Molecular Mechanisms of Cadmium Stress Resistance in Vegetable Crops
Abstract
1. Introduction
2. Physiological and Biochemical Reactions of Vegetable Crops Under Cadmium Stress
2.1. Growth Inhibition
2.2. Impaired Photosynthesis
2.3. Oxidative Stress Response and Altered Enzyme Activity
2.4. Disordered Nutrient Element Absorption
2.5. Changes in Phytohormone Levels
2.6. Changes in Gene Expression Regulation
3. Molecular Mechanisms of Multiple Genes in Regulating Vegetable Crops’ Response to Cadmium Stress
3.1. Transporter-Related Genes
3.2. Transcription Factors
3.3. Enzyme-Encoding Genes
3.4. MicroRNAs and Other Types of Genes
3.5. Putative Functional Genes
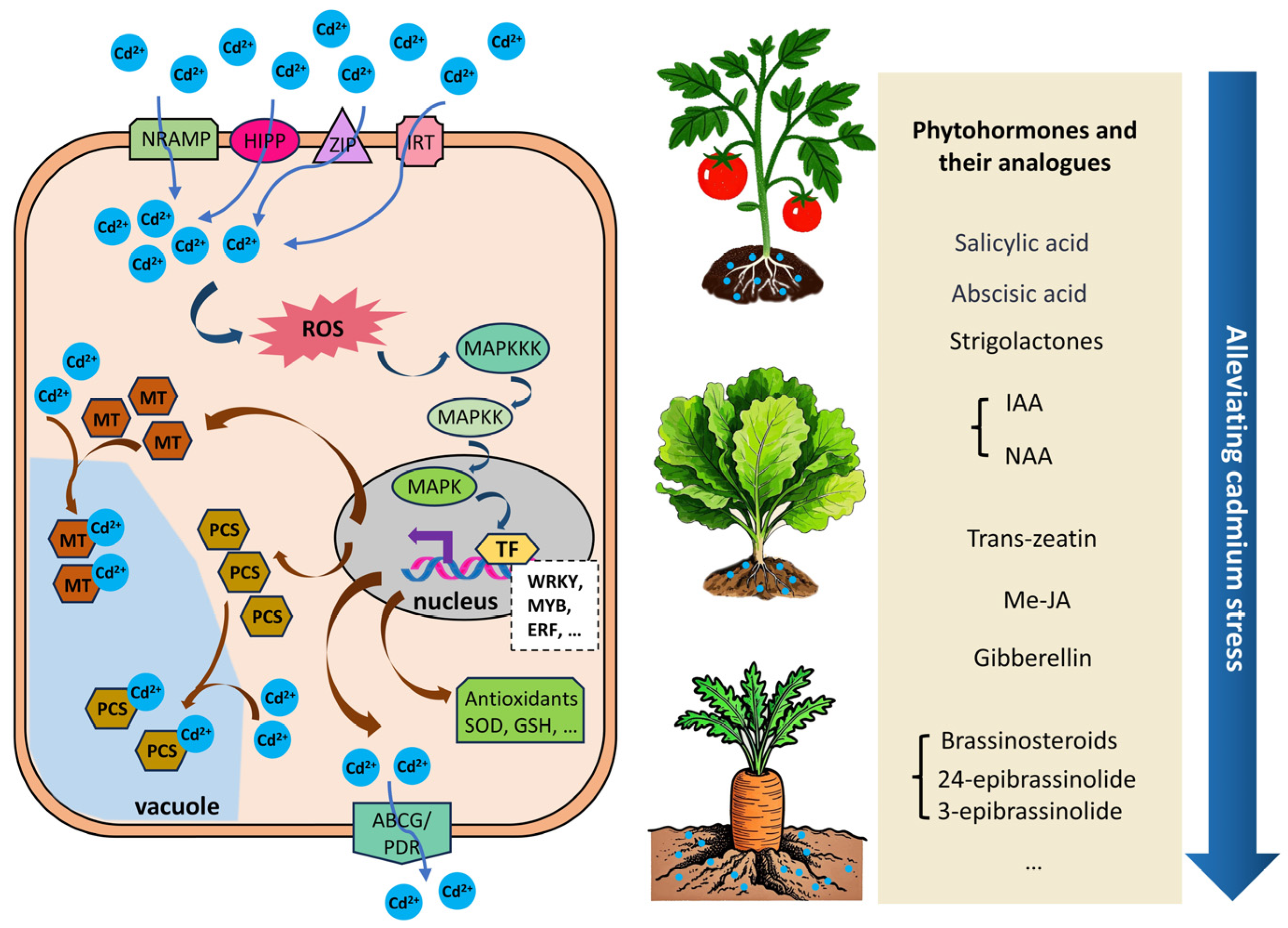
4. Molecular Mechanisms of Phytohormones and Their Analogues in Regulating Vegetable Crops’ Response to Cadmium Stress
5. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Huang, Y.; He, C.; Shen, C.; Guo, J.; Mubeen, S.; Yuan, J.; Yang, Z. Toxicity of cadmium and its health risks from leafy vegetable consumption. Food Funct. 2017, 8, 1373–1401. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Alseekh, S.; Zhu, Z.H.; Zhou, K.J.; Fernie, A.R. Multiomics and biotechnologies for understanding and influencing cadmium accumulation and stress response in plants. Plant Biotechnol. J. 2024, 22, 2641–2659. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Bao, X.Q.; Qiu, G.Y.; Li, H.; Wang, Y.; Chen, X.D.; Fu, Q.L.; Guo, B. Genome-Wide identification and expression analysis of SlNRAMP genes in tomato under nutrient deficiency and cadmium stress during arbuscular mycorrhizal symbiosis. Int. J. Mol. Sci. 2024, 25, 8269. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Yuan, C.; Bu, T.; Liu, Q.; Li, J.J.; Wang, F.D.; Zhang, Y.H.; He, L.L.; Gao, J.W. Mediates Cd tolerance via activation of the cis element of VHS1 in yeast and enhances Cd tolerance in Chinese cabbage. Int. J. Mol. Sci. 2024, 25, 11026. [Google Scholar] [CrossRef]
- Zhang, Y.L.; He, T.B.; Tian, W.J.; Xia, Y.B.; He, Y.Q.; Su, M.M.; He, G.D. The expression of the StNRAMP2 gene determined the accumulation of cadmium in different tissues of potato. Int. J. Mol. Sci. 2023, 24, 9322. [Google Scholar] [CrossRef]
- Niu, M.Y.; Bao, C.J.; Zhan, J.Y.; Yue, X.M.; Zou, J.N.; Su, N.A.; Cui, J. Plasma membrane-localized protein BcHIPP16 promotes the uptake of copper and cadmium in planta. Ecotox. Environ. Safe 2021, 227, 112920. [Google Scholar] [CrossRef]
- Yan, G.C.; Hua, Y.C.; Jin, H.; Huang, Q.Y.; Zhou, G.F.; Xu, Y.M.; He, Y.; Zhu, Z.J. Sly-miR398 participates in cadmium stress acclimation by regulating antioxidant system and cadmium transport in tomato (Solanum lycopersicum). Int. J. Mol. Sci. 2023, 24, 1953. [Google Scholar] [CrossRef]
- Dong, Y.L.; Ma, Y.F.; Li, Q.; Cao, Y.L.; Dong, D.X.; Chen, C.; Zhang, X.X.; Fan, Y.W.; Jin, X.X. Overexpression of histone demethylase gene SlJMJ18 and SlJMJ23 from tomato confers cadmium tolerance by regulating metal transport and hormone content in Arabidopsis. Plant Sci. 2024, 346, 112169. [Google Scholar] [CrossRef]
- Pan, X.X.; Zheng, Y.; Lei, K.R.; Tao, W.L.; Zhou, N. Systematic analysis of Heat Shock Protein 70 (HSP70) gene family in radish and potential roles in stress tolerance. BMC Plant Biol. 2024, 24, 2. [Google Scholar] [CrossRef]
- Huang, L.P.; Liu, X.; Liu, Y.; Tanveer, M.; Chen, W.; Fu, W.X.; Wang, Q.Q.; Guo, Y.J.; Shabala, S. Revealing mechanistic basis of ameliorating detrimental effects of cadmium in cherry tomatoes by exogenous application of melatonin and brassinosteroids. Ecotox. Environ. Safe 2024, 283, 116768. [Google Scholar] [CrossRef]
- Tang, W.; Liang, L.; Xie, Y.D.; Li, X.M.; Lin, L.J.; Huang, Z.; Sun, B.; Sun, G.C.; Tu, L.H.; Li, H.X.; et al. Foliar application of salicylic acid inhibits the cadmium uptake and accumulation in lettuce (Lactuca sativa L.). Front. Plant Sci. 2023, 14, 1200106. [Google Scholar] [CrossRef] [PubMed]
- Waris, Z.; Noreen, Z.; Shah, A.A.; Usman, S.; Shah, A.N.; Rizwan, M.; Casini, R.; Elansary, H.O. Efficacy of γ-aminobutyric acid (GABA) on physio-biochemical attributes of lettuce (Lactuca sativa L.) under cadmium toxicity. J. Plant Growth Regul. 2023, 42, 5041–5057. [Google Scholar] [CrossRef]
- Meena, M.; Aamir, M.; Kumar, V.; Swapnil, P.; Upadhyay, R.S. Evaluation of morpho-physiological growth parameters of tomato in response to Cd induced toxicity and characterization of metal sensitive NRAMP3 transporter protein. Environ. Exp. Bot. 2018, 148, 144–167. [Google Scholar] [CrossRef]
- Sun, M.; Qiao, H.X.; Yang, T.; Zhao, P.; Zhao, J.H.; Luo, J.M.; Luan, H.Y.; Li, X.; Wu, S.C.; Xiong, A.S. Hydrogen sulfide alleviates cadmium stress in germinating carrot seeds by promoting the accumulation of proline. J. Plant Physiol. 2024, 303, 154357. [Google Scholar] [CrossRef]
- Hédiji, H.; Djebali, W.; Belkadhi, A.; Cabasson, C.; Moing, A.; Rolin, D.; Brouquisse, R.; Gallusci, P.; Chaïbi, W. Impact of long-term cadmium exposure on mineral content of Solanum lycopersicum plants: Consequences on fruit production. S. Afr. J. Bot. 2015, 97, 176–181. [Google Scholar] [CrossRef]
