Allah, E.F. Biochar production and characteristics, its impacts on soil health, crop production, and yield enhancement: A review. Plants 2024, 13, 166. [Google Scholar] [CrossRef] [PubMed]
- Ondrasek, G.; Rengel, Z. Environmental salinization processes: Detection, implications & solutions. Sci. Total Environ. 2021, 754, 142432. [Google Scholar] [PubMed]
- Andrade Foronda, D. Reclamation of a saline-sodic soil with organic amendments and leaching. Environ. Sci. Proc. 2022, 16, 56. [Google Scholar]
- Mockeviciene, I.; Repsiene, R.; Amaleviciute-Volunge, K.; Karcauskiene, D.; Slepetiene, A.; Lepane, V. Effect of long-term application of organic fertilizers on improving organic matter quality in acid soil. Arch. Agron. Soil Sci. 2022, 68, 1192–1204. [Google Scholar] [CrossRef]
- Irshad, I.; Anwar-ul-Haq, M.; Akhtar, J.; Maqsood, M. Effects of different organic amendments on maize (Zea mays L.) growth in salt affected soil. J. Glob. Innov. Agric. Sci. 2022, 10, 121–130. [Google Scholar] [CrossRef]
- Yang, L.; Bian, X.; Yang, R.; Zhou, C.; Tang, B. Assessment of organic amendments for improving coastal saline soil. Land Degrad. Dev. 2018, 29, 3204–3211. [Google Scholar] [CrossRef]
- Gupta, A.; Tiwari, R.K.; Shukla, R.; Singh, A.N.; Sahu, P.K. Salinity alleviator bacteria in rice (Oryza sativa L.), their colonization efficacy, and synergism with melatonin. Front. Plant Sci. 2023, 13, 1060287. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Gaurav, A.K.; Srivastava, S.; Verma, J.P. Plant growth-promoting bacteria: Biological tools for the mitigation of salinity stress in plants. Front. Microbiol. 2020, 11, 1216. [Google Scholar] [CrossRef]
- Gupta, A.; Mishra, R.; Rai, S.; Bano, A.; Pathak, N.; Fujita, M.; Kumar, M.; Hasanuzzaman, M. Mechanistic insights of plant growth promoting bacteria mediated drought and salt stress tolerance in plants for sustainable agriculture. Int. J. Mol. Sci. 2022, 23, 3741. [Google Scholar] [CrossRef]
- Ansari, F.A.; Ahmad, I.; Pichtel, J. Growth stimulation and alleviation of salinity stress to wheat by the biofilm forming Bacillus pumilus strain FAB10. Appl. Soil Ecol. 2019, 143, 45–54. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.; Jha, P.N. The plant-growth-promoting bacterium Klebsiella sp. SBP-8 confers induced systemic tolerance in wheat (Triticum aestivum) under salt stress. J. Plant Physiol. 2015, 184, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L.; Khan, A.; Ahmed, A.-H. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: A review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef]
- Khan, M.A.; Asaf, S.; Khan, A.L.; Adhikari, A.; Jan, R.; Ali, S.; Imran, M.; Kim, K.M.; Lee, I.J. Plant growth-promoting endophytic bacteria augment growth and salinity tolerance in rice plants. Plant Biol. 2020, 22, 850–862. [Google Scholar] [CrossRef] [PubMed]
- Ouziad, F.; Wilde, P.; Schmelzer, E.; Hildebrandt, U.; Bothe, H. Analysis of expression of aquaporins and Na+/H+ transporters in tomato colonized by arbuscular mycorrhizal fungi and affected by salt stress. Environ. Exp. Bot. 2006, 57, 177–186. [Google Scholar] [CrossRef]
- Zuccarini, P. Mycorrhizal infection ameliorates chlorophyll content and nutrient uptake of lettuce exposed to saline irrigation. Plant Soil Environ. 2007, 53, 283. [Google Scholar] [CrossRef]
- Evelin, H.; Kapoor, R.; Giri, B. Arbuscular mycorrhizal fungi in alleviation of salt stress: A review. Ann. Bot. 2009, 104, 1263–1280. [Google Scholar] [CrossRef]
