Alternative Splicing of Functional Genes in Plant Growth, Development, and Stress Responses
Abstract
1. Introduction
2. Key Genes Undergoing Alternative Splicing During Plant Growth and Development
2.1. Seed Development
2.2. Vegetative Growth and Morphogenesis
2.3. Flowering and Development
2.4. Maturation and Senescence
3. Alternative Splicing of Stress-Regulatory Genes During Plant Adaptation
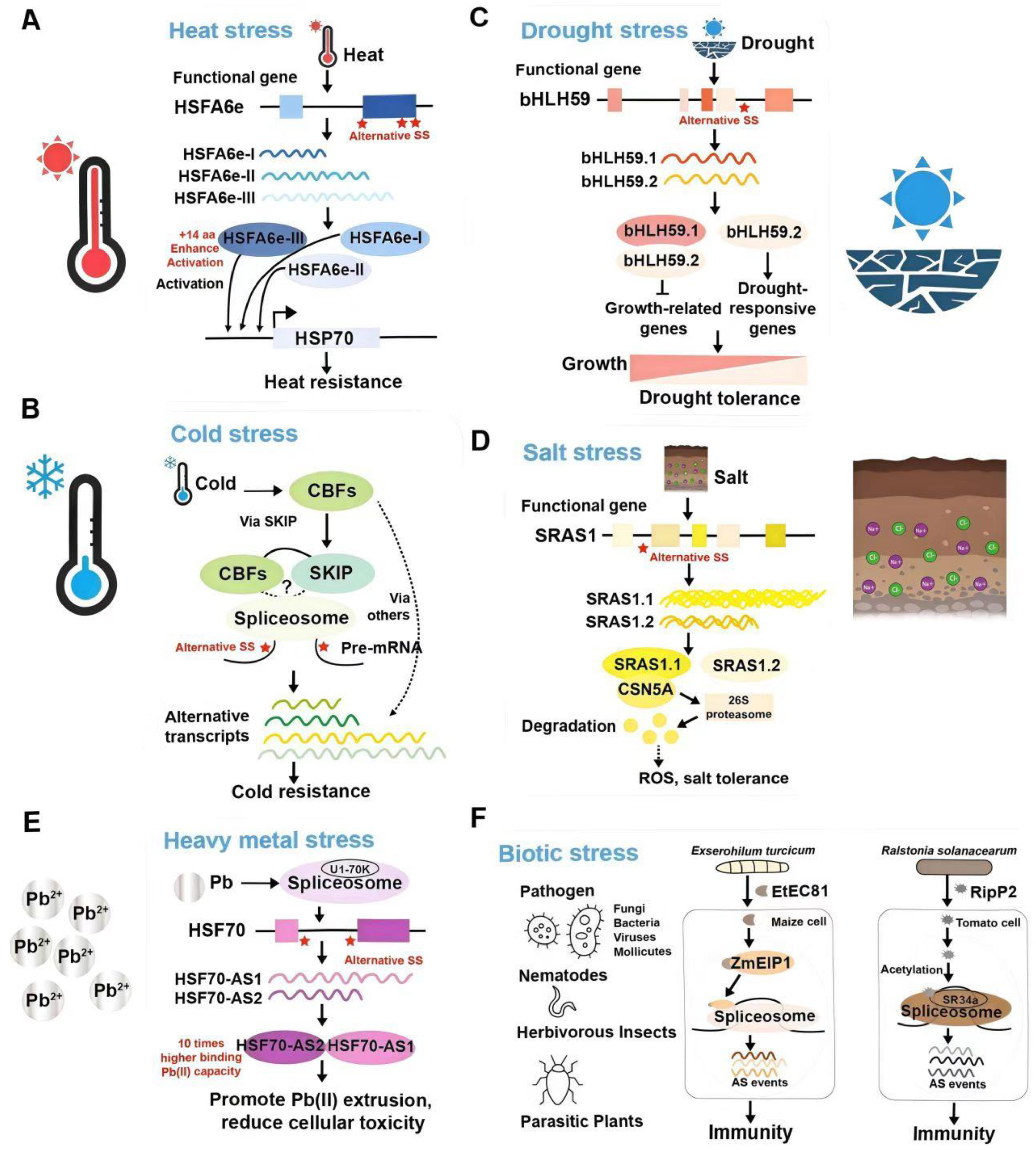
3.1. Temperature Stresses
3.2. Drought Stress
3.3. Salt Stress
3.4. Biotic Stress
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic Acid |
| AS | Alternative splicing |
| BPS | Branch point sequence |
| BR | Brassinosteroid |
| CBC | Cap-Binding Complex |
| CO | CONSTANS |
| COL | CONSTANS-LIKE |
| COR | Cold-Regulated |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| DAS | Differentially Alternative Splicing |
| DOG | DELAY OF GERMINATION |
| DRT | DNA-DAMAGE REPAIR/TOLERATION PROTEIN |
| ELF | EARLY FLOWERING |
| ERF | Ethylene Response Factor |
| ESE | Exonic Splicing Enhance |
| ESS | Exonic Splicing Silencer |
| FCA | FLOWERING CONTROL LOCUS A |
| FLC | FLOWERING LOCUS C |
| FLM | FLOWERING LOCUS M |
| FT | FLOWERING LOCUS T |
| HAB1 | HYPERSENSITIVE TO ABA1 |
| hnRNP | Heterogeneous Nuclear Ribonucleoprotein |
| HSF | Heat Shock Transcription Factor |
| HSP | Heat Shock Protein |
| ICE | Inducer of CBF Expression |
| IR | Intron Retention |
| ISE | Intronic Splicing Enhancer |
| ISS | Intronic Splicing Silencer |
| MAF | MADS AFFECTING FLOWERING |
| NMD | Nonsense-Mediated Decay |
| NTC | NineTeen Complex |
| NTR | NTC-Related Complex |
| Pb | Lead |
| PIN | PIN-FORMED |
| PP2C | Protein Phosphatase 2C |
| pre-mRNA | precursor messenger RNA |
| RBP | RNA Binding Protein |
| ROS | Reactive Oxygen Species |
| SF | Splicing Factor |
| SKIP | SKI-INTERACTING PROTEIN |
| snRNA | Small Nuclear RNA |
| snRNP | Small Nuclear Ribonucleoprotein |
| SR protein | Serine/arginine-rich protein |
| SS | Splice Site |
| SUA | SUPPRESSOR OF ABI3-5 |
| SVP | SHORT VEGETATIVE PHASE |
| TFL1 | TERMINAL FLOWER 1 |
| TT8 | TRANSPARENT TESTA 8 |
References
- Werneke, J.M.; Chatfield, J.M.; Ogren, W.L. Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell 1989, 1, 815–825. [Google Scholar]
- Hu, X.; Wang, H.; Li, K.; Liu, X.; Liu, Z.; Wu, Y.; Li, S.; Huang, C. Genome-wide alternative splicing variation and its potential contribution to maize immature-ear heterosis. Crop J. 2021, 9, 476–486. [Google Scholar] [CrossRef]
- Miao, T.; Bao, H.; Ling, H.; Li, P.; Zhang, Y.; He, Y.; Hu, X.; Ling, C.; Liu, Y.; Tang, W.; et al. Comparative Transcriptomic Analysis Revealed the Suppression and Alternative Splicing of Kiwifruit (Actinidia latifolia) NAP1 Gene Mediating Trichome Development. Int. J. Mol. Sci. 2023, 24, 4481. [Google Scholar] [CrossRef]
- Shen, Y.; Qin, Z.; Ren, G.; Deng, P.; Ji, W.; Jiao, C.; Wu, L. Complexity and regulation of age-dependent alternative splicing in Brachypodium distachyon. Plant Physiol. 2023, 192, 2703–2722. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, L.; Tang, Q.; Zhang, L.; Li, X.; Chen, S.; Zhang, K.; Li, Y.; Hou, X.; Cheng, F. Improved genome annotation of Brassica oleracea highlights the importance of alternative splicing. Hortic. Plant J. 2024, 10, 961–970. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, W.; Zhu, D.; Zhang, B.; Xu, Q.; Shi, C.; He, H.; Dai, X.; Li, Y.; He, W.; et al. Population-level exploration of alternative splicing and its unique role in controlling agronomic traits of rice. Plant Cell 2024, 36, 4372–4387. [Google Scholar] [CrossRef]