- Huang, Y.F.; Chen, J.H.; Sun, Y.M.; Wang, H.X.; Zhan, J.Y.; Huang, Y.N.; Zou, J.W.; Wang, L.; Su, N.N.; Cui, J. Mechanisms of calcium sulfate in alleviating cadmium toxicity and accumulation in pak choi seedlings. Sci. Total Environ. 2022, 805, 150115. [Google Scholar] [CrossRef]
- Shahzad, K.; Danish, S.; Mubeen, S.; Dawar, K.; Fahad, S.; Hasnain, Z.; Ansari, M.J.; Almoallim, H.S. Minimization of heavy metal toxicity in radish (Raphanus sativus) by strigolactone and biochar. Sci. Rep. 2024, 14, 13616. [Google Scholar] [CrossRef]
- Dong, J.; Wu, F.; Zhang, G. Influence of cadmium on antioxidant capacity and four microelement concentrations in tomato seedlings (Lycopersicon esculentum). Chemosphere 2006, 64, 1659–1666. [Google Scholar] [CrossRef]
- Cui, Q.L.; Liu, D.D.; Chen, H.S.; Qiu, T.Y.; Zhao, S.L.; Duan, C.J.; Cui, Y.X.; Zhu, X.Z.; Chao, H.R.; Wang, Y.H.; et al. Synergistic interplay between Azospirillum brasilense and exogenous signaling molecule H₂S promotes Cd stress resistance and growth in pak choi (Brassica chinensis L.). J. Hazard. Mater. 2023, 444, 130425. [Google Scholar] [CrossRef]
- Ma, Y.F.; Huang, Y.D.; Zhang, W.L.; Dong, J.H.; Zhang, X.L.; Zhu, Y.L.; Wang, Y.; Liu, L.W.; Xu, L. RsNRAMP5, a major metal transporter, promotes cadmium influx and ROS accumulation in radish (Raphanus sativus L.). Plant Physiol. Bioch. 2025, 218, 109323. [Google Scholar] [CrossRef]
- Wu, X.; Su, N.N.; Yue, X.M.; Fang, B.; Zou, J.W.; Chen, Y.H.; Shen, Z.G.; Cui, J. IRT1 and ZIP2 were involved in exogenous hydrogen-rich water-reduced cadmium accumulation in Brassica chinensis and Arabidopsis thaliana. J. Hazard. Mater. 2021, 407, 124599. [Google Scholar] [CrossRef] [PubMed]
- Asaf, S.; Jan, R.H.T.L.; Khan, M.A.; Lubna; Khan, A.L.; Asif, S.; Bilal, S.; Ahmad, W.; Waqas, M.; Kim, K.M.; et al. Unraveling the mutualistic interaction between endophytic Curvularia lunata CSL1 and tomato to mitigate cadmium (Cd) toxicity via transcriptomic insights. Sci. Total Environ. 2023, 861, 160542. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.F.; Yang, S.W.; Wang, W.D.; Wei, X.L.; Lou, F.; He, G.D.; He, T.B. Multiomics combined with expression pattern analysis reveals the regulatory response of key genes in potato jasmonic acid signaling pathways to cadmium stress. J. Agr. Food Chem. 2024, 72, 22369–22384. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, H.S.; Song, W.Y.; Choi, K.S.; Hur, Y. Chloroplast-targeted BrMT1 (Brassica rapa type-1 metallothionein) enhances resistance to Cadmium and ROS in transgenic Arabidopsis plants. J. Plant Biol. 2007, 50, 1–7. [Google Scholar] [CrossRef]
- Sun, M.; Qiao, H.X.; Yang, T.; Zhao, P.; Zhao, J.H.; Luo, J.M.; Liu, F.F.; Xiong, A.S. DcMYB62, a transcription factor from carrot, enhanced cadmium tolerance of Arabidopsis by inducing the accumulation of carotenoids and hydrogen sulfide. Plant Physiol. Bioch. 2024, 216, 109114. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Ma, Y.F.; Lai, D.Q.; He, M.; Zhang, X.L.; Zhang, W.L.; Ji, M.M.; Zhu, Y.L.; Wang, Y.; Liu, L.W.; et al. RsPDR8, a member of ABCG subfamily, plays a positive role in regulating cadmium efflux and tolerance in radish (Raphanus sativus L.). Plant Physiol. Bioch. 2023, 205, 108149. [Google Scholar] [CrossRef]
- Yue, X.M.; Song, J.X.; Fang, B.; Wang, L.; Zou, J.W.; Su, N.N.; Cui, J. BcNRAMP1 promotes the absorption of cadmium and manganese in Arabidopsis. Chemosphere 2021, 283, 131113. [Google Scholar] [CrossRef]
- Cai, S.Y.; Zhang, Y.; Xu, Y.P.; Qi, Z.Y.; Li, M.Q.; Ahammed, G.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Reiter, R.J.; et al. HsfA1a upregulates melatonin biosynthesis to confer cadmium tolerance in tomato plants. J. Pineal Res. 2017, 62, e12387. [Google Scholar] [CrossRef]
- Dang, F.F.; Lin, J.H.; Chen, Y.P.; Li, G.X.; Guan, D.Y.; Zheng, S.J.; He, S.L. A feedback loop between CaWRKY41 and H2O2 coordinates the response to Ralstonia solanacearum and excess cadmium in pepper. J. Exp. Bot. 2019, 70, 1581–1595. [Google Scholar] [CrossRef]
- Lin, T.T.; Yang, W.N.; Lu, W.; Wang, Y.; Qi, X.T. Transcription factors PvERF15 and PvMTF-1 form a cadmium stress transcriptional pathway. Plant Physiol. 2017, 173, 1565–1573. [Google Scholar] [CrossRef]
- Chen, W.W.; Jin, J.F.; Lou, H.Q.; Liu, L.; Kochian, L.V.; Yang, J.L. LeSPL-CNR negatively regulates Cd acquisition through repressing nitrate reductase-mediated nitric oxide production in tomato. Planta 2018, 248, 893–907. [Google Scholar] [CrossRef] [PubMed]
- He, G.D.; Saleem, M.; Deng, T.F.; Zhong, Z.Y.; He, T.B.; Wu, J.H. Unraveling the mechanism of StWRKY6 in potato (Solanum tuberosum)'s cadmium tolerance for ensuring food safety. Foods 2023, 12, 2303. [Google Scholar] [CrossRef] [PubMed]