- Giri, B.; Mukerji, K.G. Mycorrhizal inoculant alleviates salt stress in Sesbania aegyptiaca and Sesbania grandiflora under field conditions: Evidence for reduced sodium and improved magnesium uptake. Mycorrhiza 2004, 14, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Al-Karaki, G.N. Nursery inoculation of tomato with arbuscular mycorrhizal fungi and subsequent performance under irrigation with saline water. Sci. Hortic. 2006, 109, 1–7. [Google Scholar] [CrossRef]
- Sharifi, M.; Ghorbanli, M.; Ebrahimzadeh, H. Improved growth of salinity-stressed soybean after inoculation with salt pre-treated mycorrhizal fungi. J. Plant Physiol. 2007, 164, 1144–1151. [Google Scholar] [CrossRef]
- Takahashi, F.; Hanada, K.; Kondo, T.; Shinozaki, K. Hormone-like peptides and small coding genes in plant stress signaling and development. Curr. Opin. Plant Biol. 2019, 51, 88–95. [Google Scholar] [CrossRef]
- Quamruzzaman, M.; Manik, S.M.N.; Shabala, S.; Zhou, M. Improving Performance of Salt-Grown Crops by Exogenous Application of Plant Growth Regulators. Biomolecules 2021, 11, 788. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Du, Y.; Feng, N.; Zheng, D.; Zhou, H.; Huo, J. Exogenous Uniconazole promotes physiological metabolism and grain yield of rice under salt stress. Front. Plant Sci. 2024, 15, 1459121. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for plant improvement: Status and prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar]
- Gong, Q.; Li, P.; Ma, S.; Indu Rupassara, S.; Bohnert, H.J. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005, 44, 826–839. [Google Scholar] [CrossRef]
- Widodo; Patterson, J.H.; Newbigin, E.; Tester, M.; Bacic, A.; Roessner, U. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J. Exp. Bot. 2009, 60, 4089–4103. [Google Scholar] [CrossRef]
- Yancey, P.H. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 2005, 208, 2819–2830. [Google Scholar] [CrossRef]
- Kulkarni, J.; Sharma, S.; Sahoo, S.A.; Mishra, S.; Nikam, T.D.; Borde, M.; Penna, S.; Srivastava, A.K. Resilience in primary metabolism contributes to salt stress adaptation in Sesuvium portulacastrum (L.). Plant Growth Regul. 2022, 98, 385–398. [Google Scholar] [CrossRef]
- Akula, R.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [CrossRef]
- Wink, M. Modes of action of herbal medicines and plant secondary metabolites. Medicines 2015, 2, 251–286. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Shukla, S. Impact of various factors responsible for fluctuation in plant secondary metabolites. J. Appl. Res. Med. Aromat. Plants 2015, 2, 105–113. [Google Scholar] [CrossRef]
- Sarri, E.; Termentzi, A.; Abraham, E.M.; Papadopoulos, G.K.; Baira, E.; Machera, K.; Loukas, V.; Komaitis, F.; Tani, E. Salinity stress alters the secondary metabolic profile of M. sativa, M. arborea and their hybrid (Alborea). Int. J. Mol. Sci. 2021, 22, 4882. [Google Scholar] [CrossRef]
- Mukhamejanova, A.; Alikulov, Z.; Shapekova, N.; Aubakirova, K.; Mukhtarov, A. The effect of antioxidants on xanthine oxidase activity in fresh ovine milk. Slovak J. Food Sci. 2021, 15, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.A.; Jiménez, A.; Mullineaux, P.; Sevilia, F. Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ. 2000, 23, 853–862. [Google Scholar] [CrossRef]
- Demiral, T.; Türkan, I. Comparative lipid peroxidation, antioxidant defense systems and proline content in roots of two rice cultivars differing in salt tolerance. Environ. Exp. Bot. 2005, 53, 247–257. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. The response of salinity stress-induced A. tricolor to growth, anatomy, physiology, non-enzymatic and enzymatic antioxidants. Front. Plant Sci. 2020, 11, 559876. [Google Scholar] [CrossRef] [PubMed]