- Zhu, F.Y.; Chen, X.; Song, Y.C.; Lam, L.P.Y.; Tobimatsu, Y.; Gao, B.; Chen, M.X.; Cao, F.L. SWATH-MS-based proteogenomic analysis reveals the involvement of alternative splicing in poplar upon lead stress. Genome Res. 2023, 33, 371–385. [Google Scholar] [CrossRef]
- Alhabsi, A.; Ling, Y.; Crespi, M.; Reddy, A.S.N.; Mahfouz, M. Alternative Splicing Dynamics in Plant Adaptive Responses to Stress. Annu. Rev. Plant Biol. 2025, 76, 687–717. [Google Scholar] [CrossRef]
- Liu, X.X.; Guo, Q.H.; Xu, W.B.; Liu, P.; Yan, K. Rapid Regulation of Alternative Splicing in Response to Environmental Stresses. Front. Plant Sci. 2022, 13, 832177. [Google Scholar] [CrossRef]
- Guo, Y.; Shang, X.; Ma, L.; Cao, Y. RNA-Binding Protein-Mediated Alternative Splicing Regulates Abiotic Stress Responses in Plants. Int. J. Mol. Sci. 2024, 25, 10548. [Google Scholar] [CrossRef]
- Yu, H.; Shi, X.; Ning, N.; Wu, H.; Mei, J.; Gu, X.; Ruan, H.; Zhang, M.; Li, Z.; Ma, S.; et al. The Exserohilum turcicum effector EtEC81 reprograms alternative splicing in maize and activates immunity. Cell Rep. 2025, 44, 115501. [Google Scholar] [CrossRef]
- Ning, M.; Li, Q.; Wang, Y.; Li, Q.; Tao, Y.; Zhang, F.; Hu, F.; Huang, L. Alternative splicing drives the functional diversification of a bHLH transcription factor in the control of growth and drought tolerance in rice. Sci. Bull. 2025, 70, 153–156. [Google Scholar] [CrossRef]
- Fu, D.; Song, Y.; Wu, S.; Peng, Y.; Ming, Y.; Li, Z.; Zhang, X.; Song, W.; Su, Z.; Gong, Z.; et al. Regulation of alternative splicing by CBF-mediated protein condensation in plant response to cold stress. Nat. Plants 2025, 11, 505–517. [Google Scholar] [CrossRef]
- Deng, Q.; Lu, H.; Liu, D.; Huang, Y.; Feng, J.; Wei, D.; Wang, Z.; Tang, Q. Modulation of flowering by an alternatively spliced AGL18-1 transcript in Brassica juncea. Crop J. 2025, 13, 456–467. [Google Scholar] [CrossRef]
- Ye, L.X.; Wu, Y.M.; Zhang, J.X.; Zhang, J.X.; Zhou, H.; Zeng, R.F.; Zheng, W.X.; Qiu, M.Q.; Zhou, J.J.; Xie, Z.Z.; et al. A bZIP transcription factor (CiFD) regulates drought- and low-temperature-induced flowering by alternative splicing in citrus. J. Integr. Plant Biol. 2023, 65, 674–691. [Google Scholar] [CrossRef]
- Wen, J.; Qin, Z.; Sun, L.; Zhang, Y.; Wang, D.; Peng, H.; Yao, Y.; Hu, Z.; Ni, Z.; Sun, Q.; et al. Alternative splicing of TaHSFA6e modulates heat shock protein-mediated translational regulation in response to heat stress in wheat. New Phytol. 2023, 239, 2235–2247. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Z.; Tian, H.; Wu, Y.; Kong, Y.; Wang, X.; Sui, N. Alternative Splicing Plays a Crucial Role in the Salt Tolerance of Foxtail Millet. J. Agric. Food Chem. 2024, 72, 10814–10827. [Google Scholar] [CrossRef]
- Laloum, T.; Martin, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef]
- Yang, X.; Jia, Z.; Pu, Q.; Tian, Y.; Zhu, F.; Liu, Y. ABA Mediates Plant Development and Abiotic Stress via Alternative Splicing. Int. J. Mol. Sci. 2022, 23, 3796. [Google Scholar] [CrossRef]