- Youm, J.W.; Jeon, J.H.; Choi, D.; Yi, S.Y.; Joung, H.; Kim, H.S. Ectopic expression of pepper CaPF1 in potato enhances multiple stresses tolerance and delays initiation of in vitro tuberization. Planta 2008, 228, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Qiao, F.; Wang, M.Q.; Wang, Y.; Xu, Y.; Qi, X.T. PvERF104 confers cadmium tolerance in Arabidopsis: Evidence for metal-responsive element-binding transcription factors. Environ. Exp. Bot. 2023, 206, 105167. [Google Scholar] [CrossRef]
- Sun, N.; Liu, M.; Zhang, W.T.; Yang, W.N.; Bei, X.J.; Ma, H.; Qiao, F.; Qi, X.T. Bean metal-responsive element-binding transcription factor confers cadmium resistance in tobacco. Plant Physiol. 2015, 167, 1136–1148. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Li, C.X.; Li, X.; Liu, A.R.; Chen, S.C.; Zhou, J. Overexpression of tomato RING E3 ubiquitin ligase gene SlRING1 confers cadmium tolerance by attenuating cadmium accumulation and oxidative stress. Physiol. Plant. 2021, 173, 449–459. [Google Scholar] [CrossRef]
- Qi, Z.Y.; Ahammed, G.J.; Jiang, C.Y.; Li, C.X.; Zhou, J. The E3 ubiquitin ligase gene SlRING1 is essential for plant tolerance to cadmium stress in Solanum lycopersicum. J. Biotechnol. 2020, 324, 239–247. [Google Scholar] [CrossRef]
- Xu, W.H.; Huang, H.; Li, X.D.; Yang, M.; Chi, S.L.; Pan, Y.; Li, N.N.; Paterson, A.H.; Chai, Y.R.; Lu, K. CaHMA1 promotes Cd accumulation in pepper fruit. J. Hazard. Mater. 2023, 460, 132480. [Google Scholar] [CrossRef]
- Han, Y.Y.; Du, Y.L.; Wang, J.W.; Wu, T. Overexpression of Chinese flowering cabbage BpPMSR3 enhances the tolerance of Arabidopsis thaliana to cadmium. J. Plant Nutr. Soil Sci. 2018, 181, 787–794. [Google Scholar] [CrossRef]
- Liu, C.X.; Yang, T.; Zhou, H.; Ahammed, G.J.; Qi, Z.Y.; Zhou, J. The E3 ubiquitin ligase gene Sl1 is critical for cadmium tolerance in Solanum lycopersicum L. Antioxidants 2022, 11, 456. Antioxidants 2022, 11, 456. [Google Scholar] [CrossRef]
- Zhong, M.; Wang, Y.; Hou, K.; Shu, S.; Sun, J.; Guo, S. TGase positively regulates photosynthesis via activation of Calvin cycle enzymes in tomato. Hortic. Res. 2019, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Yue, L.Q.; Qin, H.Y.; Wang, G.H.; Xiao, L.W.; Cheng, Q.Q.; Lei, B.F.; Huang, R.M.; Yang, X.; Kang, Y.Y. TGase-induced Cd tolerance by boosting polyamine, nitric oxide, cell wall composition and phytochelatin synthesis in tomato. Ecotox. Environ. Safe 2023, 259, 115023. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.Y.; Ma, Q.F.; Xu, X.; Li, G.Z.; Hao, L. Tomato jasmonic acid-deficient mutant spr2 seedling response to cadmium stress. J. Plant Growth Regul. 2016, 35, 603–610. [Google Scholar] [CrossRef]
- Muhammad, T.; Zhang, J.; Ma, Y.L.; Li, Y.S.; Zhang, F.; Zhang, Y.; Liang, Y. Overexpression of a mitogen-activated protein kinase SlMAPK3 positively regulates tomato tolerance to cadmium and drought stress. Molecules 2019, 24, 556. [Google Scholar] [CrossRef]
- Li, Q.; Sun, W.Y.; Chen, C.; Dong, D.X.; Cao, Y.L.; Dong, Y.L.; Yu, L.J.; Yue, Z.H.; Jin, X.X. Overexpression of histone demethylase gene SlJMJ524 from tomato confers Cd tolerance by regulating metal transport-related protein genes and flavonoid content in Arabidopsis. Plant Sci. 2022, 318, 111205. [Google Scholar] [CrossRef]
- Luo, W.; Long, Y.M.; Feng, Z.L.; Li, R.; Huang, X.J.; Zhong, J.X.; Liu, D.Y.; Zhao, H.B. A γ-glutamylcysteine ligase AcGCL alleviates cadmium-inhibited fructooligosaccharides metabolism by modulating glutathione level in Allium cepa L. J. Hazard. Mater. 2021, 419, 126255. [Google Scholar] [CrossRef]
- Shen, C.; Huang, Y.Y.; Xin, J.L.; He, C.T.; Yang, Z.Y. A novel microRNA IamiR-4-3p from water spinach (Ipomoea aquatica Forsk.) increased Cd uptake and translocation in Arabidopsis thaliana. Environ. Sci. Pollut. R. 2022, 29, 41375–41385. [Google Scholar] [CrossRef]
- Manara, A.; Fasani, E.; Molesini, B.; DalCorso, G.; Pennisi, F.; Pandolfini, T.; Furini, A. The tomato metallocarboxypeptidase inhibitor I, which Interacts with a heavy metal-associated isoprenylated protein, is implicated in plant response to cadmium. Molecules 2020, 25, 700. [Google Scholar] [CrossRef]
- Ma, J.Y.; Dong, X.M.; Yu, L.J.; Zhang, Y.H. Response and function of Solanum lycopersicum L. SlSGR2 Gene under cadmium stress. Horticulturae 2022, 8, 1002. [Google Scholar] [CrossRef]
- Feng, S.J.; Shen, Y.H.; Xu, H.A.; Dong, J.Y.; Chen, K.X.; Xiang, Y.; Jiang, X.D.; Yao, C.J.; Lu, T.; Huan, W.W.; et al. RNA-seq identification of Cd responsive transporters provides insights into the association of oxidation resistance and Cd accumulation in Cucumis sativus L. Antioxidants 2021, 10, 1973. [Google Scholar] [CrossRef]
- Shen, C.; Huang, B.F.; Hu, L.; Yuan, H.W.; Huang, Y.Y.; Wang, Y.B.; Sun, Y.F.; Li, Y.; Zhang, J.R.; Xin, J.L. Comparative transcriptome analysis and Arabidopsis thaliana overexpression reveal key genes associated with cadmium transport and distribution in root of two Capsicum annuum cultivars. J. Hazard. Mater. 2024, 465, 133365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.X.; Chai, T.Y.; Dong, J.; Zhao, W.M.; An, C.C.; Chen, Z.L.; Burkard, G. Cloning and expression analysis of the heavy-metal responsive gene PvSR2 from bean. Plant Sci. 2001, 161, 783–790. [Google Scholar] [CrossRef]
