Refining Criteria for Choosing the First-Line Treatment for Real-World Patients with Advanced ALK-Rearranged NSCLC
Abstract
1. Introduction
2. How Do Diagnostic Intricacies Affect Interpretation of the Treatment Outcome?
2.1. ALTA-1L Trial
2.2. ALEX Trial
2.3. CROWN Trial
2.4. NGS
3. The Importance of Intrinsic Resistance to ALK-TKIs for the First-Line Treatment
| Reported Factors of ALK-TKI Intrinsic Resistance | References | |
|---|---|---|
| 1 | Histology: LUAD, SqCC, LCNEC, SCLC | [27,28,29,30,31,32,33,34,35,36,37,38,39] |
| 2 | ALK fusion partners and EML4-ALK variants, double/triple ALK fusions | [40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59] |
| 3 | De novo on-target mutations in ALK | [60,61] |
| 4 | De novo off-target mutations in other oncogenes: EGFR, KRAS, MET | [67,68,70] |
| 5 | De novo MET amplification | [69] |
| 6 | De novo alterations in tumor suppressor genes: CDKN2A/B, NF2, TP53 | [40,41,57,58,59,62,70,110] |
| 7 | (Genotypes of CYPs and P-glycoprotein transporter) * | [117,118,119,120] |
4. Different ALK-TKIs May Provide Long-Term Responses, Including Crizotinib
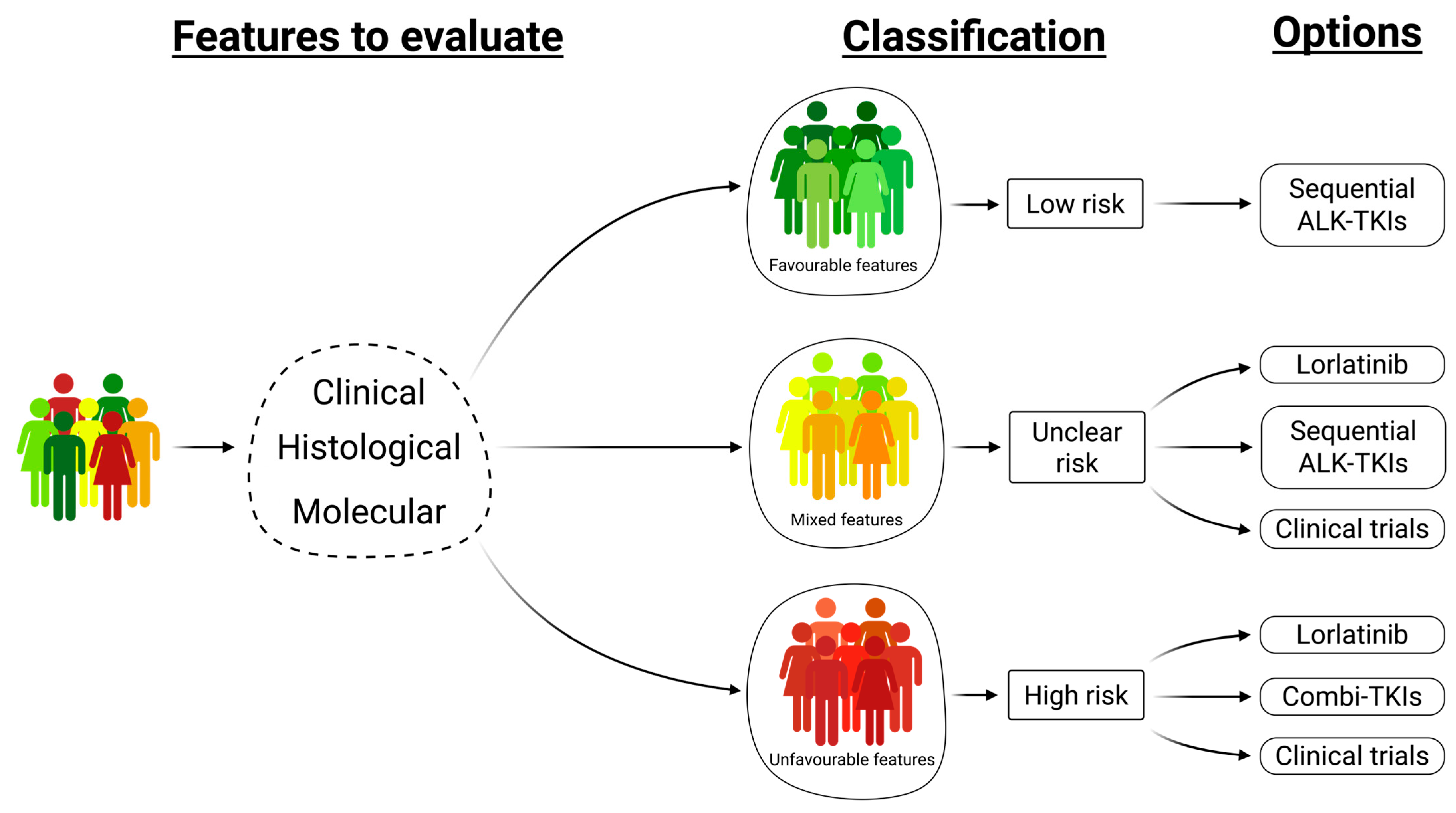
5. Future Perspectives—Favorable and Unfavorable Factors Should Be Included in Therapeutic Decision
6. Conclusions
- It is clinically important to better characterize a heterogenous disease such as ALK-rearranged NSCLC while choosing the first-line treatment.
- An optimized diagnostic approach should complementarily inform on the alterations in ALK and other genes.
- For this purpose, diagnostic biopsies of tissue should be examined comprehensively with histological assessment followed by ALK-IHC, ALK-FISH, and NGS (both DNA/RNA). Concordance among these methods should be considered. CtDNA at baseline should also be included.
- Up-front brain metastases, PS > 1, and/or presence of ctDNA represent unfavorable features linked to poorer outcomes.