- Hussain, N.; Sohail, Y.; Shakeel, N.; Javed, M.; Bano, H.; Gul, H.S.; Zafar, Z.U.; Frahat Zaky Hassan, I.; Ghaffar, A.; Athar, H.-u.-R. Role of mineral nutrients, antioxidants, osmotic adjustment and PSII stability in salt tolerance of contrasting wheat genotypes. Sci. Rep. 2022, 12, 12677. [Google Scholar] [CrossRef] [PubMed]
- Ali Solangi, K.; Wu, Y.; Xing, D.; Ahmed Qureshi, W.; Hussain Tunio, M.; Ali Sheikh, S.; Shabbir, A. Can electrophysiological information reflect the response of mangrove species to salt stress? A case study of rewatering and Sodium nitroprusside application. Plant Signal. Behav. 2022, 17, 2073420. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Yuan, D.; Chu, C.; Duan, M. Salt tolerance in rice: Physiological responses and molecular mechanisms. Crop J. 2022, 10, 13–25. [Google Scholar] [CrossRef]
| Gene Name | Gene Function | Changes in Mutants/Validation | Reference |
|---|---|---|---|
| OsBGE3 | Cytokinin transport | Reduced grain length; salt hypersensitivity | [75] |
| OsDST | Zinc finger TF | Broad leaves; decreased stomatal density; improved salt tolerance | [76] |
| OsFLN2 | Sucrose metabolism | Salt sensitivity due to inadequate assimilated supply | [77] |
| OsGTy-2 | Trihelix TF | Increased root Na+/K+ and high Na+/K+ ratio; salt hypersensitivity. | [78] |
| OsPIL14 | Basic helix-loop-helix TF | Reduced coleoptile and root elongation | [79] |
| OsPQR3 | E3 ubiquitin ligase | Enhanced oxidative/salt tolerance; upregulated OsGPX1, OsAPX1, OsSOD1 | [80] |
| OsRR9, OsRR10 | Cytokinin signaling | High salt tolerance; upregulated ion transporters | [81] |
| OsDOF15 | Transcription factor | Short roots; impaired meristem activity; salt hypersensitivity | [82] |
| OsSPL10 | Transcription factor | Glabrous leaves; enhanced seedling survival under salt | [83] |
| OsOTS1 | SUMOylation | Reduced chlorophyll/root biomass; salt sensitivity | [84] |
| OsRR22 | Cytokinin signaling TF | Increased shoot biomass; improved salt tolerance | [85] |
| OsNCA1a, OsNCA1b | – | Cell death; salt sensitivity | [86] |
| OsNAC041 | – | Reduced germination; high ROS/MDA; salt sensitivity | [87] |
| OsBBS1 | Receptor-like kinase | Early senescence; reduced root length/tillers; salt hypersensitivity | [88] |
| OsMIR528 | miRNA | Delayed branching; chlorosis | [89] |
| OsRAV2 | Brassinosteroid-response TF | Loss of salt-induced expression | [90] |
| OsHAK3 | Potassium transporter involved in K+/Na+ balance | Candidate gene for salt tolerance at germination; mutants show salt sensitivity; important for ionic homeostasis | [91] |
| OsITPK5 | Inositol trisphosphate kinase involved in stress signaling | Identified as candidate gene for salt tolerance at germination stage; likely role in signaling and adaptation | [91] |
| OsWRKY53 | Transcription factor regulating salt response gene | Binds promoters of OsHKT1;5 and OsMKK10.2; elite haplotypes associated with improved salt tolerance | [92] |
| SKC1/OsHKT1;5 | Sodium transporter maintaining Na+ exclusion from shoots | Salt tolerance locus; expression positively correlated with OsWRKY53; key for ionic homeostasis | [92] |
| miR396b/GRF6 module | Regulatory module controlling salt stress response via MYB3R TF | Enhances salt tolerance by increasing ROS scavenging enzymes; MYB3R is direct target; overexpression improves survival | [93] |
| OsFLP | R2R3 MYB-like TF regulating stomatal development | Identified as salt tolerance candidate via trans-eQTL analysis; involved in adaptive response to salinity | [94] |
| OsCYP2 | Cyclophilin protein confers salt tolerance | Identified as beneficial under saline conditions; involved in protein folding and stress response | [94] |
| DMS3/OsITPK2 | Stress signaling and chromatin remodeling | Critical in salt tolerance; related to inositol phosphate metabolism | [91] |