- Nee, G.; Kramer, K.; Nakabayashi, K.; Yuan, B.; Xiang, Y.; Miatton, E.; Finkemeier, I.; Soppe, W.J.J. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 2017, 8, 72. [Google Scholar] [CrossRef]
- Hong, Y.; Yao, J.; Shi, H.; Chen, Y.; Zhu, J.K.; Wang, Z. The Arabidopsis spliceosomal protein SmEb modulates ABA responses by maintaining proper alternative splicing of HAB1. Stress. Biol. 2021, 1, 4. [Google Scholar] [CrossRef]
- Sugliani, M.; Brambilla, V.; Clerkx, E.J.; Koornneef, M.; Soppe, W.J. The conserved splicing factor SUA controls alternative splicing of the developmental regulator ABI3 in Arabidopsis. Plant Cell 2010, 22, 1936–1946. [Google Scholar] [CrossRef]
- Punzo, P.; Ruggiero, A.; Possenti, M.; Perrella, G.; Nurcato, R.; Costa, A.; Morelli, G.; Grillo, S.; Batelli, G. DRT111/SFPS Splicing Factor Controls Abscisic Acid Sensitivity during Seed Development and Germination. Plant Physiol. 2020, 183, 793–807. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, X.; Wu, Y.; Wang, R.; Zhang, Y.; Shi, F.; Zhao, H.; Yu, P.; Wang, Y.; Chen, M.; et al. TaPP2C-a5 fine-tunes wheat seed dormancy and germination with a Triticeae-specific, alternatively spliced transcript. J. Adv. Res. 2025; in press. [Google Scholar] [CrossRef]
- Sybilska, E.; Collin, A.; Sadat Haddadi, B.; Mur, L.A.J.; Beckmann, M.; Guo, W.; Simpson, C.G.; Daszkowska-Golec, A. The cap-binding complex modulates ABA-responsive transcript splicing during germination in barley (Hordeum vulgare). Sci. Rep. 2024, 14, 18278. [Google Scholar] [CrossRef]
- Adamowski, M.; Friml, J. PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef]
- Kashkan, I.; Hrtyan, M.; Retzer, K.; Humpolickova, J.; Jayasree, A.; Filepova, R.; Vondrakova, Z.; Simon, S.; Rombaut, D.; Jacobs, T.B.; et al. Mutually opposing activity of PIN7 splicing isoforms is required for auxin-mediated tropic responses in Arabidopsis thaliana. New Phytol. 2022, 233, 329–343. [Google Scholar] [CrossRef]
- Cho, L.H.; Yoon, J.; An, G. The control of flowering time by environmental factors. Plant J. 2017, 90, 708–719. [Google Scholar] [CrossRef]
- Scortecci, K.C.; Michaels, S.D.; Amasino, R.M. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001, 26, 229–236. [Google Scholar] [CrossRef]
- Capovilla, G.; Symeonidi, E.; Wu, R.; Schmid, M. Contribution of major FLM isoforms to temperature-dependent flowering in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 5117–5127. [Google Scholar] [CrossRef]
- Jin, S.; Kim, S.Y.; Susila, H.; Nasim, Z.; Youn, G.; Ahn, J.H. FLOWERING LOCUS M isoforms differentially affect the subcellular localization and stability of SHORT VEGETATIVE PHASE to regulate temperature-responsive flowering in Arabidopsis. Mol. Plant 2022, 15, 1696–1709. [Google Scholar] [CrossRef]
- Macknight, R.; Duroux, M.; Laurie, R.; Dijkwel, P.; Simpson, G.; Dean, C. Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 2002, 14, 877–888. [Google Scholar] [CrossRef]
- Gil, K.E.; Park, M.J.; Lee, H.J.; Park, Y.J.; Han, S.H.; Kwon, Y.J.; Seo, P.J.; Jung, J.H.; Park, C.M. Alternative splicing provides a proactive mechanism for the diurnal CONSTANS dynamics in Arabidopsis photoperiodic flowering. Plant J. 2017, 89, 128–140. [Google Scholar] [CrossRef]
