Genome-Wide Identification of Luffa Sucrose Synthase Genes Reveals LaSUS1-Mediated Sugar Metabolism Boosting Drought Tolerance
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification and Characterization of LaSUS
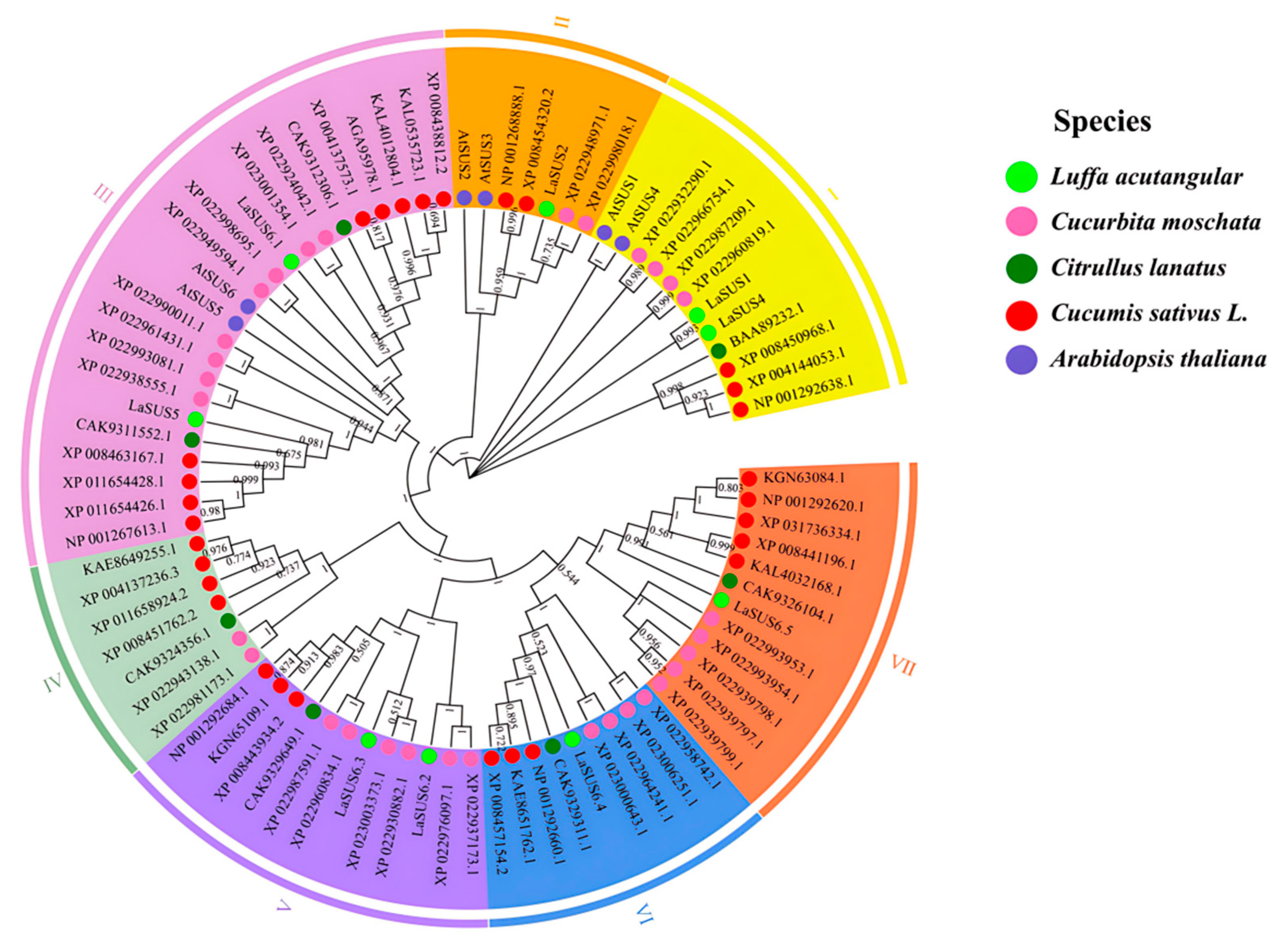
2.2. Phylogenetic Analysis of SUS Genes in Four Cucurbitaceae Species
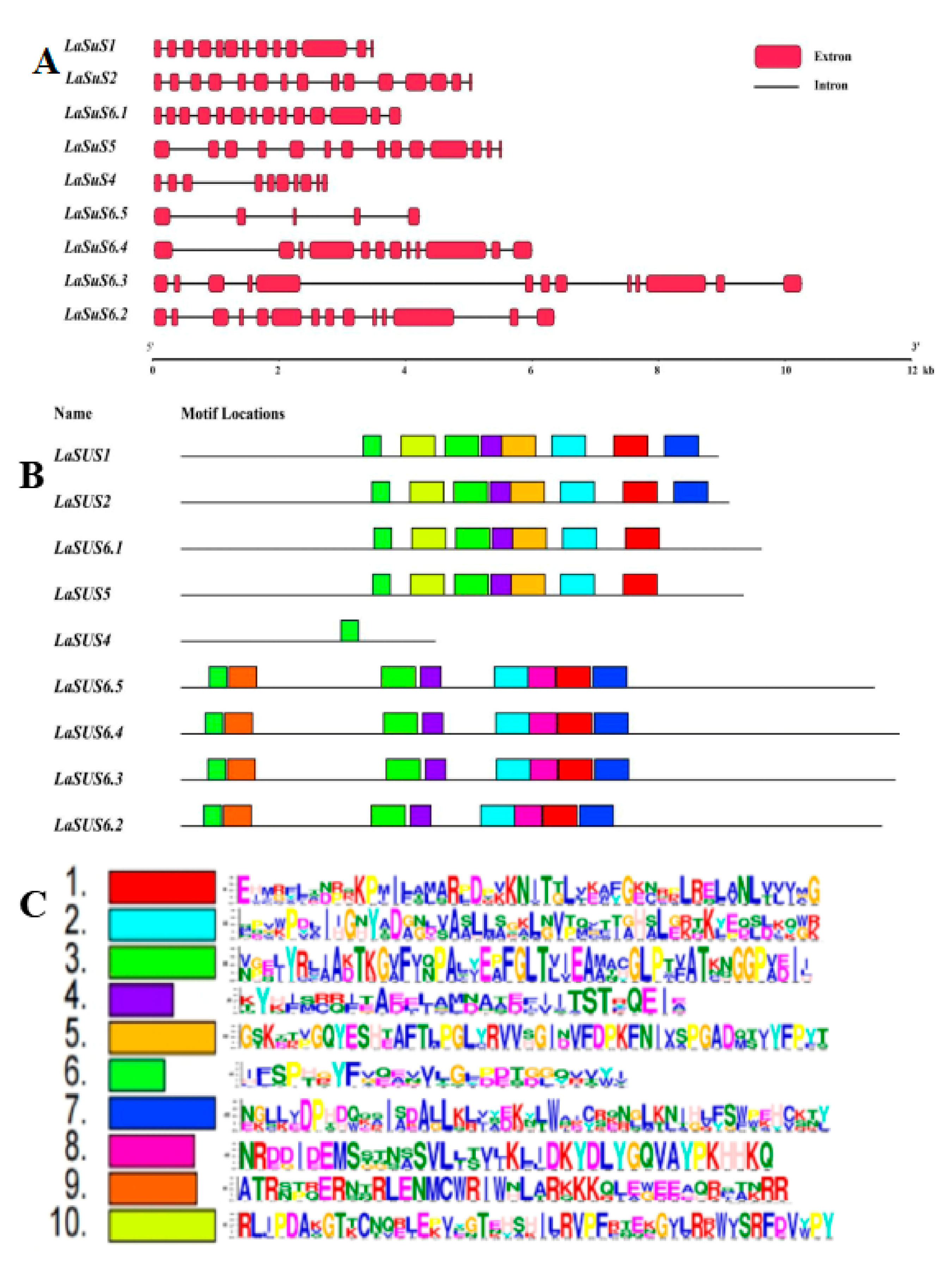