- Qi, X.T.; Zhang, Y.X.; Chai, T.Y. Characterization of a novel plant promoter specifically induced by heavy metal and identification of the promoter regions conferring heavy metal responsiveness. Plant Physiol. 2007, 143, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Li, J.L.; Cao, Y.Y.; Huang, J.B.; Duan, Q.H. Genome-wide identification and expression analysis under abiotic stress of BrAHL genes in Brassica rapa. Int. J. Mol. Sci. 2023, 24, 12447. [Google Scholar] [CrossRef]
- Jin, Q.J.; Yuan, X.X.; Cui, W.T.; Han, B.; Feng, J.F.; Xu, S.; Shen, W.B. Isolation and characterization of a heme oxygenase-1 gene from Chinese cabbage. Mol. Biotechnol. 2012, 50, 8–17. [Google Scholar] [CrossRef]
- Anwar, A.; Yuan, C.; Cui, B.; Wang, L.X.; He, L.L.; Gao, J.W. BrMYB116 transcription factor enhances Cd stress tolerance by activating FIT3 in yeast and Chinese cabbage. Front. Plant Sci. 2024, 15, 1388924. [Google Scholar] [CrossRef]
- Janicka, M.; Reda, M.; Napieraj, N.; Michalak, A.; Jakubowska, D.; Kabala, K. Involvement of diamine oxidase in modification of plasma membrane proton pump activity in Cucumis sativus L. seedlings under cadmium stress. Int. J. Mol. Sci. 2023, 24, 262. [Google Scholar] [CrossRef]
- Duan, L.; Yu, J.W.; Xu, L.; Tian, P.; Hu, X.; Song, X.M.; Pan, Y. Functional characterization of a type 4 metallothionein gene (CsMT4) in cucumber. Hortic. Plant J. 2019, 5, 120–128. [Google Scholar] [CrossRef]
- Liu, J.Y.; Zhang, J.; Kim, S.H.; Lee, H.S.; Marinoia, E.; Song, W.Y. Characterization of Brassica rapa metallothionein and phytochelatin synthase genes potentially involved in heavy metal detoxification. PLoS ONE 2021, 16, e0252899. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Xu, W.Z.; Guo, J.B.; He, Z.Y.; Ma, M. Coordinated responses of phytochelatins and metallothioneins to heavy metals in garlic seedlings. Plant Sci. 2005, 169, 1059–1065. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Xu, W.Z.; Dai, W.T.; He, Z.Y.; Ma, M. Functional characterization of cadmium-responsive garlic gene AsMT2b: A new member of metallothionein family. Chinese Sci. Bull. 2006, 51, 409–416. [Google Scholar] [CrossRef]
- Sutkovic, J.; Karic, A.; Yildirim, A. In silico identification and expression analysis of Metal-nicotianamine transporter (YSL3) and Oligopeptide transporter 3 (OPT3) under Cd stress in Brassica oleracea var. acephala. Bot. Sci. 2020, 98, 516–523. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, A.X.; Yang, L.; Wei, J.P.; Bei, J.L.; Xu, Z.J.; Wang, X.F.; Chen, B.X. Identification of XTH family genes and expression analysis of endosperm weakening in lettuce (Lactuca sativa L.). Agronomy 2024, 14, 324. [Google Scholar] [CrossRef]
- Kolahi, M.; Kazemi, E.M.; Yazdi, M.; Goldson-Barnaby, A. Oxidative stress induced by cadmium in lettuce (Lactuca sativa Linn.): Oxidative stress indicators and prediction of their genes. Plant Physiol. Bioch. 2020, 146, 71–89. [Google Scholar] [CrossRef]
- Gao, F.; Li, J.; Zhang, J.; Li, N.H.; Tang, C.A.; Bakpa, E.P.; Xie, J.M. Genome-wide identification of the ZIP gene family in lettuce (Lactuca sativa L.) and expression analysis under different element stress. PLoS ONE 2022, 17, e0274319. [Google Scholar] [CrossRef]
- Cheng, H.; Kun, W.P.; Liu, D.S.; Su, Y.Q.; He, Q.W. Molecular cloning and expression analysis of CmMlo1 in melon. Mol. Biol. Rep. 2012, 39, 1903–1907. [Google Scholar] [CrossRef]
- Leng, Y.; Niu, Z.B.; Liu, S.H.; Qiao, F.J.; Liu, G.F.; Cheng, B.; Li, S.W. Characterisation of cytochrome c oxidase-coding genes from mung bean and their response to cadmium stress based on genome-wide identification and transcriptome analysis. Mol. Biol. Rep. 2025, 52, 17. [Google Scholar] [CrossRef]
- Yu, R.G.; Tang, Y.S.; Liu, C.F.; Du, X.L.; Miao, C.M.; Shi, G.R. Comparative transcriptomic analysis reveals the roles of ROS scavenging genes in response to cadmium in two pak choi cultivars. Sci. Rep. 2017, 7, 9217. [Google Scholar] [CrossRef]
- Wu, X.; Chen, J.H.; Yue, X.M.; Wei, X.N.; Zou, J.W.; Chen, Y.H.; Su, N.N.; Cui, J. The zinc-regulated protein (ZIP) family genes and glutathione s-transferase (GST) family genes play roles in Cd resistance and accumulation of pak choi (Brassica campestris ssp. chinensis). Ecotox. Environ. Safe 2019, 183, 109571. [Google Scholar] [CrossRef]
- Yang, X.Y.; Lv, H.P.; Zhang, W.; Liang, H.J.; Gao, Y.P.; Kang, Y.C.; Wu, Y.B.; Wang, F.F.; Xi, C.Y. Genome-wide identification and expression patterns in response to signals from cadmium of gene family in potato (Solanum tuberosum L.). Chem. Biol. Technol. Agric. 2024, 11, 27. [Google Scholar] [CrossRef]