- Wang, P.; Li, Y.; Liu, Z.; Li, X.; Wang, Y.; Liu, W.; Li, X.; Hu, J.; Zhu, W.; Wang, C.; et al. Reciprocal regulation of flower induction by ELF3alpha and ELF3beta generated via alternative promoter usage. Plant Cell 2023, 35, 2095–2113. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, G.; Guo, C.; He, Y.; Li, Z.; Ning, G.; Shi, X.; Bao, M. The FLOWERING LOCUS T orthologous gene of Platanus acerifolia is expressed as alternatively spliced forms with distinct spatial and temporal patterns. Plant Biol. 2011, 13, 809–820. [Google Scholar] [CrossRef]
- Ai, X.-Y.; Zhang, J.-Z.; Liu, T.-J.; Hu, C.G. PtFCA from precocious trifoliate orange is regulated by alternative splicing and affects flowering time and root development in transgenic Arabidopsis. Tree Genet. Genomes 2016, 12, 85. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Li, Z.M.; Mei, L.; Yao, J.L.; Hu, C.G. PtFLC homolog from trifoliate orange (Poncirus trifoliata) is regulated by alternative splicing and experiences seasonal fluctuation in expression level. Planta 2009, 229, 847–859. [Google Scholar] [CrossRef]
- Ma, H.; Pei, J.; Zhuo, J.; Tang, Q.; Hou, D.; Lin, X. The CONSTANS-LIKE gene PeCOL13 regulates flowering through intron-retained alternative splicing in Phyllostachys edulis. Int. J. Biol. Macromol. 2024, 274 Pt 1, 133393. [Google Scholar] [CrossRef]
- Liu, J.; Miao, P.; Qin, W.; Hu, W.; Wei, Z.; Ding, W.; Zhang, H.; Wang, Z. A novel single nucleotide mutation of TFL1 alters the plant architecture of Gossypium arboreum through changing the pre-mRNA splicing. Plant Cell Rep. 2023, 43, 26. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Y.; Chen, T.; Qin, G.; Tian, S. Current insights into posttranscriptional regulation of fleshy fruit ripening. Plant Physiol. 2023, 192, 1785–1798. [Google Scholar] [CrossRef]
- Jiang, G.; Zhang, D.; Li, Z.; Liang, H.; Deng, R.; Su, X.; Jiang, Y.; Duan, X. Alternative splicing of MaMYB16L regulates starch degradation in banana fruit during ripening. J. Integr. Plant Biol. 2021, 63, 1341–1352. [Google Scholar] [CrossRef]
- Wang, J.; Xu, R.; Qiu, S.; Wang, W.; Zheng, F. CsTT8 regulates anthocyanin accumulation in blood orange through alternative splicing transcription. Hortic. Res. 2023, 10, uhad190. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Deng, L.; Guo, S.; Yuan, G.; Li, C.; Li, C. Alternative transcription and feedback regulation suggest that SlIDI1 is involved in tomato carotenoid synthesis in a complex way. Hortic. Res. 2022, 9, uhab045. [Google Scholar] [CrossRef]
- Chen, L.; Li, W.; Li, Y.; Feng, X.; Du, K.; Wang, G.; Zhao, L. Identified trans-splicing of YELLOW-FRUITED TOMATO 2 encoding the PHYTOENE SYNTHASE 1 protein alters fruit color by map-based cloning, functional complementation and RACE. Plant Mol. Biol. 2019, 100, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Bi, X.; Li, Z.; Fu, X.; Li, Y.; Li, Y.; Yang, Y.; Liu, D.; Li, G.; Dong, W.; et al. Transcriptomic Analysis of Alternative Splicing Events during Different Fruit Ripening Stages of Coffea arabica L. Genes 2024, 15, 459. [Google Scholar] [CrossRef] [PubMed]
- Rawoof, A.; Ahmad, I.; Islam, K.; Momo, J.; Kumar, A.; Jaiswal, V.; Ramchiary, N. Integrated omics analysis identified genes and their splice variants involved in fruit development and metabolites production in Capsicum species. Funct. Integr. Genom. 2022, 22, 1189–1209. [Google Scholar] [CrossRef]