- Merging the clinical, histological, and molecular features helps to define risk groups of ALK-rearranged NSCLC patients and direct treatment choice.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Solomon, B.J.; Liu, G.; Felip, E.; Mok, T.S.K.; Soo, R.A.; Mazieres, J.; Shaw, A.T.; de Marinis, F.; Goto, Y.; Wu, Y.L.; et al. Lorlatinib Versus Crizotinib in Patients with Advanced ALK-Positive Non-Small Cell Lung Cancer: 5-Year Outcomes from the Phase III CROWN Study. J. Clin. Oncol. 2024, 42, 3400–3409. [Google Scholar] [CrossRef]
- Drilon, A.; Lin, J.J.; Filleron, T.; Ni, A.; Milia, J.; Bergagnini, I.; Hatzoglou, V.; Velcheti, V.; Offin, M.; Li, B.; et al. Frequency of Brain Metastases and Multikinase Inhibitor Outcomes in Patients with RET-Rearranged Lung Cancers. J. Thorac. Oncol. 2018, 13, 1595–1601. [Google Scholar] [CrossRef] [PubMed]
- Hoe, H.J.; Solomon, B.J. Based on the CROWN Findings, Lorlatinib Should Be the Preferred First-Line Treatment for Patients with Advanced ALK-Positive NSCLC. J. Thorac. Oncol. 2025, 20, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Lam, V.K.; Gadgeel, S.M. Even with the CROWN Findings, There Remain Multiple First-Line Treatment Options for Patients with Advanced ALK-Positive NSCLC. J. Thorac. Oncol. 2025, 20, 150–153, Corrigendum in J. Thorac. Oncol. 2025, 20, 680. [Google Scholar] [CrossRef]
- Urbanska, E.M.; Sørensen, J.B.; Santoni-Rugiu, E. Can we refine criteria for the first line-treatment for patients with advanced ALK-positive NSCLC in the real world? J. Thorac. Oncol. 2025, 20, e71–e72. [Google Scholar] [CrossRef] [PubMed]
- Camidge, D.R.; Kim, H.R.; Ahn, M.J.; Yang, J.C.; Han, J.Y.; Lee, J.S.; Hochmair, M.; Li, J.Y.; Chang, G.C.; Lee, K.H.; et al. Brigatinib versus Crizotinib in ALK-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 22, 2027–2039. [Google Scholar] [CrossRef]
- Peters, S.; Camidge, D.R.; Shaw, A.T.; Gadgeel, S.; Ahn, J.S.; Kim, D.W.; Ou, S.I.; Pérol, M.; Dziadziuszko, R.; Rosell, R.; et al. ALEX Trial Investigators. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 829–838. [Google Scholar] [CrossRef]
- Camidge, D.R.; Kim, H.R.; Ahn, M.J.; Yang, J.C.H.; Han, J.Y.; Hochmair, M.J.; Lee, K.H.; Delmonte, A.; Garcia Campelo, M.R.; Kim, D.; et al. Brigatinib Versus Crizotinib in ALK Inhibitor-Naive Advanced ALK-Positive NSCLC: Final Results of Phase 3 ALTA-1L Trial. J. Thorac. Oncol. 2021, 16, 2091–2108. [Google Scholar] [CrossRef]
- Cabillic, F.; Gros, A.; Dugay, F.; Begueret, H.; Mesturoux, L.; Chiforeanu, D.C.; Dufrenot, L.; Jauffret, V.; Dachary, D.; Corre, R.; et al. Parallel FISH and Immunohistochemical Studies of ALK Status in 3244 Non–Small-Cell Lung Cancers Reveal Major Discordances. J. Thorac. Oncol. 2014, 9, 295–306. [Google Scholar] [CrossRef]
- Scattone, A.; Catino, A.; Schirosi, L.; Caldarola, L.; Tommasi, S.; Lacalamita, R.; Montagna, E.S.; Galetta, D.; Serio, G.; Zito, F.A.; et al. Discordance between FISH, IHC, and NGS Analysis of ALK Status in Advanced Non-Small Cell Lung Cancer (NSCLC): A Brief Report of 7 Cases. Transl. Oncol. 2019, 2, 389–395. [Google Scholar] [CrossRef]
- Batra, U.; Nathany, S.; Sharma, M.; Pasricha, S.; Bansal, A.; Jain, P.; Mehta, A. IHC versus FISH versus NGS to detect ALK gene rearrangement in NSCLC: All questions answered? J. Clin. Pathol. 2022, 75, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Nambirajan, A.; Sood, R.; Katoon, W.; Malik, P.S.; Mohan, A.; Jain, D. Concordance of Immunohistochemistry and Fluorescence in Situ Hybridization in the Detection of Anaplastic Lymphoma Kinase (ALK) and Ros Proto-oncogene 1 (ROS1) Gene Rearrangements in Non–Small Cell Lung Carcinoma-A 4.5-Year Experience Highlighting Challenges and Pitfalls. Arch. Pathol. Lab. Med. 2024, 14, 928–937. [Google Scholar] [CrossRef]
- Jiang, F.; Wang, C.; Yang, P.; Sun, P.; Liu, J. Pathological cytomorphologic features and the percentage of ALK FISH-positive cells predict pulmonary adenocarcinoma prognosis: A prospective cohort study. World J. Surg. Oncol. 2021, 19, 278. [Google Scholar] [CrossRef] [PubMed]
- Soria, J.C.; Ho, S.N.; Varella-Garcia, M.; Iafrate, A.J.; Solomon, B.J.; Shaw, A.T.; Blackhall, F.; Mok, T.S.K.; Wu, Y.L.; Pestova, K.; et al. Correlation of extent of ALK FISH positivity and crizotinib efficacy in three prospective studies of ALK-positive patients with non-small-cell lung cancer. Ann. Oncol. 2018, 29, 1964–1971. [Google Scholar] [CrossRef]
- Toruner, G.A.; Tang, Z.; Tang, G.; Medeiros, L.J.; Hu, S. Low ALK FISH positive metastatic non-small cell lung cancer (NSCLC) patients have shorter progression-free survival after treatment with ALK inhibitors. Cancer Genet. 2020, 241, 57–60. [Google Scholar] [CrossRef]
- Mok, T.; Peters, S.; Camidge, D.R.; Noé, J.; Gadgeel, S.; Ou, S.I.; Kim, D.W.; Konopa, K.; Pozzi, E.; Liu, T.; et al. Outcomes According to ALK Status Determined by Central Immunohistochemistry or Fluorescence In Situ Hybridization in Patients with ALK-Positive NSCLC Enrolled in the Phase 3 ALEX Study. J. Thorac. Oncol. 2021, 16, 259–268. [Google Scholar] [CrossRef]
- Canterbury, C.R.; Fernandes, H.; Crapanzano, J.P.; Murty, V.V.; Mansukhani, M.M.; Shu, C.A.; Szabolcs, M.; Saqi, A. ALK Gene Rearrangements in Lung Adenocarcinomas: Concordance of Immunohistochemistry, Fluorescence In Situ Hybridization, RNA In Situ Hybridization, and RNA Next-Generation Sequencing Testing. JTO Clin. Res. Rep. 2021, 2, 100223. [Google Scholar] [CrossRef]
- Gendarme, S.; Matton, L.; Antoine, M.; Kerrou, K.; Ruppert, A.M.; Epaud, C.; Cadranel, J.; Fallet, V. Strong ALK and PD-L1 positive IHC expression related ALK amplification in an advanced lung sarcomatoid carcinoma: A therapeutic trap? Lung Cancer 2021, 152, 94–97. [Google Scholar] [CrossRef]
- Salido, M.; Pijuan, L.; Martínez-Avilés, L.; Galván, A.B.; Cañadas, I.; Rovira, A.; Zanui, M.; Martínez, A.; Longarón, R.; Sole, F.; et al. Increased ALK gene copy number and amplification are frequent in non-small cell lung cancer. J. Thorac. Oncol. 2011, 6, 21–27. [Google Scholar] [CrossRef]
- Zito Marino, F.; Botti, G.; Aquino, G.; Ferrero, S.; Gaudioso, G.; Palleschi, A.; Rocco, D.; Salvi, R.; Micheli, M.C.; Micheli, P.; et al. Unproductive Effects of ALK Gene Amplification and Copy Number Gain in Non-Small-Cell Lung Cancer. ALK Gene Amplification and Copy Gain in NSCLC. Int. J. Mol. Sci. 2020, 21, 4927. [Google Scholar] [CrossRef]