2.3. Gene Structure and Conserved Motif of LaSUS
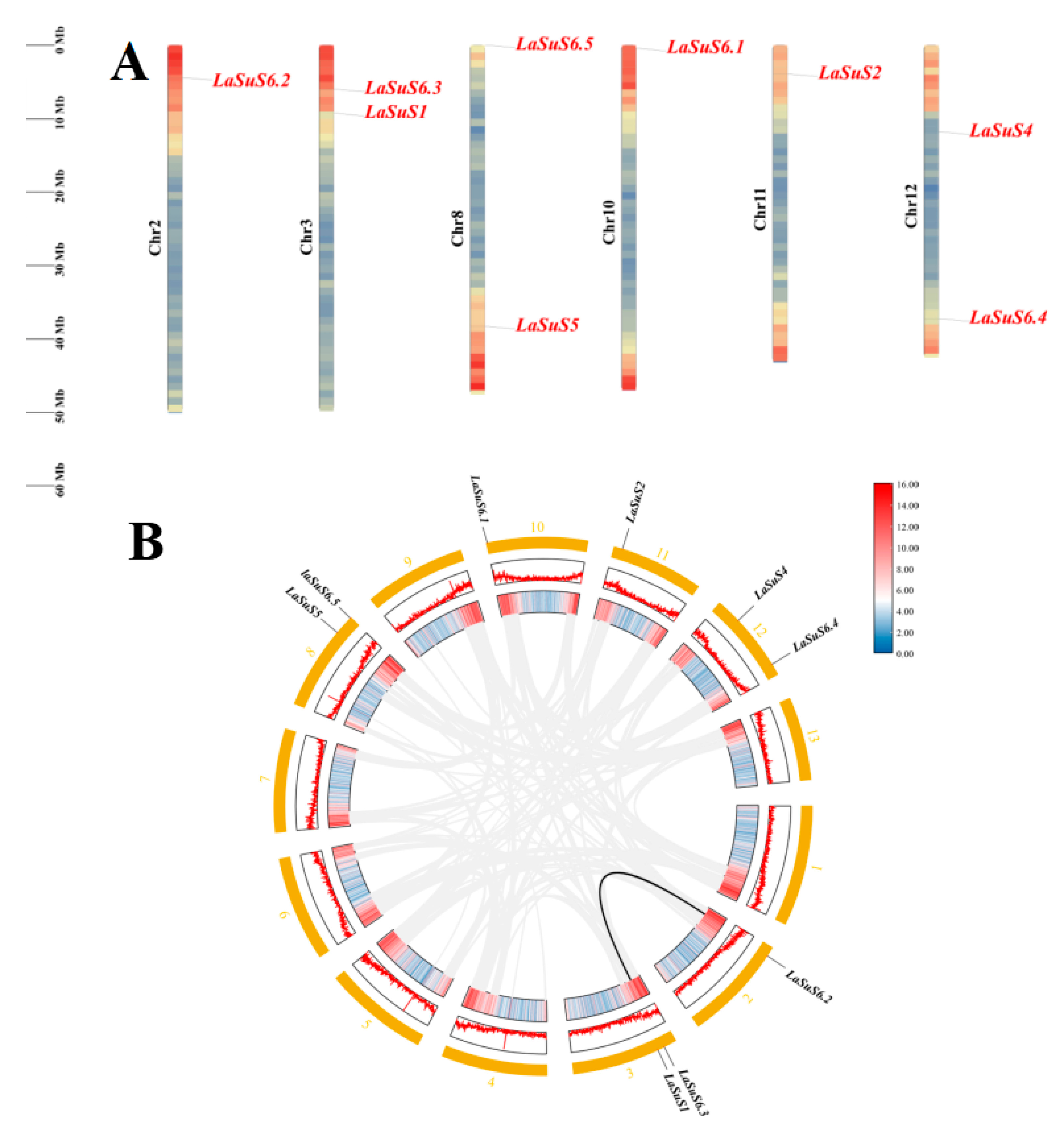
2.4. Chromosomal Location and Homology Analysis of LaSUS Genes