- Zuo, D.-D.; He, G.; Sun, H.-T.; Guo, D.-l.J.S.H. Methylation-related alternative splicing events in H2O2-treated Kyoho grape berries during development. Sci. Hortic. 2023, 321, 112255. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf Senescence: Systems and Dynamics Aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, D.; Han, S.H.; Kim, S.H.; Piao, W.; Yanagisawa, S.; An, G.; Paek, N.C. Multilayered Regulation of Membrane-Bound ONAC054 Is Essential for Abscisic Acid-Induced Leaf Senescence in Rice. Plant Cell 2020, 32, 630–649. [Google Scholar] [CrossRef]
- Riester, L.; Koster-Hofmann, S.; Doll, J.; Berendzen, K.W.; Zentgraf, U. Impact of Alternatively Polyadenylated Isoforms of ETHYLENE RESPONSE FACTOR4 with Activator and Repressor Function on Senescence in Arabidopsis thaliana L. Genes 2019, 10, 91. Genes 2019, 10, 91. [Google Scholar] [CrossRef]
- Wang, H.L.; Zhang, Y.; Wang, T.; Yang, Q.; Yang, Y.; Li, Z.; Li, B.; Wen, X.; Li, W.; Yin, W.; et al. An alternative splicing variant of PtRD26 delays leaf senescence by regulating multiple NAC transcription factors in Populus. Plant Cell 2021, 33, 1594–1614. [Google Scholar] [CrossRef]
- Ma, J.; Li, S.; Wang, T.; Tao, Z.; Huang, S.; Lin, N.; Zhao, Y.; Wang, C.; Li, P. Cooperative condensation of RNA-DIRECTED DNA METHYLATION 16 splicing isoforms enhances heat tolerance in Arabidopsis. Nat. Commun. 2025, 16, 433. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Zhao, X.; Liu, H.; Ku, L.; Wang, S.; Han, Z.; Wu, L.; Shi, Y.; Song, X.; Chen, Y. Alternative splicing of ZmCCA1 mediates drought response in tropical maize. PLoS ONE 2019, 14, e0211623. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Luo, T.; Zhao, H.; Su, Y.; Ji, W.; Li, H. Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene 2020, 740, 144514. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, X.H.; Guo, Q.H.; Liu, P.; Li, Y.; Wu, C.A.; Yang, G.D.; Huang, J.G.; Zhang, S.Z.; Zheng, C.C.; et al. Salt responsive alternative splicing of a RING finger E3 ligase modulates the salt stress tolerance by fine-tuning the balance of COP9 signalosome subunit 5A. PLoS Genet. 2021, 17, e1009898. [Google Scholar] [CrossRef]
- Zhang, D.; Lv, A.; Yang, T.; Cheng, X.; Zhao, E.; Zhou, P. Protective functions of alternative splicing transcripts (CdDHN4-L and CdDHN4-S) of CdDHN4 from bermudagrass under multiple abiotic stresses. Gene 2020, 763, 100033. [Google Scholar] [CrossRef]
- Li, Y.; Kou, S. A Ralstonia solanacearum Effector Targets Splicing Factor SR34a to Reprogram Alternative Splicing and Regulate Plant Immunity. Plants 2025, 14, 534. [Google Scholar] [CrossRef]
- Zeng, X.; Wu, C.; Zhang, L.; Lan, L.; Fu, W.; Wang, S. Molecular Mechanism of Resistance to Alternaria alternata Apple Pathotype in Apple by Alternative Splicing of Transcription Factor MdMYB6-like. Int. J. Mol. Sci. 2024, 25, 4353. [Google Scholar] [CrossRef] [PubMed]
- Amano, M.; Iida, S.; Kosuge, K. Comparative studies of thermotolerance: Different modes of heat acclimation between tolerant and intolerant aquatic plants of the genus Potamogeton. Ann. Bot. 2012, 109, 443–452. [Google Scholar] [CrossRef]
- Sugio, A.; Dreos, R.; Aparicio, F.; Maule, A.J. The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 2009, 21, 642–654. [Google Scholar] [CrossRef]