- Tobiášová, K.; Barthová, M.; Janáková, Ľ.; Lešková, K.; Farkašová, A.; Loderer, D.; Grendár, M.; Plank, L. Discordant ALK Status in Non-Small Cell Lung Carcinoma: A Detailed Reevaluation Comparing IHC, FISH, and NGS Analyses. Int. J. Mol. Sci. 2024, 25, 8168. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Chuang, J.C.; Berry, G.J.; Wakelee, H.A. Anaplastic Lymphoma Kinase Testing: IHC vs. FISH vs. NGS. Curr. Treat. Options Oncol. 2017, 18, 71. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Lu, C.; Xu, C.W.; Zheng, Z. Noncanonical Gene Fusions Detected at the DNA Level Necessitate Orthogonal Diagnosis Methods Before Targeted Therapy. J. Thorac. Oncol. 2021, 16, 344–348. [Google Scholar] [CrossRef]
- Li, W.; Guo, L.; Liu, Y.; Dong, L.; Yang, L.; Chen, L.; Liu, K.; Shao, Y.; Ying, J. Potential Unreliability of Uncommon ALK, ROS1, and RET Genomic Breakpoints in Predicting the Efficacy of Targeted Therapy in NSCLC. J. Thorac. Oncol. 2021, 16, 404–418. [Google Scholar] [CrossRef]
- Benayed, R.; Offin, M.; Mullaney, K.; Sukhadia, P.; Rios, K.; Desmeules, P.; Ptashkin, R.; Won, H.; Chang, J.; Halpenny, D.; et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin. Cancer Res. 2019, 25, 4712–4722. [Google Scholar] [CrossRef]
- Vollbrecht, C.; Lenze, D.; Hummel, M.; Lehmann, A.; Moebs, M.; Frost, N.; Jurmeister, P.; Schweizer, L.; Kellner, U.; Dietel, M.; et al. RNA-based analysis of ALK fusions in non-small cell lung cancer cases showing IHC/FISH discordance. BMC Cancer 2018, 18, 1158. [Google Scholar] [CrossRef]
- Chen, C.; Wang, L.; Gu, C.; Wang, Y.; Pan, X.; Fu, S.; Yang, J.; Wang, R. Survival analyses and immunohistochemical study of primary signet ring cell carcinoma of the lung adenocarcinoma. Transl. Cancer Res. 2020, 9, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Huang, Y.H.; Xue, J.W.; Zhang, R.; Liu, R.; Wang, Y.; Feng, Z.B. Clinicopathological features and prognostic significance of pulmonary adenocarcinoma with signet ring cell components: Meta-analysis and SEER analysis. Clin. Exp. Med. 2023, 23, 4341–4354. [Google Scholar] [CrossRef]
- Popat, S.; Gonzalez, D.; Min, T.; Swansbury, J.; Dainton, M.; Croud, J.G.; Rice, A.J.; Nicholson, A.G. ALK translocation is associated with ALK immunoreactivity and extensive signet-ring morphology in primary lung adenocarcinoma. Lung Cancer 2012, 75, 300–305. [Google Scholar] [CrossRef]
- Nishino, M.; Klepeis, V.E.; Yeap, B.Y.; Bergethon, K.; Morales-Oyarvide, V.; Dias-Santagata, D.; Yagi, Y.; Mark, E.J.; Iafrate, A.J.; Mino-Kenudson, M. Histologic and cytomorphologic features of ALK-rearranged lung adenocarcinomas. Mod. Pathol. 2012, 25, 1462–1472. [Google Scholar] [CrossRef]
- Possidente, L.; Landriscina, M.; Patitucci, G.; Borgia, L.; Lalinga, V.; Vita, G. ALK rearrangement in specific subtypes of lung adenocarcinoma: Immunophenotypic and morphological features. Med. Oncol. 2017, 34, 76. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Yuan, X.; Wang, Y.; Yang, M.; Wu, P.; Chen, H.; Yun, Y.; Shen, Z.; Ji, D.; Ma, Y. Ensartinib in the treatment of anaplastic lymphoma kinase-positive locally advanced or metastatic patients with lung squamous or adenosquamous carcinoma: A real-world, retrospective study. Asia Pac. J. Clin. Oncol. 2024, 20, 700–706. [Google Scholar] [CrossRef]
- Tamiya, A.; Shimizu, S.; Atagi, S.A. Case of Squamous Cell Carcinoma Harboring an EML4-ALK Rearrangement that Was Unsuccessfully Treated with the ALK Inhibitor Alectinib. J. Thorac. Oncol. 2015, 10, e74–e75. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, J.; Wang, X.; Zong, W.; Sun, L.; Qin, J.; Yin, Y. Two case reports: EML4-ALK rearrangement large cell neuroendocrine carcinoma and literature review. Front. Oncol. 2023, 13, 1227980. [Google Scholar] [CrossRef]
- Akhoundova, D.; Haberecker, M.; Fritsch, R.; Höller, S.; Kiessling, M.K.; Rechsteiner, M.; Rüschoff, J.H.; Curioni-Fontecedro, A. Targeting ALK in Neuroendocrine Tumors of the Lung. Front. Oncol. 2022, 12, 911294. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Uehara, Y.; Watanabe, K.; Hishima, T.; Hosomi, Y. Successful Treatment of ALK-Positive Large-Cell Neuroendocrine Carcinoma of the Lung with Sequential ALK Inhibitors: A Case Report. JTO Clin. Res. Rep. 2023, 4, 100538. [Google Scholar] [CrossRef] [PubMed]
- Kondoh, C.; Horio, Y.; Hayashi, Y.; Ebi, H.; Hida, T.; Hasegawa, Y.; Yatabe, Y. Anaplastic lymphoma kinase expression in small-cell lung cancer. Histopathology 2019, 75, 20–28. [Google Scholar] [CrossRef]
- Wang, M.; Liu, H.; Zhang, R.; Li, R.; Qin, X.; Ning, F.; Tian, L. Small cell lung cancer with EML4-ALK fusion: Report of a case responding to ALK TKI and literature review. J. Cancer Res. Clin. Oncol. 2025, 151, 62. [Google Scholar] [CrossRef]
- Jiang, H.; Zhu, T.; Chang, Z.; Liu, Z.; Ou, W.; Wang, S. A Recurrent Small Cell Lung Carcinoma Harboring an EML4-ALK Fusion Mutation with Sustained Response to Ensartinib: A Case Report. Curr. Oncol. 2025, 32, 163. [Google Scholar] [CrossRef]
- Lee, A.T.M.; Ou, S.I. Overcoming Central β-Sheet #6 (Cβ6) ALK Mutation (L1256F), TP53 Mutations and Short Forms of EML4-ALK v3/b and v5a/b Splice Variants are the Unmet Need That a Re-Imagined 5th-Generation (5G) ALK TKI Must Deliver. Lung Cancer Targets Ther. 2024, 15, 19–27. [Google Scholar] [CrossRef]
- Parikh, K.; Dimou, A.; Leventakos, K.; Mansfield, A.S.; Shanshal, M.; Wan, Y.; Lin, H.M.; Vincent, S.; Elliott, J.; Bonta, I.R. Impact of EML4-ALK Variants and Co-Occurring TP53 Mutations on Duration of First-Line ALK Tyrosine Kinase Inhibitor Treatment and Overall Survival in ALK Fusion-Positive NSCLC: Real-World Outcomes from the GuardantINFORM database. J. Thorac. Oncol. 2024, 19, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Elshatlawy, M.; Sampson, J.; Clarke, K.; Bayliss, R. EML4-ALK biology and drug resistance in non-small cell lung cancer: A new phase of discoveries. Mol. Oncol. 2023, 17, 950–963. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.I.; Nagasaka, M. Catalog of 5’ Fusion Partners in ALK-positive NSCLC Circa 2020. JTO Clin. Res. Rep. 2020, 1, 100015. [Google Scholar] [CrossRef]