2.5. Detection of Positive Selection Among SUS Genes in Four Cucurbitaceae Species
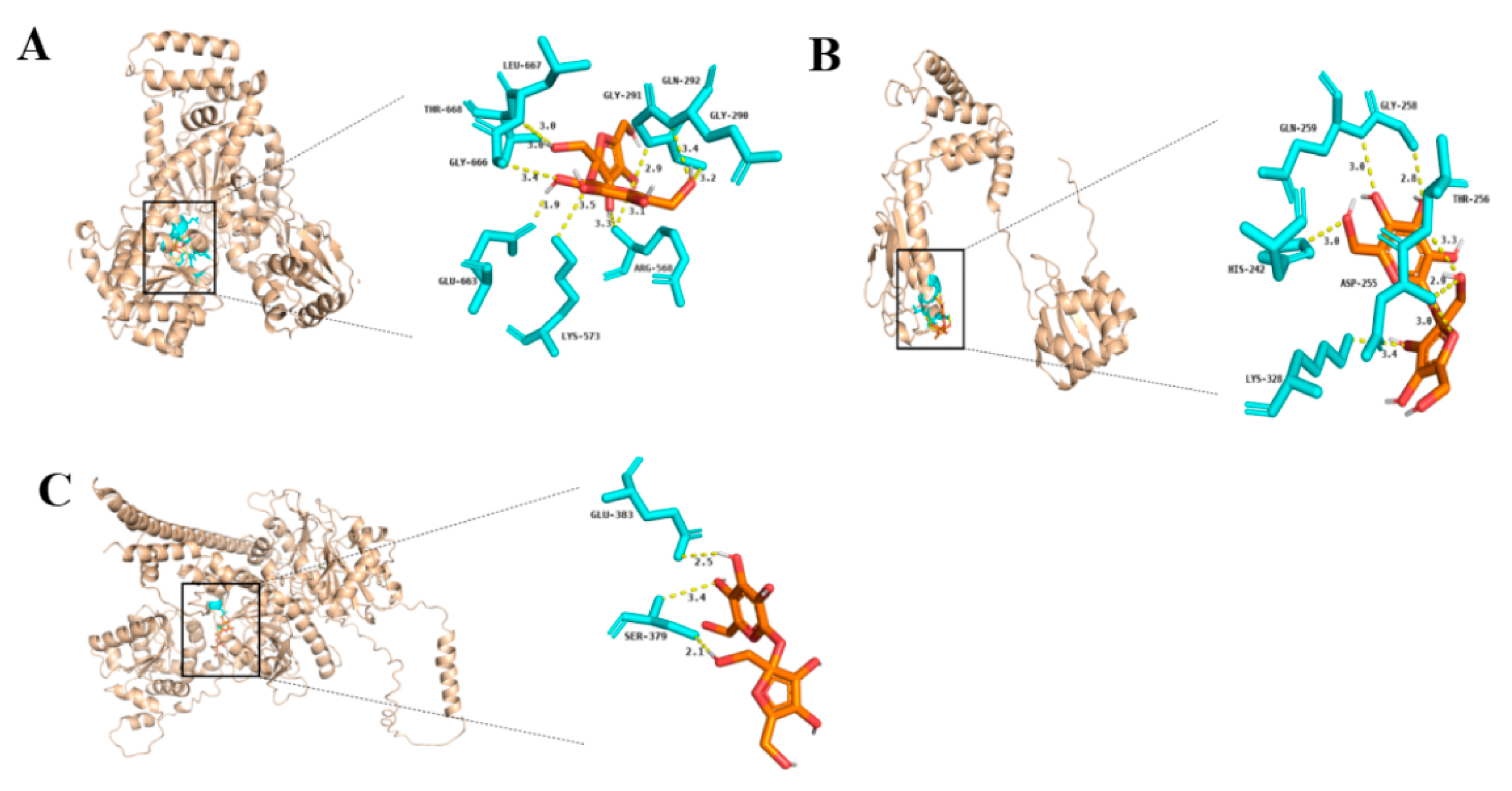
2.6. Molecular Docking Reveals the Function of LaSUS in Sucrose Metabolism