- He, Z.S.; Xie, R.; Zou, H.S.; Wang, Y.Z.; Zhu, J.B.; Yu, G.Q. Structure and alternative splicing of a heat shock transcription factor gene, MsHSF1, in Medicago sativa. Biochem. Biophys. Res. Commun. 2007, 364, 1056–1061. [Google Scholar] [CrossRef]
- Cheng, Q.; Zhou, Y.; Liu, Z.; Zhang, L.; Song, G.; Guo, Z.; Wang, W.; Qu, X.; Zhu, Y.; Yang, D. An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol. 2015, 17, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liang, J.; Wang, C.; Ding, L.; Zhao, X.; Cao, X.; Xu, S.; Teng, N.; Yi, M. Alternative Splicing Provides a Mechanism to Regulate LlHSFA3 Function in Response to Heat Stress in Lily. Plant Physiol. 2019, 181, 1651–1667. [Google Scholar] [CrossRef]
- Ling, Y.; Mahfouz, M.M.; Zhou, S. Pre-mRNA alternative splicing as a modulator for heat stress response in plants. Trends Plant Sci. 2021, 26, 1153–1170. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Cheng, X.; Li, C.; Li, Y.; Pan, C.; Lu, G. Decoding plant thermosensors: Mechanism of temperature perception and stress adaption. Front. Plant Sci. 2025, 16, 1560204. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, T.; Liu, J. Full-length single-molecule sequencing uncovers novel insight into the global landscape of the cold stress response in trifoliate orange (Citrus trifoliata). Front. Plant Sci. 2024, 15, 1506414. [Google Scholar] [CrossRef]
- Song, L.; Pan, Z.; Chen, L.; Dai, Y.; Wan, J.; Ye, H.; Nguyen, H.T.; Zhang, G.; Chen, H. Analysis of Whole Transcriptome RNA-seq Data Reveals Many Alternative Splicing Events in Soybean Roots under Drought Stress Conditions. Genes 2020, 11, 1520. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.; Tan, M.; Wang, L.; Zhao, W.; You, J.; Wang, L.; Yan, X.; Wang, W. The pattern of alternative splicing and DNA methylation alteration and their interaction in linseed (Linum usitatissimum L.) response to repeated drought stresses. Biol. Res. 2023, 56, 12. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Yu, K.; Han, L.; Li, X.; Wang, H.; Liu, Y.; Zhang, Y. Global profiling of alternative splicing landscape responsive to salt stress in wheat (Triticum aestivum L.). Plant Growth Regul. 2020, 92, 107–116. [Google Scholar] [CrossRef]
- Punzo, P.; Grillo, S.; Batelli, G. Alternative splicing in plant abiotic stress responses. Biochem. Soc. Trans. 2020, 48, 2117–2126. [Google Scholar] [CrossRef]
- Tognacca, R.S.; Rodriguez, F.S.; Aballay, F.E.; Cartagena, C.M.; Servi, L.; Petrillo, E. Alternative splicing in plants: Current knowledge and future directions for assessing the biological relevance of splice variants. J. Exp. Bot. 2023, 74, 2251–2272. [Google Scholar] [CrossRef]
- Liu, L.; Tang, Z.; Liu, F.; Mao, F.; Yujuan, G.; Wang, Z.; Zhao, X. Normal, novel or none: Versatile regulation from alternative splicing. Plant Signal. Behav. 2021, 16, 1917170. [Google Scholar] [CrossRef] [PubMed]
- Kashkan, I.; Timofeyenko, K.; Ruzicka, K. How alternative splicing changes the properties of plant proteins. Quant. Plant Biol. 2022, 3, e14. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Cao, L.; Wang, S.; Guo, L.; Tan, L.; Liu, H.; Feng, Y.; Wu, W. Differences in alternative splicing and their potential underlying factors between animals and plants. J. Adv. Res. 2024, 64, 83–98. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Species | Stage/Stress | Type | Isoform | Function | Reference |
|---|---|---|---|---|---|---|