- Bearz, A.; Bertoli, E.; Stanzione, B.; De Carlo, E.; Del Conte, A.; Bortolot, M.; Torresan, S.; Berto, E.; Da Ros, V.; Pelin, G.M.; et al. EML4-ALK: Update on ALK Inhibitors. Int. J. Mol. Sci. 2025, 26, 308. [Google Scholar] [CrossRef]
- Childress, M.A.; Himmelberg, S.M.; Chen, H.; Deng, W.; Davies, M.A.; Lovly, C.M. ALK Fusion Partners Impact Response to ALK Inhibition: Differential Effects on Sensitivity, Cellular Phenotypes, and Biochemical Properties. Mol. Cancer Res. 2018, 16, 1724–1736. [Google Scholar] [CrossRef] [PubMed]
- Tabbò, F.; Muscarella, L.A.; Gobbini, E.; Trombetta, D.; Castellana, S.; Rigutto, A.; Galetta, D.; Maiello, E.; Martelli, O.; Tiseo, M.; et al. Detection of ALK fusion variants by RNA-based NGS and clinical outcome correlation in NSCLC patients treated with ALK-TKI sequences. Eur. J. Cancer. 2022, 174, 200–211. [Google Scholar] [CrossRef]
- Barberis, M.; Rappa, A.; de Marinis, F.; Pelosi, G.; Guerini Rocco, E.; Zhan, Y.; Tiana, G. A rationale for the poor response to alectinib in a patient with adenocarcinoma of the lung harboring a STRN-ALK fusion by artificial intelligence and molecular modelling: A case report. Transl. Lung Cancer Res. 2024, 13, 3807–3814. [Google Scholar] [CrossRef]
- Su, C.; Jiang, Y.; Jiang, W.; Wang, H.; Liu, S.; Shao, Y.; Zhao, W.; Ning, R.; Yu, Q. STRN-ALK Fusion in Lung Adenocarcinoma with Excellent Response upon Alectinib Treatment: A Case Report and Literature Review. Onco Targets Ther. 2020, 13, 12515–12519. [Google Scholar] [CrossRef]
- Siblini, L.; Schott, R.; Trensz, P.; Pencreach, E.; Bender, L. Primary resistance to ALK inhibitors in KLC1/ALK-rearranged pleural metastatic lung adenocarcinoma: A case report. Transl. Lung Cancer Res. 2023, 12, 2342–2346. [Google Scholar] [CrossRef]
- Xiang, Y.; Zhang, S.; Fang, X.; Jiang, Y.; Fang, T.; Liu, J.; Lu, K. Therapeutic Advances of Rare ALK Fusions in Non-Small Cell Lung Cancer. Curr. Oncol. 2022, 29, 7816–7831. [Google Scholar] [CrossRef]
- Li, M.; An, Z.; Tang, Q.; Ma, Y.; Yan, J.; Chen, S.; Wang, Y. Mixed responses to first-line alectinib in non-small cell lung cancer patients with rare ALK gene fusions: A case series and literature review. J. Cell Mol. Med. 2021, 25, 9476–9481. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Gu, D.; Lu, H.; Liu, S.; Kong, J. Coexistence of a Novel PRKCB-ALK, EML4-ALK Double-Fusion in a Lung Adenocarcinoma Patient and Response to Crizotinib. J. Thorac. Oncol. 2019, 14, e266–e268. [Google Scholar] [CrossRef]
- Ning, S.; Shi, C.; Zhang, H.; Li, J. Identification of triple gene fusion ALK-LRRN2, LTBP1-ALK, and HIP1-ALK in advanced lung adenocarcinoma and response to alectinib: A case report. Medicine 2021, 100, e27999. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, M.; Bai, L.; Niu, Y.; Wang, X.; Jiang, R.; Wang, Y.; Feng, Q.; Wang, B.; Dai, T.; et al. Coexistence of a novel SV2B-ALK, EML4-ALK double-fusion in a lung poorly differentiated adenocarcinoma patient and response to alectinib: A case report and literature review. Front. Oncol. 2024, 14, 1453259. [Google Scholar] [CrossRef]
- Zhu, L.; Qin, J. Coexistence of a novel SETD2-ALK, EML4-ALK double-fusion in an advanced lung adenocarcinoma patient after alectinib resistant and response to immunotherapy combined with chemotherapy: A case report. Discov. Oncol. 2023, 14, 44. [Google Scholar] [CrossRef]
- Wang, Z.; Luo, Y.; Gong, H.; Chen, Y.; Tang, H. A novel double fusion of EML4-ALK and PLEKHA7-ALK contribute to rapid progression of lung adenocarcinoma: A case report and literature review. Discov. Oncol. 2024, 15, 638. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, M.; Harada, G.; Ghanem, P.; Bubie, A.; Kiedrowski, L.A.; Murray, J.C.; Marrone, K.A.; Scott, S.C.; Houseknecht, S.; Falcon, C.J.; et al. Impact of Tumor-intrinsic Molecular Features on Survival and Acquired Tyrosine Kinase Inhibitor Resistance in ALK-positive NSCLC. Cancer Res. Commun. 2024, 4, 786–795. [Google Scholar] [CrossRef]
- Christopoulos, P.; Kirchner, M.; Bozorgmehr, F.; Endris, V.; Elsayed, M.; Budczies, J.; Ristau, J.; Penzel, R.; Herth, F.J.; Heussel, C.P.; et al. Identification of a highly lethal V3+ TP53+ subset in ALK+ lung adenocarcinoma. Int. J. Cancer 2018, 144, 190–199. [Google Scholar] [CrossRef]
- Bearz, A.; Martini, J.F.; Jassem, J.; Kim, S.W.; Chang, G.C.; Shaw, A.T.; Shepard, D.A.; Dall’O’, E.; Polli, A.; Thurm, H.; et al. Efficacy of Lorlatinib in Treatment-Naive Patients with ALK-Positive Advanced NSCLC in Relation to EML4::ALK Variant Type and ALK With or Without TP53 Mutations. J. Thorac. Oncol. 2023, 18, 1581–1593. [Google Scholar] [CrossRef]
- Lucena-Araujo, A.R.; Moran, J.P.; VanderLaan, P.A.; Dias-Santagata, D.; Folch, E.; Majid, A.; Kent, M.S.; Gangadharan, S.P.; Rangachari, D.; Huberman, M.S.; et al. De novo ALK kinase domain mutations are uncommon in kinase inhibitor-naïve ALK rearranged lung cancers. Lung Cancer 2016, 99, 17–22. [Google Scholar] [CrossRef]
- Zhao, J.; Li, X.; Fan, R.; Qin, Y.; Wang, Z.; Wang, B.; Li, S.; Fan, J.; Wu, X.; Liu, H.; et al. Primary resistance to first- and second-generation ALK inhibitors in a non-small cell lung cancer patient with coexisting ALK rearrangement and an ALK F1174L-cis-S1189C de novo mutation: A case report. Front. Pharmacol. 2022, 13, 1060460. [Google Scholar] [CrossRef] [PubMed]
- Lara-Mejía, L.; Cardona, A.F.; Mas, L.; Martin, C.; Samtani, S.; Corrales, L.; Cruz-Rico, G.; Remon, J.; Galvez-Nino, M.; Ruiz, R.; et al. Impact of Concurrent Genomic Alterations on Clinical Outcomes in Patients with ALK-Rearranged NSCLC. J. Thorac. Oncol. 2024, 19, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhu, Y.; Sun, K.; Shen, Q.; Wang, Y.; Cao, H.; Lizaso, A.; Yu, B.; Lin, J.; Chen, S.; et al. Investigation on the prognostic impact of concurrent genomic alterations in crizotinib-treated EML4-ALK-rearranged advanced non-small cell lung cancer patients. Lung Cancer 2020, 146, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, K.; Ji, H.; Qian, J.; Lu, H.; Zhang, Y.; Russo, A.; Romero, A.; Urbanska, E.M.; Tabbò, F.; et al. Efficacy of alectinib in lung adenocarcinoma patients with different anaplastic lymphoma kinase (ALK) rearrangements and co-existing alterations-a retrospective cohort study. Transl. Lung Cancer Res. 2023, 12, 2505–2519. [Google Scholar] [CrossRef]