- Tian, W.J.; Huang, Y.; Li, D.D.; Meng, L.L.; He, T.B.; He, G.D. Identification of StAP2/ERF genes of potato (Solanum tuberosum) and their multiple functions in detoxification and accumulation of cadmium in yeast: Implication for Genetic-based phytoremediation. Sci. Total Environ. 2022, 810, 152322. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.J.; He, G.D.; Qin, L.J.; Li, D.D.; Meng, L.L.; Huang, Y.; He, T.B. Genome-wide analysis of the NRAMP gene family in potato (Solanum tuberosum): Identification, expression analysis and response to five heavy metals stress. Ecotox. Environ. Safe 2021, 208, 111661. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, G.D.; He, Y.Q.; Tang, Y.Y.; Zhao, F.L.; He, T.B. Discovery of cadmium-tolerant biomacromolecule (StCAX1/4 transportproteins) in potato and its potential regulatory relationship with WRKY transcription factors. Int. J. Biol. Macromol. 2023, 228, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; He, G.D.; Tian, W.J.; Saleem, M.; Huang, Y.; Meng, L.L.; Wu, D.X.; He, T.B. Comparative and systematic omics revealed low Cd accumulation of potato StMTP9 in yeast: Suggesting a new mechanism for heavy metal detoxification. Int. J. Mol. Sci. 2021, 22, 10478. [Google Scholar] [CrossRef]
- He, Y.Q.; He, G.D.; Lou, F.; Zhou, Z.; Liu, Y.; Zhang, Y.L.; He, T.B. Identification of the major effector StSROs in potato: A potential StWRKY-SRO6 regulatory pathway enhances plant tolerance to cadmium stress. Int. J. Mol. Sci. 2022, 23, 14318. [Google Scholar] [CrossRef]
- He, G.D.; Tian, W.J.; Qin, L.J.; Meng, L.L.; Wu, D.X.; Huang, Y.; Li, D.D.; Zhao, D.G.; He, T.B. Identification of novel heavy metal detoxification proteins in Solanum tuberosum: Insights to improve food security protection from metal ion stress. Sci. Total Environ. 2021, 779, 146197. [Google Scholar] [CrossRef]
- Charfeddine, M.; Charfeddine, S.; Bouaziz, D.; Ben Messaoud, R.; Bouzid, R.G. The effect of cadmium on transgenic potato (Solanum tuberosum) plants overexpressing the StDREB transcription factors. Plant Cell Tissue Organ Cult. 2017, 128, 521–541. [Google Scholar] [CrossRef]
- Zheng, Z.F.; Gao, J.; Wang, C.Y.; Peng, H.; Zeng, J.; Chen, F.B. Genome-wide identification and expression pattern analysis of the MATE gene family in carmine radish (Raphanus sativus L.). Gene 2023, 887, 147734. [Google Scholar] [CrossRef]
- Tang, M.J.; Zhang, X.L.; Xu, L.; Wang, Y.; Chen, S.; Dong, J.H.; Liu, L.W. Genome- and transcriptome-wide characterization of ZIP gene family reveals their potential role in radish (Raphanus sativus) response to heavy metal stresses. Sci. Hortic. 2024, 324, 112564. [Google Scholar] [CrossRef]
- Peng, H.; Gao, J.; Song, X. Identification of heavy metal-responsive genes in radish (Raphanus sativus L.) through RNA-Seq meta-analysis. Sci. Hortic. 2021, 288, 110402. [Google Scholar] [CrossRef]
- Chen, L.; Wu, M.; Jin, W.; Lei, T.; Li, Y.; Wu, X.; Fu, S. Gene identification and transcriptome analysis of cadmium stress in tomato. Front. Sustain. Food Syst. 2023, 7, 1303753. [Google Scholar] [CrossRef]
- Xing, Q.F.; Hasan, M.K.; Li, Z.C.; Yang, T.; Jin, W.D.; Qi, Z.Y.; Yang, P.; Wang, G.H.; Ahammed, G.J.; Zhou, J. Melatonin-induced plant adaptation to cadmium stress involves enhanced phytochelatin synthesis and nutrient homeostasis in Solanum lycopersicum L. J. Hazard. Mater. 2023, 456, 131670. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.J.; Liu, Z.S.; Feng, L.; Wang, C.L.; Liu, C.; Li, A.L.; Yao, K.D.; Liao, W.B. The response of DNA methyltransferase and demethylase genes to abiotic stresses in tomato seedling. Plant Physiol. Bioch. 2024, 217, 109276. [Google Scholar] [CrossRef] [PubMed]
- Kisa, D.; Öztürk, L.; Tekin, S. Gene expression analysis of metallothionein and mineral elements uptake in tomato (Solanum lycopersicum) exposed to cadmium. J. Plant Res. 2016, 129, 989–995. [Google Scholar] [CrossRef]
- Khan, I.; Asaf, S.; Jan, R.H.T.L.; Bilal, S.; Lubna; Khan, A.L.; Kim, K.M.; Al-Harrasi, A. Genome-wide annotation and expression analysis of WRKY and bHLH transcriptional factor families reveal their involvement under cadmium stress in tomato (Solanum lycopersicum L.). Front. Plant Sci. 2023, 14, 1100895. [Google Scholar] [CrossRef]
- He, L.; Wang, H.T.; Zhao, Q.; Cheng, Z.B.; Tai, P.D.; Liu, W. Tomato grafting onto Torubamu (Solanum melongena): miR166a and miR395b reduce scion Cd accumulation by regulating sulfur transport. Plant Soil 2020, 452, 267–279. [Google Scholar] [CrossRef]
- Beauchet, A.; Gévaudant, F.; Gonzalez, N.; Chevalier, C. In search of the still unknown function of FW2.2/CELL NUMBER REGULATOR, a major regulator of fruit size in tomato. J. Exp. Bot. 2021, 72, 5300–5311. [Google Scholar] [CrossRef]
- Xu, J.R.; Cui, J.; He, Q.Y.; Liu, Y.Z.; Lu, X.F.; Qi, J.; Xiong, J.L.; Yu, W.J.; Li, C.X. Genome-wide identification of HIPP and mechanism of SlHIPP4/7/9/21/26/32 mediated phytohormones response to Cd, osmotic, and salt stresses in tomato. Plant Physiol. Bioch. 2024, 217, 109220. [Google Scholar] [CrossRef]
- Anwar, A.; Wang, Y.D.; Chen, M.Q.; Zhang, S.W.; Wang, J.M.; Feng, Y.Q.; Xue, Y.X.; Zhao, M.F.; Su, W.; Chen, R.Y.; et al. Zero-valent iron (nZVI) nanoparticles mediate SlERF1 expression to enhance cadmium stress tolerance in tomato. J. Hazard. Mater. 2024, 468, 133829. [Google Scholar] [CrossRef]