| DOG1 | Arabidopsis thaliana | Seed development | Alternative splicing site | 5 isoforms | Regulates depth of seed dormancy | [20] |
| HAB1 | Arabidopsis thaliana | Seed development | Intron retention | HAB1α/HAB1β | Balances ABA responses during seed germination | [21] |
| ABI3 | Arabidopsis thaliana | Seed development | Exon skipping | ABI3-α/ABI3-β | Functional vs. non-functional; affects seed maturation | [22,23] |
| TaPP2C-a5 | Triticum aestivum | Seed development | Exon skipping | a5.1/a5.2 | a5.2 is seed-specific; regulates ABA-DOG1 coordination | [24] |
| PIN7 | Arabidopsis thaliana | Vegetative growth | Alternative 5′ splice site | PIN7a/PIN7b | Different membrane dynamics; fine-tune auxin transport | [27] |
| FLM | Arabidopsis thaliana | Flowering | Exon inclusion/skipping | FLM-β/FLM-δ | Competes for SVP binding, temperature-dependent isoforms modulate flowering via SVP interaction | [30,31] |
| CO | Arabidopsis thaliana | Flowering | Truncated transcript | CO-α/CO-β | CO-β forms non-functional dimers to inhibit CO-α activity | [33] |
| ELF3 | Pyrus sp. | Flowering | Intron retention & alt. promoter | ELF3α/ELF3β | ELF3β alleviates repression of flowering by ELF3α | [34] |
| FD | Citrus spp. | Flowering | Exon skipping | CiFDα/CiFDβ | Responds to cold/drought to regulate flowering time | [15] |
| FT | Platanus acerifolia | Flowering | Alternative splicing site | 21 isoforms | Exhibits conserved AS pattern in flowering regulation | [35] |
| FCA/FLC | Poncirus trifoliata | Flowering | Alternative splicing site | PtFCA1-3, PtFLC1-5 | Modulate flowering time | [36,37] |
| COL13 | Phyllostachys edulis | Flowering | Intron retention | PeCOL13α/PeCOL13β | Regulates photoperiod-responsive flowering | [38] |
| AGL18-1 | Brassica juncea | Flowering | Truncated transcript | BjuAGL18-1L/BjuAGL18-1S | Involved in flowering regulation | |
| TFL1 | Gossypium arboreum | Vegetative/Floral development | Splice site mutation | Full-length/Truncated | Loss-of-function mutant affects meristem fate | [39] |
| MYB16 | Musa acuminata | Fruit maturation | Truncated transcript | MaMYB16L/MaMYB16S | Modulates starch degradation via isoform ratio | [41] |
| TT8 | Citrus sinensis | Fruit pigmentation | Exon skipping | TT8/Δ15-TT8 | Fine-tunes anthocyanin biosynthesis via feedback regulation | [42] |
| IDI1 | Solanum lycopersicum | Fruit maturation | Alternative transcription initiation and alternative splicing | Long/short isoforms | Only the long isoform is functional in carotenoid synthesis | [43] |
| PSY1 | S. lycopersicum var. cerasiforme | Fruit pigmentation | Alternative trans-splicing | LT-YFT2/YFT2 | Alters carotenoid content in yellow-fruited variants | [44] |
| ERF4 | Arabidopsis thaliana | Leaf senescence | Alternative splicing site | ERF4-R/ERF4-A | ERF4-R promotes, ERF4-A suppresses senescence | [50] |
| NAC054 | Oryza sativa | Leaf senescence | Exon skipping | ONAC054α/ONAC054β | Both isoforms regulate ABA-induced senescence | [49] |
| RD26 | Populus tomentosa | Leaf senescence | Intron retention | PtRD26/PtRD26IR | Negatively regulates senescence in aging leaves | [51] |
| HSFA6e | Triticum aestivum | Heat stress | Exon extension | TaHSFA6e-II/TaHSFA6e-III | AS generates TaHSFA6e-III with enhanced transactivation ability on TaHSP70s, improving thermotolerance | [16] |