- Zhang, S.S.; Nagasaka, M.; Zhu, V.W.; Ou, S.I. Going beneath the tip of the iceberg. Identifying and understanding EML4-ALK variants and TP53 mutations to optimize treatment of ALK fusion positive (ALK+) NSCLC. Lung Cancer 2021, 158, 126–136. [Google Scholar] [CrossRef]
- Tanimoto, A.; Matsumoto, S.; Takeuchi, S.; Arai, S.; Fukuda, K.; Nishiyama, A.; Yoh, K.; Ikeda, T.; Furuya, N.; Nishino, K.; et al. Proteasome Inhibition Overcomes ALK-TKI Resistance in ALK-Rearranged/TP53-Mutant NSCLC via Noxa Expression. Clin. Cancer Res. 2021, 27, 1410–1420. [Google Scholar] [CrossRef]
- Cipriano, É.; Magalhães, H.; Tavares, C.; Pinto, J.; Cirnes, L.; Estevinho, F. Concurrent EGFR mutation and ALK rearrangement in stage IV lung adenocarcinoma-a case report and a literature review. Porto Biomed. J. 2021, 11, e124. [Google Scholar] [CrossRef]
- Schmid, S.; Gautschi, O.; Rothschild, S.; Mark, M.; Froesch, P.; Klingbiel, D.; Reichegger, H.; Jochum, W.; Diebold, J.; Früh, M. Clinical Outcome of ALK-Positive Non-Small Cell Lung Cancer (NSCLC) Patients with De Novo EGFR or KRAS Co-Mutations Receiving Tyrosine Kinase Inhibitors (TKIs). J. Thorac. Oncol. 2017, 12, 681–688. [Google Scholar] [CrossRef]
- Urbanska, E.M.; Grauslund, M.; Berger, S.M.S.; Costa, J.C.; Koffeldt, P.R.; Sørensen, J.B.; Santoni-Rugiu, E. ALK-tyrosine kinase inhibitor intrinsic resistance due to de novo MET-amplification in metastatic ALK-rearranged non-small cell lung cancer effectively treated by alectinib-crizotinib combination-case report. Transl. Lung Cancer Res. 2024, 13, 2453–2462. [Google Scholar] [CrossRef]
- Hu, J.; Ding, N.; Xu, X.; Chen, Y.; Zhang, Y.; Liu, J.; Zhou, J.; Bao, H.; Zhang, D.; Song, Y.; et al. MET and NF2 alterations confer primary and early resistance to first-line alectinib treatment in ALK-positive non-small-cell lung cancer. Mol. Oncol. 2025. [Google Scholar] [CrossRef]
- Solomon, B.J.; Bauer, T.M.; Mok, T.S.K.; Liu, G.; Mazieres, J.; de Marinis, F.; Goto, Y.; Kim, D.W.; Wu, Y.L.; Jassem, J.; et al. Efficacy and safety of first-line lorlatinib versus crizotinib in patients with advanced, ALK-positive non-small-cell lung cancer: Updated analysis of data from the phase 3, randomised, open-label CROWN study. Lancet Respir. Med. 2023, 11, 354–366. [Google Scholar] [CrossRef]
- Dziadziuszko, R.; Peters, S.; Mok, T.; Camidge, D.R.; Gadgeel, S.M.; Ou, S.I.; Konopa, K.; Noé, J.; Nowicka, M.; Bordogna, W.; et al. Circulating Cell-free DNA as a Prognostic Biomarker in Patients with Advanced ALK+ Non-small Cell Lung Cancer in the Global Phase III ALEX Trial. Clin. Cancer Res. 2022, 28, 1800–1808. [Google Scholar] [CrossRef] [PubMed]
- Camidge, D.R.; Dziadziuszko, R.; Peters, S.; Mok, T.; Noe, J.; Nowicka, M.; Gadgeel, S.M.; Cheema, P.; Pavlakis, N.; de Marinis, F.; et al. Updated Efficacy and Safety Data and Impact of the EML4-ALK Fusion Variant on the Efficacy of Alectinib in Untreated ALK-Positive Advanced Non-Small Cell Lung Cancer in the Global Phase III ALEX Study. J. Thorac. Oncol. 2019, 14, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Noé, J.; Bordogna, W.; Archer, V.; Smoljanovic, V.; Hilton, M.; Woodhouse, R.; Mocci, S.; Gadgeel, S.M. Concordance Between Tissue ALK Detection by Immunohistochemistry and Plasma ALK Detection by Next-Generation Sequencing in the Randomized Phase 3 ALEX Study in Patients with Treatment-Naive Advanced ALK-Positive NSCLC. JTO Clin. Res. Rep. 2022, 3, 100341. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.B.; Ou, S.-H.I. Plasma Genotyping from the CROWN, ALTA-1L, and ALEX Trials: Can We Speak with One Voice on What to Test, How to Test, When to Test, and for What Purpose? J. Thorac. Oncol. 2023, 18, 1434–1442. [Google Scholar] [CrossRef]
- McCoach, C.E.; Blakely, C.M.; Banks, K.C.; Levy, B.; Chue, B.M.; Raymond, V.M.; Le, A.T.; Lee, C.E.; Diaz, J.; Waqar, S.N.; et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 2758–2770. [Google Scholar] [CrossRef]
- Kwon, M.; Ku, B.M.; Olsen, S.; Park, S.; Lefterova, M.; Odegaard, J.; Jung, H.A.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; et al. Longitudinal monitoring by next-generation sequencing of plasma cell-free DNA in ALK rearranged NSCLC patients treated with ALK tyrosine kinase inhibitors. Cancer Med. 2022, 11, 2944–2956. [Google Scholar] [CrossRef]
- Choudhury, Y.; Cher, C.Y.; Ho, J.M.; Chan, C.; Teh, M.W.; Ngeow, K.C.; Pek, M. A cell-free RNA-based next-generation sequencing (NGS) assay for the detection of actionable gene fusions in patients with non–small cell lung cancer (NSCLC). J. Clin. Oncol. 2022, 40, 3040. [Google Scholar] [CrossRef]
- Heeke, S.; Gandhi, S.; Tran, H.T.; Lam, V.K.; Byers, L.A.; Gibbons, D.L.; Gay, C.M.; Altan, M.; Antonoff, M.B.; Le, X.; et al. Longitudinal Tracking of ALK-Rearranged NSCLC from Plasma Using Circulating Tumor RNA and Circulating Tumor DNA. JTO Clin. Res. Rep. 2025, 6, 100795. [Google Scholar] [CrossRef]
- Hotta, K.; Hida, T.; Nokihara, H.; Morise, M.; Kim, Y.H.; Azuma, K.; Seto, T.; Takiguchi, Y.; Nishio, M.; Yoshioka, H.; et al. Final overall survival analysis from the phase III J-ALEX study of alectinib versus crizotinib in ALK inhibitor-naïve Japanese patients with ALK-positive non-small-cell lung cancer. ESMO Open 2022, 7, 100527. [Google Scholar] [CrossRef]
- Ou, S.H.; Kilvert, H.; Candlish, J.; Lee, B.; Polli, A.; Thomaidou, D.; Le, H. Systematic review and network meta-analysis of lorlatinib with comparison to other anaplastic lymphoma kinase (ALK) tyrosine kinase inhibitors (TKIs) as first-line treatment for ALK-positive advanced non-small cell lung cancer (NSCLC). Lung Cancer 2024, 197, 107968. [Google Scholar] [CrossRef]
- Gibson, A.J.W.; Box, A.; Dean, M.L.; Elegbede, A.A.; Hao, D.; Sangha, R.; Bebb, D.G. Retrospective Real-World Outcomes for Patients with ALK-Rearranged Lung Cancer Receiving ALK Receptor Tyrosine Kinase Inhibitors. JTO Clin. Res. Rep. 2021, 2, 100157. [Google Scholar] [CrossRef]
- Wang, M.; Slatter, S.; Sussell, J.; Lin, C.W.; Ogale, S.; Datta, D.; Butte, A.J.; Bazhenova, L.; Rudrapatna, V.A. ALK Inhibitor Treatment Patterns and Outcomes in Real-World Patients with ALK-Positive Non-Small-Cell Lung Cancer: A Retrospective Cohort Study. Target. Oncol. 2023, 18, 571–583. [Google Scholar] [CrossRef]