- Wu, Y.S.; Li, X.; Chen, D.; Han, X.; Li, B.Q.; Yang, Y.H.; Yang, Y.P. Comparative expression analysis of heavy metal ATPase subfamily genes between Cd-tolerant and Cd-sensitive turnip landraces. Plant Divers. 2019, 41, 275–283. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Gong, F.Y.; Shen, C.; He, C.T.; Fu, H.L.; Wang, X.S.; Tan, X.; Xu, P.L.; Yang, Z.Y. Cloning, characterization and expression analysis of metallothioneins from Ipomoea aquatica and their cultivar-dependent roles in Cd accumulation and detoxification. Ecotox. Environ. Safe 2018, 165, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.A.; Ahmed, S.; Yasin, N.A. 24-epibrassinolide triggers cadmium stress mitigation in Cucumis sativus through intonation of antioxidant system. S. Afr. J. Bot. 2019, 127, 349–360. [Google Scholar] [CrossRef]
- Shah, A.A.; Ahmed, S.; Abbas, M.; Yasin, N.A. Seed priming with 3-epibrassinolide alleviates cadmium stress in Cucumis sativus through modulation of antioxidative system and gene expression. Sci. Hortic. 2020, 265, 109203. [Google Scholar] [CrossRef]
- Chen, H.; Yang, R.X.; Zhang, X.; Chen, Y.H.; Xia, Y.; Xu, X.M. Foliar application of gibberellin inhibits the cadmium uptake and xylem transport in lettuce (Lactuca sativa L.). Sci. Hortic. 2021, 288, 110410. [Google Scholar] [CrossRef]
- Feng, L.; Wei, L.J.; Liu, Y.Y.; Hu, D.L.; Gong, W.T.; Liao, W.B. Hydrogen peroxide is involved in methyl jasmonate-induced adventitious rooting in cucumber under cadmium stress. Sci. Hortic. 2023, 309, 111666. [Google Scholar] [CrossRef]
- Chen, X.M.; Shi, X.Y.; Ai, Q.; Han, J.Y.; Wang, H.S.; Fu, Q.S. Transcriptomic and metabolomic analyses reveal that exogenous strigolactones alleviate the response of melon root to cadmium stress. Hortic. Plant J. 2022, 8, 637–649. [Google Scholar] [CrossRef]
- Leng, Y.; Wu, P.M.; Li, S.W.; Zhang, X.J. Foliar application of abscisic acid alleviates cadmium stress by modulating differential physiological and transcriptome response in leaves, stems, and roots of mung bean seedlings. J. Plant Growth Regul. 2024, 43, 4904–4922. [Google Scholar] [CrossRef]
- Waheed, A.; Zhang, Q.; Xu, H.; Dou, H.; Muhammad, M.; Aili, A.; Alshaharni, M.O. Mitigation of cadmium stress by salicylic acid: Physiological and biochemical responses in NM-2006, NM-92, and Mash-88 mung bean varieties. J. Hazard. Mater. 2024, 485, 136878. [Google Scholar] [CrossRef]
- Rady, M.M.; Alshallash, K.S.; Desoky, E.M.; Taie, H.A.A.; Mohamed, I.A.A.; El-Badri, A.M.; Howladar, S.M.; Abdelkhalik, A. Synergistic effect of trans-zeatin and silymarin on mitigation of cadmium stress in chili pepper through modulating the activity of antioxidant enzymes and gene expressions. J. Appl. Res. Med. Aroma. 2023, 35, 100498. [Google Scholar] [CrossRef]
- Khan, M.Y.; Prakash, V.; Yadav, V.; Chauhan, D.K.; Prasad, S.M.; Ramawat, N.; Singh, V.P.; Tripathi, D.K.; Sharma, S. Regulation of cadmium toxicity in roots of tomato by indole acetic acid with special emphasis on reactive oxygen species production and their scavenging. Plant Physiol. Bioch. 2019, 142, 193–201. [Google Scholar] [CrossRef]
- Guan, X.X.; Sui, C.L.; Luo, K.C.; Chen, Z.F.; Feng, C.Y.; Dong, X.F.; Zeng, B.P.; Dong, X.; Liu, X.F. Effects of α-naphthylacetic acid on cadmium stress and related factors of tomato by regulation of gene expression. Agronomy 2022, 12, 2141. [Google Scholar] [CrossRef]
- Roche, M.C.; Liu, W.; Hernández, R. An improved method for agrobacterium-mediated genetic transformation of three types of lettuce. Plants 2025, 14, 620. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Wang, C.; Yuan, X.; Zhang, M.; Zhang, C.; Qin, T.; Wang, H.; Xu, L.; Liu, L.; Wang, Y. Exploring an economic and highly efficient genetic transformation and genome-editing system for radish through developmental regulators and visible reporter. Plant J. 2024, 120, 1682–1692. [Google Scholar] [CrossRef] [PubMed]

| Vegetable Name | Gene | Tissue Specificity a | Localization b | Core Mechanism Summary f | Key Functional Impact | Ref. |
|---|---|---|---|---|---|---|
| Tomato | LeNRAMP3 | - | PM | Interacts with NAS/FRO/IRT1 complex; mediates Fe/Cd transport and signaling | ↑ heavy metal translocation | [13] |
| Potato | StNRAMP2 | - | - | Regulates Cd partitioning: silencing ↑ Cd in tubers, overexpression ↑ Cd in shoots | Modulates Cd accumulation patterns | [5] |
| Pak choi | BcHIPP16 | Whole seedling | PM | Direct Cd2+ influx promoter in roots | ↑ Cd uptake efficiency | [6] |
| Chinese cabbage | BcNRAMP1 | Whole plant | PM | Activated by Cd/Mn deficiency; ↑ Cd/Mn root uptake | Dual regulation under metal stress | [27] |
| Chinese cabbage | BrMT1 | Roots/flowers c | Cyto/Chl d | Cysteine-rich Cd chelation; chloroplast targeting ↑ ROS scavenging | Confers Cd resistance via subcellular targeting | [24] |
| Radish | RsPDR8 | Vascular tissues e | PM | ↑ Cd efflux via PM transport; ↑ ROS scavenging & membrane stability | ↓ cellular Cd accumulation | [26] |
| Radish | RsNRAMP5 | Vascular cambium | PM | ↑ Cd influx; differential regulation of ROS/proline genes | Mediates Cd uptake-toxicity balance | [20] |