| OsbHLH024 | Negative regulator of salt stress; affects ion balance and antioxidant activity | Knockout mutants exhibited improved salt resistance and upregulation of ion transporter genes | [95] |
| OsRR22 | Main effect gene for salt tolerance; loss-of-function increases tolerance | CRISPR/Cas9 generates knockout lines with enhanced salt tolerance | [96] |
| OsSPL10 | Influences rhizosphere microbiota and ion accumulation under salt stress | CRISPR/Cas9-edited lines with loss of function showed better adaptation to salt stress | [97] |
| OsDSG1 | Involved in ubiquitination pathway; regulates biochemical reactions under salt stress | CRISPR/Cas9-induced mutants displayed enhanced salt tolerance at germination and seedling stages | [98] |
| OsSHMT3 | Photorespiration | Overexpression | [99] |
| SIDP361 | Proline | Overexpression | [100] |
| OsNHX1 | Compartmentalization of Na+ into vacuoles | Overexpression | [101] |
| OsCIPK15 | Enhanced salt tolerance | Overexpression | [102] |
| CYP94C2b | Deactivation of jasmonate | Overexpression | [103] |
| Oshkt1;1 | Sensitive to salt stress | Mutant studies | [104] |
| OsHKT2:1 | Na+ accumulation under low K+ supply | Overexpression | [105] |
| OsHAK5 | Root K acquisition and transport to shoot at low K levels | Overexpression | [106] |
| OsSOS1 | Improved salt tolerance | Transformation | [107] |
| SAPK4 | Improved salt tolerance | Transgenic | [108] |
| AKT1 | Enhances K+ uptake | Overexpression | [109] |
| OsHAK21 | Na+/K+ homeostasis | Quantitative expression | [110] |
| OsPP1a | Enhanced tolerance to high salinity/upregulation of SOD | Transgenic | [111] |
| PtCYP714A3 | Shoot response to salt toxicity | Transgenics (ectopic expression) | [112] |
| OsSUV3 | Salinity tolerance by maintaining photosynthesis and antioxidant machinery | Transgenic | [113] |
| OsRMC | Negative regulation of salt tolerance | Knock down expression | [114] |
| OsCPK4 | Enhances salt and drought tolerance | Overexpression | [115] |
| P5CS | High accumulation of proline | Transgenic | [116] |
| codA | Promotes synthesis of glycine betaine | Transgenic | [117] |
| OsTPS1 | Enhances salt tolerance | Overexpression | [118] |
| PINO1 | Allows growth of transgenic plants in salt environment | Introgression and expression | [119] |
| OsTIP1;1 | Upregulation in salt stress | Overexpression | [120] |
| HvPIP2;1 | Enhances sensitivity to salinity | Overexpression | [121] |
| OsDREB1A, OsDREB1F, OsDREB2A | Improved salt tolerance | Transgenic | [122] |
| OsCDPK7 | Enhances salt tolerance | Transgenic | [123] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saleem, M.A.; Khan, A.; Tu, J.; Huang, W.; Liu, Y.; Feng, N.; Zheng, D.; Xue, Y. Salinity Stress in Rice: Multilayered Approaches for Sustainable Tolerance. Int. J. Mol. Sci. 2025, 26, 6025. https://doi.org/10.3390/ijms26136025
Saleem MA, Khan A, Tu J, Huang W, Liu Y, Feng N, Zheng D, Xue Y. Salinity Stress in Rice: Multilayered Approaches for Sustainable Tolerance. International Journal of Molecular Sciences. 2025; 26(13):6025. https://doi.org/10.3390/ijms26136025
Chicago/Turabian StyleSaleem, Muhammad Ahmad, Ahmad Khan, Jinji Tu, Wenkang Huang, Ying Liu, Naijie Feng, Dianfeng Zheng, and Yingbin Xue. 2025. "Salinity Stress in Rice: Multilayered Approaches for Sustainable Tolerance" International Journal of Molecular Sciences 26, no. 13: 6025. https://doi.org/10.3390/ijms26136025
APA StyleSaleem, M. A., Khan, A., Tu, J., Huang, W., Liu, Y., Feng, N., Zheng, D., & Xue, Y. (2025). Salinity Stress in Rice: Multilayered Approaches for Sustainable Tolerance. International Journal of Molecular Sciences, 26(13), 6025. https://doi.org/10.3390/ijms26136025