- Dhamelincourt, E.; Descourt, R.; Rousseau-Bussac, G.; Doubre, H.; Decroisette, C.; Demontrond, P.; Le Garff, G.; Falchero, L.; Huchot, E.; Vieillot, S.; et al. Clinical Characteristics of Patients with Advanced ALK-Translocated Non-small Cell Lung Cancers and Long-Term Responses to Crizotinib (CRIZOLONG GFPC 05-19 Study). Target. Oncol. 2023, 18, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, T.; Yajima, T.; Yamaki, E.; Nakazawa, S.; Tomizawa, K.; Onozato, R.; Yamazaki, A.; Hirato, J.; Yatabe, Y.; Shimizu, K.; et al. Long-term complete response in a patient with postoperative recurrent ALK-rearranged lung adenocarcinoma treated with crizotinib: A case report. Mol. Clin. Oncol. 2019, 11, 309–312. [Google Scholar] [CrossRef]
- Emirzeoglu, L.; Olmez, O. ALK-positive locally advanced lung cancer in a patient who achieved long-term complete response with crizotinib: A case report. Exp. Ther. Med. 2022, 24, 650. [Google Scholar] [CrossRef] [PubMed]
- Jimenez Munarriz, B.E.; Khan, S.; Li, Y.; Ghazali, N.; Liu, G. Update advances in anaplastic lymphoma kinase-positive non-small cell lung cancer treatment. Cancer 2025, 131 (Suppl. S1), e35786. [Google Scholar] [CrossRef] [PubMed]
- Chayab, L.; Leighl, N.B.; Tadrous, M.; Warren, C.M.; Wong, W.W.L. Trends in Real-World Clinical Outcomes of Patients with Anaplastic Lymphoma Kinase (ALK) Rearranged Non-Small Cell Lung Cancer (NSCLC) Receiving One or More ALK Tyrosine Kinase Inhibitors (TKIs): A Cohort Study in Ontario, Canada. Curr. Oncol. 2024, 32, 13. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kapoor, A.; Noronha, V.; Patil, V.; Menon, N.; Mahajan, A.; Janu, A.; Purandare, N.; Kaushal, R.; Prabhash, K. ALK-positive advanced non-small cell lung cancer patients with poor performance status: Outcomes in a real-world scenario. Ecancermedicalscience 2022, 16, 1407. [Google Scholar] [CrossRef]
- Cameron, L.B.; Hitchen, N.; Chandran, E.; Morris, T.; Manser, R.; Solomon, B.J.; Jordan, V. Targeted therapy for advanced anaplastic lymphoma kinase (<I>ALK</I>)-rearranged non-small cell lung cancer. Cochrane Database Syst. Rev. 2022, 1, CD013453. [Google Scholar] [CrossRef]
- Mok, T.; Camidge, D.R.; Gadgeel, S.M.; Rosell, R.; Dziadziuszko, R.; Kim, D.W.; Pérol, M.; Ou, S.I.; Ahn, J.S.; Shaw, A.T.; et al. Updated overall survival and final progression-free survival data for patients with treatment-naive advanced ALK-positive non-small-cell lung cancer in the ALEX study. Ann. Oncol. 2020, 31, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Rooney, M.; Lin, J.J.; Nagy, R.J.; Yeap, B.Y.; Hubbeling, H.; Chin, E.; Ackil, J.; Farago, A.F.; Hata, A.N.; et al. Treatment with Next-Generation ALK Inhibitors Fuels Plasma ALK Mutation Diversity. Clin. Cancer Res. 2019, 25, 6662–6670. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.J.; Horan, J.C.; Tangpeerachaikul, A.; Swalduz, A.; Valdivia, A.; Johnson, M.L.; Besse, B.; Camidge, D.R.; Fujino, T.; Yoda, S.; et al. NVL-655 Is a Selective and Brain-Penetrant Inhibitor of Diverse ALK-Mutant Oncoproteins, Including Lorlatinib-Resistant Compound Mutations. Cancer Disc. 2024, 14, 2367–2386. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Thorac. Oncol. 2018, 13, 323–358. [Google Scholar] [CrossRef]
- Kalemkerian, G.P.; Narula, N.; Kennedy, E.B.; Biermann, W.A.; Donington, J.; Leighl, N.B.; Lew, M.; Pantelas, J.; Ramalingam, S.S.; Reck, M.; et al. Molecular Testing Guideline for the Selection of Patients with Lung Cancer for Treatment with Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J. Clin. Oncol. 2018, 36, 911–919. [Google Scholar] [CrossRef]
- Pennell, N.A.; Arcila, M.E.; Gandara, D.R.; Howard, W. Biomarker Testing for Patients with Advanced Non-Small Cell Lung Cancer: Real-World Issues and Tough Choices. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 531–542. [Google Scholar] [CrossRef]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef]
- Hallberg, B.; Palmer, R.H. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat. Rev. Cancer. 2013, 13, 685–700. [Google Scholar] [CrossRef]
- GTEx Portal. Available online: https://www.gtexportal.org/home/gene/ALK (accessed on 30 April 2025).
- Vilachã, J.F.; Wassenaar, T.A.; Marrink, S.J. Structural Aspects of the ROS1 Kinase Domain and Oncogenic Mutations. Crystals 2024, 14, 106. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. ROS1 protein-tyrosine kinase inhibitors in the treatment of ROS1 fusion protein-driven non-small cell lung cancers. Pharmacol. Res. 2017, 12, 202–212. [Google Scholar] [CrossRef]
- Mescam-Mancini, L.; Lantuéjoul, S.; Moro-Sibilot, D.; Rouquette, I.; Souquet, P.J.; Audigier-Valette, C.; Sabourin, J.C.; Decroisette, C.; Sakhri, L.; Brambilla, E.; et al. On the relevance of a testing algorithm for the detection of ROS1-rearranged lung adenocarcinomas. Lung Cancer 2014, 83, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Nambirajan, A.; Singh, V.; Rana, D.; Malik, P.; Mohan, A.; Jain, D. P59.27 Complementary Utility of Combined ALK/ROS1 FISH with Immunohistochemistry for ALK/ROS1 Rearrangement Testing in Lung Cancer. J. Thorac. Oncol. 2021, 6, S1160. [Google Scholar] [CrossRef]
- Xia, P.; Zhang, L.; Li, P.; Liu, E.; Li, W.; Zhang, J.; Li, H.; Su, X.; Jiang, G. Molecular characteristics and clinical outcomes of complex ALK rearrangements identified by next-generation sequencing in non-small cell lung cancers. J. Transl. Med. 2021, 19, 308. [Google Scholar] [CrossRef] [PubMed]
- Smeltzer, M.P.; Wynes, M.W.; Lantuejoul, S.; Soo, R.; Ramalingam, S.S.; Varella-Garcia, M.; Meadows Taylor, M.; Richeimer, K.; Wood, K.; Howell, K.E.; et al. The international association for the study of lung cancer (IASLC) global survey of molecular testing in lung cancer. J. Thor. Oncol. 2020, 15, 1434–1448. [Google Scholar] [CrossRef]
- Ferreira, C.G. Lung cancer in developing countries: Access to molecular testing. Am. Soc. Clin. Oncol. Educ. Book 2013, 33, 327–331. [Google Scholar] [CrossRef]
- Hendriks, L.E.; Kerr, K.M.; Menis, J.; Mok, T.S.; Nestle, U.; Passaro, A.; Peters, S.; Planchard, D.; Smit, E.F.; Solomon, B.J.; et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 339–357. [Google Scholar] [CrossRef]
- NCCN Guidelines. Version 3.2025. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf (accessed on 30 April 2025).