| RDM16 | Arabidopsis thaliana | Heat stress | Exon skipping | RDL, RDS | RDS enhances RDL-mediated heat resistance via functional interaction | [52] |
| CBFs | Arabidopsis thaliana | Cold stress | Spliceosome | Spliceosome modulation | Interacts with SKIP to form nuclear condensates; modulates AS and cold-responsive gene expression | [13] |
| bHLH59 | Oryza sativa | Drought stress | Truncated transcript | OsbHLH59.1/OsbHLH59.2 | OsbHLH59.1 promotes growth; OsbHLH59.2 induced by ABA suppresses growth and activates drought response | [12] |
| CCA1 | Zea mays | Drought stress | Alternative splicing site | ZmCCA1.1/ZmCCA1.2/ZmCCA1.3 | Splice variants responsive to drought and photoperiod; ZmCCA1.1 enhances drought tolerance | [53] |
| DREB3 | Triticum aestivum | Drought stress | Alternative 3′/5′ splicing | TaDREB3-I/TaDREB3-II/TaDREB3-III | Only isoform I significantly improves tolerance to drought, salt, and heat in transgenic Arabidopsis | [54] |
| SRAS1 | Arabidopsis thaliana | Salt stress | Intron retention | SRAS1.1/SRAS1.2 | Salt-induced AS shifts from non-functional SRAS1.2 to SRAS1.1, enhancing CSN5A degradation and salt tolerance | [55] |
| DHN4 | Cynodon spp. | Cold/drought/salt stress | Exon skipping | CdDHN4-L/CdDHN4-S | CdDHN4-S enhances ROS scavenging and osmotic response; CdDHN4-L is more effective under cold and drought stress | [56] |
| HSP70 | Populus trichocarpa | Heavy metal (Pb) stress | Alternative splicing site | PtHSP70-AS1/PtHSP70-AS2 | PtHSP70-AS2 binds Pb(II) 10× more effectively, promotes extrusion and enhances lead tolerance | [7] |
| EIP1 | Zea mays | Biotic (fungus) stress | Alternative splicing site | Multiple AS events | Interacts with pathogen effector EtEC81, reprograms 119 AS events to activate immune response | [11] |
| SR34a | Solanum lycopersicum | Biotic (bacteria) stress | Intron retention | Intron retention in defense genes | Effector RipP2 acetylates SR34a, promotes IR in defense genes, suppressing immunity | [57] |
| MYB6-like | Malus domestica | Biotic (fungus) stress | Intron retention | MdMYB6-like-α/MdMYB6-like-β/MdMYB6-like-γ | AS isoform MdMYB6-like-β induced by Alternaria alternata, enhances lignin synthesis and disease resistance | [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Wang, H.; Gao, H.; Yu, S.; Liu, C.; Wang, Y.; Sun, Y.; Zhang, D. Alternative Splicing of Functional Genes in Plant Growth, Development, and Stress Responses. Int. J. Mol. Sci. 2025, 26, 5864. https://doi.org/10.3390/ijms26125864
Liu G, Wang H, Gao H, Yu S, Liu C, Wang Y, Sun Y, Zhang D. Alternative Splicing of Functional Genes in Plant Growth, Development, and Stress Responses. International Journal of Molecular Sciences. 2025; 26(12):5864. https://doi.org/10.3390/ijms26125864
Chicago/Turabian StyleLiu, Guan, Hanhui Wang, Huan Gao, Song Yu, Changhua Liu, Yang Wang, Yan Sun, and Dongye Zhang. 2025. "Alternative Splicing of Functional Genes in Plant Growth, Development, and Stress Responses" International Journal of Molecular Sciences 26, no. 12: 5864. https://doi.org/10.3390/ijms26125864
APA StyleLiu, G., Wang, H., Gao, H., Yu, S., Liu, C., Wang, Y., Sun, Y., & Zhang, D. (2025). Alternative Splicing of Functional Genes in Plant Growth, Development, and Stress Responses. International Journal of Molecular Sciences, 26(12), 5864. https://doi.org/10.3390/ijms26125864