| Vegetable Name | Gene | Tissue Specificity a | Localization b | Core Mechanism Summary c | Key Functional Impact | Ref. |
|---|---|---|---|---|---|---|
| Tomato | LeSPL-CNR | - | - | ↓ Cd acquisition via NO-mediated repression of nitrate reductase | ↓ Cd accumulation (via iron-uptake suppression) | [31] |
| Tomato | HsfA1a | - | - | ↑ Melatonin biosynthesis (COMT1) and HSP expression under Cd stress | ↑ Cd tolerance (via upregulating HSP) | [28] |
| Potato | StWRKY6 | - | Nu | ↑ Antioxidant enzymes and photosynthesis regulation for stress resilience | ↑ Cd tolerance; ↓ Cd accumulation | [32] |
| Pepper | CaPF1 | - | - | ↑ Antioxidant enzyme activity and ↓ lipid peroxidation under Cd stress | ↑ Cd tolerance in transgenic plants | [33] |
| Pepper | CaWRKY41 | Roots, leaves, (shoots, flowers) | Nu (Arabidopsis) | ↑ H2O2 accumulation and ↑ Zn transporters activation for Cd uptake | ↓ Cd tolerance; ↑ Cd uptake | [29] |
| Carrot | DcMYB62 | - | Nu | ↑ Carotenoid/ABA/H2S biosynthesis, ↓ ROS, and ↑ stomatal closure activation | ↓ ROS; ↑ expression of heavy metal resistance genes | [25] |
| Bean | PvERF104 | - | Nu | ↓ Cd-induced lipid peroxidation and ↑ regulation of MRE-containing Cd response genes | ↓ Cd accumulation; ↑ stress tolerance | [34] |
| Bean | PvMTF-1 | - | Nu | ↑ Tryptophan synthesis via ASA2-mediated pathway by binding ASA2 promoter under Cd stress | ↓ Cd accumulation | [35] |
| Bean | PvERF15 | - | Nu | ↑ PvMTF-1 promoter activation via ACE-binding for Cd stress response regulation | Forms transcriptional pathway for Cd response | [30] |
| Vegetable Name | Gene | Tissue Specificity a | Localization b | Core Mechanism Summary d | Key Functional Impact | Ref. |
|---|---|---|---|---|---|---|
| Tomato | SlRING1 | - | PM/Nu | E3 ligase-mediated ubiquitination ↑ antioxidant/detoxification pathways | ↓ Cd accumulation & oxidative stress | [36,37] |
| Tomato | Sl1 | Roots c | PM | E3 ligase-mediated suppression of metal transporters ↑ antioxidant activity | ↓ Root Cd uptake | [40] |
| Tomato | Tgase | Leaves/flowers | Mt/CW/Chl | ↑ Polyamine/NO accumulation & cell wall modification | ↑ Cd chelation capacity | [41,42] |
| Tomato | SlSPR2 | - | - | JA-dependent regulation ↑ Cd-responsive metabolic pathways | Maintains physiological homeostasis | [43] |
| Tomato | SlMAPK3 | Stems/roots | - | MAPK signaling ↑ antioxidant activation & root morphology regulation | ↑ Root function under stress | [44] |
| Tomato | SlJMJ524 | Leaves/flowers | - | Epigenetic regulation ↓ metal transporters & ↑ GSH-PC synthesis | ↑ Cd sequestration | [45] |
| Tomato | SlJMJ18/23 | Flowers (SlJMJ18) c, young leaves (SlJMJ23) c | - | Epigenetic regulation ↓ metal transporters (ZIP1, IRT1) & ↑ antioxidant/phenol synthesis | ↑ Antioxidant capacity & phenol synthesis | [8] |
| Pepper | CaHMA1 | - | GA/ER | Heavy metal-binding domain ↑ Cd accumulation | ↑ Cd translocation to fruits | [38] |
| Chinese cabbage | BpPMSR3 | - | - | Methionine redox regulation ↑ GSH synthesis | ↑ Cd detoxification | [39] |
| Onion | AcGCL | Roots c | - | ↑ GSH/PC synthesis & protection of FOS metabolism | Maintains carbohydrate metabolism | [46] |
| Vegetable Name | Gene | Tissue Specificity a | Localization b | Core Mechanism Summary c | Key Functional Impact | Ref. |
|---|---|---|---|---|---|---|
| Tomato | Sly-miR398 | Roots, stems | - | ↓ regulated under Cd stress; ↑ CSD1/SOD expression to ↑ antioxidant defense; ↓ Cd uptake/translocation genes. | ↓ Oxidative damage; ↑ growth recovery | [7] |
| Water spinach | IamiR-4-3p | - | - | ↓ regulates GST3 and AWPM19-like, causing ↑ oxidative damage and ↑ Cd uptake/translocation. | ↑ Cd toxicity; ↓ apoplastic barrier | [47] |
| Tomato | SlTCMP-1 | Flower buds, leaves, fruits | - | Induced by Cd stress; interacts with HIPP26 to ↑ ROS scavenging genes and ↓ Cd translocation. | ↑ Cd stress response | [48] |
| Tomato | SlSGR2 | Various tissues | Chloroplast | Inhibits chlorophyll degradation; ↓ MDA content; ↑ antioxidant enzyme activity under Cd stress. | ↑ Cd tolerance; ↓ chlorophyll loss | [49] |
| Chinese cabbage | BrSSA4c | - | - | Overexpression ↑ Cd tolerance via activation of cis elements. | ↑ Cd resilience | [4] |
| Vegetable Name | Botanical Name | Genes | Ref(s). |
|---|---|---|---|
| bean | Phaseolus vulgaris L. | PvSR3; PvSR2 | [52,53] |
| Chinese cabbage | Brassica rapa L. | BrAHL24; BrHO1; BrMYB116 | [54,55,56] |
| cucumber | Cucumis sativus L. | DAO; CsNramp1, CsNramp4, CsZIP1, CsZIP8, CsHMA5, CsHMA2, CsHMA7; CsMT4 | [50,57,58] |
| flowering Chinese cabbage | Brassica rapa L. Chinensis | BrMTs, BrPCSs | [59] |
| garlic | Allium sativum L. | AsPCS1, AsMT2a; AsMT2b | [60,61] |
| kale | Brassica oleracea var. acephala | OPT3, YSL3 | [62] |
| lettuce | Lactuca sativa L. | LsXTH6, LsXTH7, LsXTH8, LsXTH32, and LsXTH33; LsAPXs, LsSODs; LsZIP1, LsZIP3, LsZIP10, LsZIP12, LsZIP13, LsZIP17, LsZIP19 | [63,64,65] |
| melon | Cucumis melo L. | CmMlo1 | [66] |
| mung bean | Vigna radiata (L.) R. Wilczek | VrCOXs | [67] |