- Mok, T.; Solomon, B.J.; Campelo, M.R.G.; Wu, Y.-L.; Streich, G.; Zemanova, M.; Zalcman, G.; Bearz, A.; Chang, G.-C.; Setti, M.; et al. MA06.07 Patterns of Progression with Lorlatinib and Insights into Subsequent Anticancer Therapy Efficacy in Advanced ALK+ NSCLC. J. Thorac. Oncol. 2024, 19, S75. [Google Scholar] [CrossRef]
- Huang, X.; Zhou, L.; Xia, J.; Jian, H.; Liu, J.; Huang, Y.; Chen, Q. Ensartinib for EML4-ALK-positive lung adenocarcinoma with comorbid mutations in TP53, EGFR, and ERBB2: A case report. Front. Oncol. 2025, 15, 1520287. [Google Scholar] [CrossRef]
- Hill, L.; Reddy, P.; Gouffon, J.; Mertins, B.; Sanchez, E.; Selvaggi, G.; Bulow, C. Predictors of long-term ensartinib response from the eXalt3 trial. In Proceedings of the 2024 World Conference on Lung Cancer, San Diego, CA, USA, 7–10 September 2024. [Google Scholar]
- Luukkainen, M.E.K.; Koivunen, J.P. ALK Inhibitor and Chemotherapy Combinations in Models of ALK-Translocated NSCLC. Anticancer. Res. 2024, 44, 2805–2813. [Google Scholar] [CrossRef]
- Planchard, D.; Jänne, P.A.; Cheng, Y.; Yang, J.C.; Yanagitani, N.; Kim, S.W.; Sugawara, S.; Yu, Y.; Fan, Y.; Geater, S.L.; et al. Osimertinib with or without Chemotherapy in EGFR-Mutated Advanced NSCLC. N. Engl. J. Med. 2023, 389, 1935–1948. [Google Scholar] [CrossRef]
- Urbanska, E.M.; Sørensen, J.B.; Santoni-Rugiu, E. Finding One Treatment for All Advanced EGFR-positive NSCLC—An Infinite Task. J. Thorac. Oncol. 2024, 19, e11–e12. [Google Scholar] [CrossRef]
- Wakuda, K.; Kenmotsu, H.; Sato, Y.; Nakamura, A.; Akamatsu, H.; Tachihara, M.; Miura, S.; Yokoyama, T.; Mori, K.; Nakagawa, K.; et al. Randomized, open-label phase II study of brigatinib and carboplatin plus pemetrexed and brigatinib alone for chemotherapy-naive patients with ALK-rearranged non-squamous non-small cell lung cancer: Treatment rationale and protocol design of the B-DASH study (WJOG 14720 L). BMC Cancer. 2023, 23, 902. [Google Scholar] [CrossRef]
- Leporati, R.; Auclin, É.; Morchón, D.; Ferriol-Galmés, M.; Laguna, J.C.; Gorria, T.; Teixidó, C.; Aranzazu Amores, M.; Ambrosini, P.; Isla, D.; et al. Sex differences in patients with Non-Small Cell Lung Cancer harboring driver fusions treated with tyrosine kinase inhibitors: A systematic review. Ther. Adv. Med. Oncol. 2024, 16, 17588359241306940. [Google Scholar] [CrossRef] [PubMed]
- Katayama, R.; Sakashita, T.; Yanagitani, N.; Ninomiya, H.; Horiike, A.; Friboulet, L.; Gainor, J.F.; Motoi, N.; Dobashi, A.; Sakata, S.; et al. P-glycoprotein Mediates Ceritinib Resistance in Anaplastic Lymphoma Kinase-rearranged Non-small Cell Lung Cancer. eBioMedicine 2015, 3, 54–66. [Google Scholar] [CrossRef]
- Liu, Y.N.; Chen, J.; Wang, J.; Li, Q.; Hu, G.X.; Cai, J.P.; Lin, G.; Xu, R.A. Effects of drug-drug interactions and CYP3A4 variants on alectinib metabolism. Arch. Toxicol. 2023, 97, 2133–2142. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Hanley, M.J.; Griffin, R.J.; Zhang, P.; Venkatakrishnan, K.; Sinha, V. Clinical Pharmacology of Brigatinib: A Next-Generation Anaplastic Lymphoma Kinase Inhibitor. Clin. Pharmacokinet. 2023, 62, 1063–1079. [Google Scholar] [CrossRef]
- Chen, J.; Bearz, A.; Kim, D.W.; Mamdani, H.; Bauman, J.; Chiari, R.; Ou, S.I.; Solomon, B.J.; Soo, R.A.; Felip, E.; et al. Evaluation of the Effect of Lorlatinib on CYP2B6, CYP2C9, UGT, and P-Glycoprotein Substrates in Patients with Advanced Non-Small Cell Lung Cancer. Clin. Pharmacokinet. 2024, 63, 171–182. [Google Scholar] [CrossRef]
- Alexander, M.; Wei, J.; Parakh, S.; John, T.; Kao, S.; Nagrial, A.; Bowyer, S.; Warburton, L.; Moore, M.; Hughes, B.G.M.; et al. LOREALAUS: LOrlatinib REAL-World AUStralian Experience in Advanced ALK-Rearranged NSCLC. JTO Clin. Res. Rep. 2023, 4, 100490. [Google Scholar] [CrossRef]
- Baldacci, S.; Besse, B.; Avrillon, V.; Mennecier, B.; Mazieres, J.; Dubray-Longeras, P.; Cortot, A.B.; Descourt, R.; Doubre, H.; Quantin, X.; et al. Lorlatinib for advanced anaplastic lymphoma kinase-positive non-small cell lung cancer: Results of the IFCT-1803 LORLATU cohort. Eur. J. Cancer. 2022, 166, 51–59. [Google Scholar] [CrossRef]
- Nakashima, K.; Demura, Y.; Kurokawa, K.; Takeda, T.; Jikuya, N.; Oi, M.; Tada, T.; Akai, M.; Ishizuka, T. Successful treatment with lorlatinib in a patient with meningeal carcinomatosis of ALK-positive non-small cell lung cancer resistant to alectinib and brigatinib: A case report. Medicine 2021, 100, e227385. [Google Scholar] [CrossRef]
- Pan, Y.; Zeng, Y.; Peng, Y.; Liu, X.; Li, Y.; Wu, F. Dramatic response to brigatinib in a lung adenocarcinoma patient harboring EML4-ALK fusion and a G1202R de novo gene mutation. Eur. J. Cancer. 2022, 165, 154–156. [Google Scholar] [CrossRef]
- Popat, S.; Brustugun, O.T.; Cadranel, J.; Felip, E.; Garassino, M.C.; Griesinger, F.; Helland, Å.; Hochmair, M.; Pérol, M.; Bent-Ennakhil, N.; et al. Real-world treatment outcomes with brigatinib in patients with pretreated ALK+ metastatic non-small cell lung cancer. Lung Cancer 2021, 157, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Descourt, R.; Perol, M.; Rousseau-Bussac, G.; Planchard, D.; Mennecier, B.; Wislez, M.; Cortot, A.; Guisier, F.; Galland, L.; Dô, P.; et al. Brigatinib in patients with ALK-positive advanced non-small-cell lung cancer pretreated with sequential ALK inhibitors: A multicentric real-world study (BRIGALK study). Lung Cancer 2019, 136, 109–114. [Google Scholar] [CrossRef]
- Cheung, J.M.; Kang, J.; Yeap, B.Y.; Peterson, J.L.; Do, A.; Gainor, J.F.; Digumarthy, S.R.; Lin, J.J. Efficacy and Safety of Dose-Escalated Alectinib in Patients with Metastatic ALK-Positive NSCLC and Central Nervous System Relapse on Standard-Dose Alectinib. JTO Clin. Res. Rep. 2024, 5, 100645. [Google Scholar] [CrossRef]
- Ohara, G.; Sasatani, Y.; Okauchi, S.; Satoh, H. Five-year Disease Control with Alectinib in a Patient with Metastatic ALK-rearranged Lung Adenocarcinoma. Cancer Diagn. Progn. 2022, 2, 707–710. [Google Scholar] [CrossRef]
- Huang, M.; Zhu, X.; Xu, W.; Zhu, J.; Xun, X.; Su, B.; Chen, H. TTC7A-ALK, a novel ALK fusion variant identified in a patient with metastatic lung adenocarcinoma, exhibits excellent response to crizotinib. Transl. Oncol. 2025, 54, 102345. [Google Scholar] [CrossRef]
- Wang, S.; Hao, X.; Dai, L.; Lou, N.; Fan, G.; Gao, R.; Yang, M.; Xing, P.; Liu, Y.; Wang, L.; et al. Longitudinal plasma proteomic profiling of EML4-ALK positive lung cancer receiving ALK-TKIs therapy. Lung Cancer 2024, 189, 107503. [Google Scholar] [CrossRef] [PubMed]
- Christopoulos, P.; Harel, M.; McGregor, K.; Brody, Y.; Puzanov, I.; Bar, J.; Elon, Y.; Sela, I.; Yellin, B.; Lahav, C.; et al. Plasma Proteome-Based Test for First-Line Treatment Selection in Metastatic Non-Small Cell Lung Cancer. JCO Precis. Oncol. 2024, 8, e2300555. [Google Scholar] [CrossRef] [PubMed]
- Agius, R.; Brieghel, C.; Andersen, M.A.; Pearson, A.T.; Ledergerber, B.; Cozzi-Lepri, A.; Louzoun, Y.; Andersen, C.L.; Bergstedt, J.; von Stemann, J.H.; et al. Machine learning can identify newly diagnosed patients with CLL at high risk of infection. Nat. Commun. 2020, 11, 363. [Google Scholar] [CrossRef]
- Solomon, B.J.; Mok, T.; Kim, D.W.; Wu, Y.L.; Nakagawa, K.; Mekhail, T.; Felip, E.; Cappuzzo, F.; Paolini, J.; Usari, T.; et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med. 2014, 371, 2167–2177. [Google Scholar] [CrossRef]
- Qi, R.; Yu, Y.; Shen, M.; Lv, D.; He, S. Current status and challenges of immunotherapy in ALK rearranged NSCLC. Front Oncol. 2022, 12, 1016869. [Google Scholar] [CrossRef] [PubMed]
- Gainor, J.F.; Shaw, A.T.; Sequist, L.V.; Fu, X.; Azzoli, C.G.; Piotrowska, Z.; Huynh, T.G.; Zhao, L.; Fulton, L.; Schultz, K.R.; et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin. Cancer Res. 2016, 22, 4585–4593. [Google Scholar] [CrossRef] [PubMed]
- Mhanna, L.; Guibert, N.; Milia, J.; Mazieres, J. When to Consider Immune Checkpoint Inhibitors in Oncogene-Driven Non-Small Cell Lung Cancer? Curr. Treat. Options Oncol. 2019, 20, 60. [Google Scholar] [CrossRef]
- Zhai, X.; Wang, M.; Zhang, Q.; Li, D.; Wu, Y.; Liang, Z.; Liu, J.; Wang, W.; Liu, Y.; Che, G.; et al. Identifying the Intergenic ALK Fusion LOC388942-ALK as a Driver of Non-Small Cell Lung Cancer. Med. Comm. 2025, 6, e70154. [Google Scholar] [CrossRef] [PubMed]
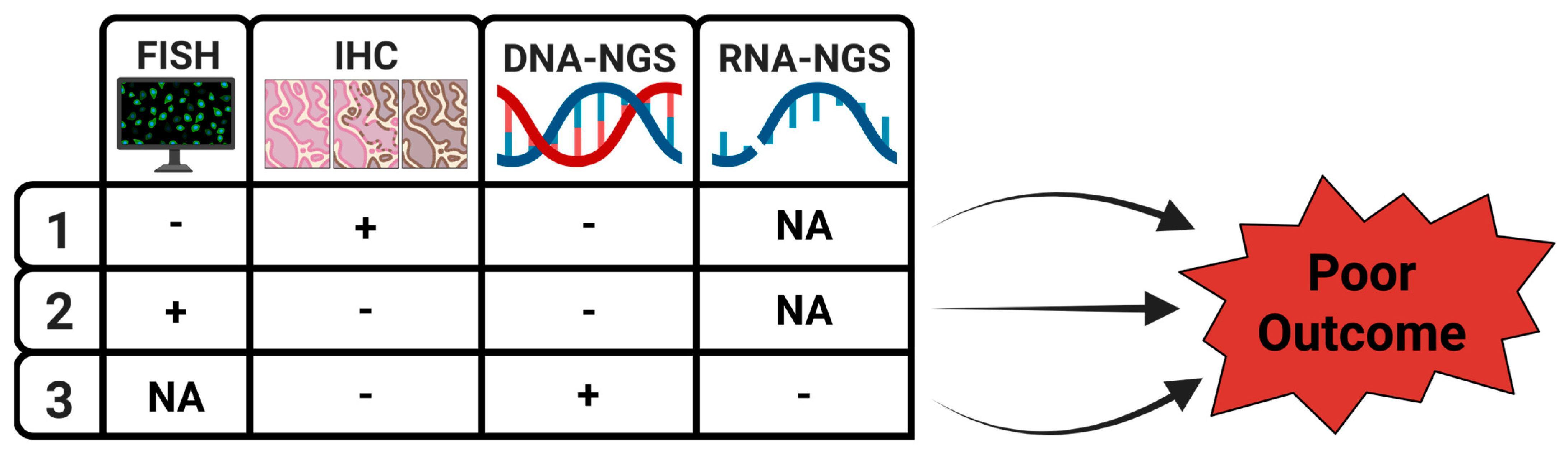
| Favorable Features | Unfavorable Features | References | |
|---|---|---|---|
| 1 | Concordant IHC, FISH, and NGS | Discordant IHC, FISH, and NGS | [9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26] |
| 2 | Pure LUAD | LUAD combined with other histological types * | [27,28,29,30,31,32,33,34,35,36,37,38,39] |
| 3 | Canonical ALK fusion (except EML4-ALK v.3) | Non-canonical ALK fusion, EML4-ALK v.3, double/triple fusions | [40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59] |
| 4 | High % of ALK-rearranged tumor cells by FISH | Low % of ALK-rearranged tumor cells by FISH | [13,14,15] |
| 5 | No de novo co-alterations | De novo co-alterations (ALK, CDKN2A/B, EGFR, KRAS, MET, NF2, TP53) | [40,41,57,58,59,60,61,62,63,64,65,66,67,68,69,70] |
| 6 | No ctDNA/ctRNA | ctDNA/ctRNA | [1,8,41,71,72,73,74,75,76,77,78,79] |
| 7 | No brain metastases | Brain metastases | [1,2,6,7,8,71,80,81,82,83,84,85,86,87,88] |
| 8 | PS 0–1 | PS ≥ 2 | [6,7,8,57,71,80,82,83,87,88,89] |
| 9 | Treatment with next-generation ALK-TKIs | Treatment with Crizotinib | [1,6,7,8,44,57,87,88,90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbanska, E.M.; Koffeldt, P.R.; Grauslund, M.; Melchior, L.C.; Sørensen, J.B.; Santoni-Rugiu, E. Refining Criteria for Choosing the First-Line Treatment for Real-World Patients with Advanced ALK-Rearranged NSCLC. Int. J. Mol. Sci. 2025, 26, 5969. https://doi.org/10.3390/ijms26135969
Urbanska EM, Koffeldt PR, Grauslund M, Melchior LC, Sørensen JB, Santoni-Rugiu E. Refining Criteria for Choosing the First-Line Treatment for Real-World Patients with Advanced ALK-Rearranged NSCLC. International Journal of Molecular Sciences. 2025; 26(13):5969. https://doi.org/10.3390/ijms26135969
Chicago/Turabian StyleUrbanska, Edyta Maria, Peter Rindom Koffeldt, Morten Grauslund, Linea Cecilie Melchior, Jens Benn Sørensen, and Eric Santoni-Rugiu. 2025. "Refining Criteria for Choosing the First-Line Treatment for Real-World Patients with Advanced ALK-Rearranged NSCLC" International Journal of Molecular Sciences 26, no. 13: 5969. https://doi.org/10.3390/ijms26135969
APA StyleUrbanska, E. M., Koffeldt, P. R., Grauslund, M., Melchior, L. C., Sørensen, J. B., & Santoni-Rugiu, E. (2025). Refining Criteria for Choosing the First-Line Treatment for Real-World Patients with Advanced ALK-Rearranged NSCLC. International Journal of Molecular Sciences, 26(13), 5969. https://doi.org/10.3390/ijms26135969