| pak choi | Brassica chinensis L. | Fe SOD1, POD A2/44/54/62, and GST1; BcIRT1, BcZIP2; BcGSTUs | [21,68,69] |
| pepper | Capsicum annuum L. | CaNRAMP5, CaCOMT1 | [51] |
| potato | Solanum tuberosum L. | StCADs; StOPR1, StJAZ14; StAP2/ERF genes; StNRAMPs; StSROs 5/6; StABCs; StDREB1, StDREB2 | [23,70,71,72,73,74,75,76,77] |
| radish | Raphanus sativus L. | RsHSP70-5, RsHSP70-14, RsHSP70-21, RsHSP70-32; RsMATE37-a, RsMATE21, RsMATE43-c, RsMATE49-b, RsMATE31-b, RsMATE33, RsMATE46-c, RsMATE13-a, RsMATE16-b, RsMATE43-a, RsMATE27, RsMATE35-a, RsMATE40-b, RsMATE13-c and RsMATE26; RsZIP genes; WRKY6, WRKY28-like, WRKY33, MYB16, bHLH143, ERF—rap2.7, PIN1, MRP, ABC transporter genes, GST and LCC4 | [9,78,79,80] |
| tomato | Solanum lycopersicum L. | Solyc05g051550, Solyc02g077370, Solyc04g009440, Solyc12g099130, Solyc04g077960, Solyc04g051690, Solyc08g078180, Solyc07g045030, Solyc05g015850, Solyc05g053330, Solyc12g013640, Solyc07g065320, Solyc11g012700, Solyc11g069735, Solyc01g104820; COMT, PCS; SlDML1, SlDML2, SlDML3, SlDML4, SlMET1, SlDRM1L, SlDRM5 and SlDRM1L1; SlNRAMP1-5; SlMT1-4; SlWRKY76, SlWRKY38, SlWRKY46, SlWRKY19, SlWRKY33, SlWRKY35, SlWRKY45, SlWRKY51, SlWRKY55, SlbHLHs; miR166a, miR395b; FW2.2/CELL NUMBER REGULATOR (CNR); SlHIPP7/21/26/32; SlERF1 | [3,81,82,83,84,85,86,87,88,89] |
| turnip | Brassica rapa L. | BrHMAs | [90] |
| water spinach | Ipomoea aquatica Forsk. | IaMT2, IaMT3 | [91] |
| Vegetable Name | Botanical Name | Substance Name | Core Mechanism Summary a | Key Functional Impact | Ref(s). |
|---|---|---|---|---|---|
| cherry tomato | Solanum lycopersicum var. cerasiforme | Melatonin and brassinosteroids (BRs) | ↓ Cd content in shoots; ↑ antioxidant enzyme activities/gene expression; regulates K+ balance | ↑ Cd detoxification | [10] |
| cucumber | Cucumis sativus L. | 24-epibrassinolide (24-EBL) | ↑ Antioxidant system; regulates ethylene/IAA biosynthesis genes; ↓ lipid peroxidation and Cd uptake | ↓ Cd toxicity | [92] |
| cucumber | Cucumis sativus L. | 3-epibrassinolide (3-EBL) | ↑ Antioxidant enzymes; regulates ethylene/auxin biosynthesis genes; improves photosynthesis/water physiology | ↑ Cd stress resilience | [93] |
| cucumber | Cucumis sativus L. | Me-JA and H2O2 | ↑ Cell cycle-related gene expression; activates adventitious rooting through H2O2 signaling (inhibited by CAT/DPI) | ↑ Root development | [95] |
| lettuce | Lactuca sativa L. | Gibberellin (GA) | ↓ IRT1/Nramp1 expression (↓ Cd uptake); ↓ HMA2/HMA4 expression (↓ root-to-shoot translocation) | ↑ Cd tolerance | [94] |
| lettuce | Lactuca sativa L. | salicylic acid (SA) | Regulates Nramp5/HMA4/SAMT expression; ↑ photosynthesis; modulates antioxidant/osmotic systems | ↓ Cd accumulation; ↑ oxidative defense | [11] |
| melon | Cucumis melo L. | SLs | ↓ Cd stress in roots by regulating redox-related genes (POD, LOX), transcription factors (MYB, AP2/ERF), and ↑ JA biosynthesis/flavonoid pathways | ↑ Antioxidant capacity; ↓ oxidative damage | [96] |
| mung bean | Vigna radiata L. | abscisic acid (ABA) | ↑ Antioxidative enzymes/IAA oxidase activity; regulates cell wall/secondary metabolism genes | ↑ Adventitious root formation | [97] |
| mung bean | Vigna radiata L. | SA | ↑ SOD/POD/CAT/APX activities; modulates phytohormones (↑ ABA/JA, ↓ ethylene); regulates osmolyte metabolism | ↑ Physiological homeostasis | [98] |
| pepper | Capsicum annuum L. | Trans-zeatin (tr-Z) and silymarin (Sm) | ↓ Cd uptake/accumulation; ↑ antioxidant enzymes/compounds; modulates stress-responsive gene expression | ↑ Antioxidant defense system | [99] |
| radish | Raphanus sativus L. | Strigolactone (SL) and acidified biochar (AB) | SL regulates root architecture and chloroplast development; AB adsorbs Cd ions and improves soil conditions; combination ↑ antioxidant activities | ↑ Cd stress tolerance | [17] |
| tomato | Solanum lycopersicum L. | IAA | ↑ NO accumulation → activates AsA-GSH cycle; ↓ Cd accumulation/oxidative markers | ↑ Photosynthetic protection | [100] |
| tomato | Solanum lycopersicum L. | α-naphthaleneacetic acid (NAA) | Regulates defense genes in shoots and oxidoreductase/auxin-response genes in roots; modulates antioxidant system/Cd transport | ↓ Cd-induced damage | [101] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Dong, C. Molecular Mechanisms of Cadmium Stress Resistance in Vegetable Crops. Int. J. Mol. Sci. 2025, 26, 5812. https://doi.org/10.3390/ijms26125812
Zhang M, Dong C. Molecular Mechanisms of Cadmium Stress Resistance in Vegetable Crops. International Journal of Molecular Sciences. 2025; 26(12):5812. https://doi.org/10.3390/ijms26125812
Chicago/Turabian StyleZhang, Mengxia, and Chunjuan Dong. 2025. "Molecular Mechanisms of Cadmium Stress Resistance in Vegetable Crops" International Journal of Molecular Sciences 26, no. 12: 5812. https://doi.org/10.3390/ijms26125812
APA StyleZhang, M., & Dong, C. (2025). Molecular Mechanisms of Cadmium Stress Resistance in Vegetable Crops. International Journal of Molecular Sciences, 26(12), 5812. https://doi.org/10.3390/ijms26125812