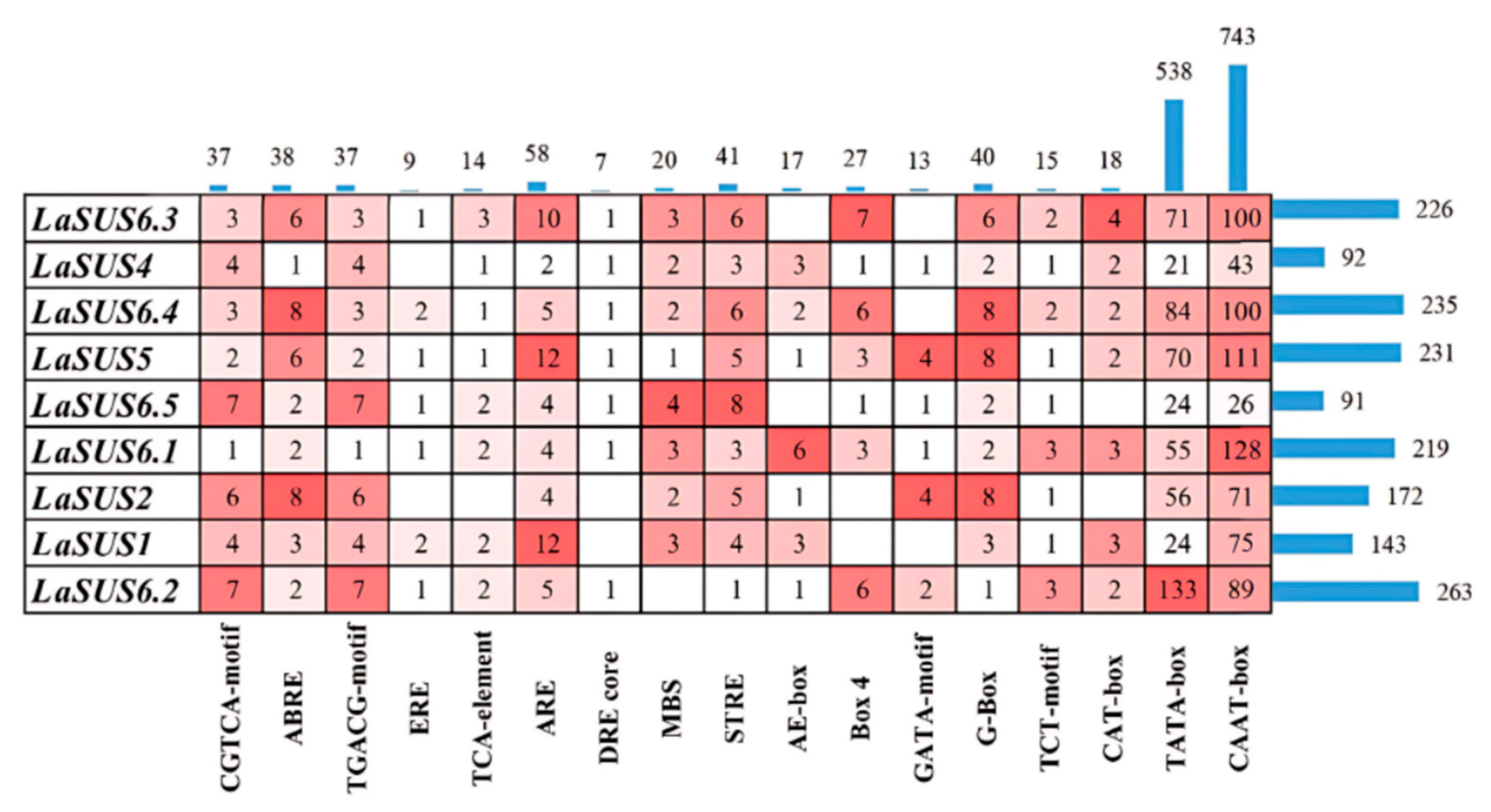
2.7. Distribution of Cis Elements in the LaSUS Promoters
2.8. Expression Levels of LaSUS in Four Postharvest Periods in Luffa Fruit
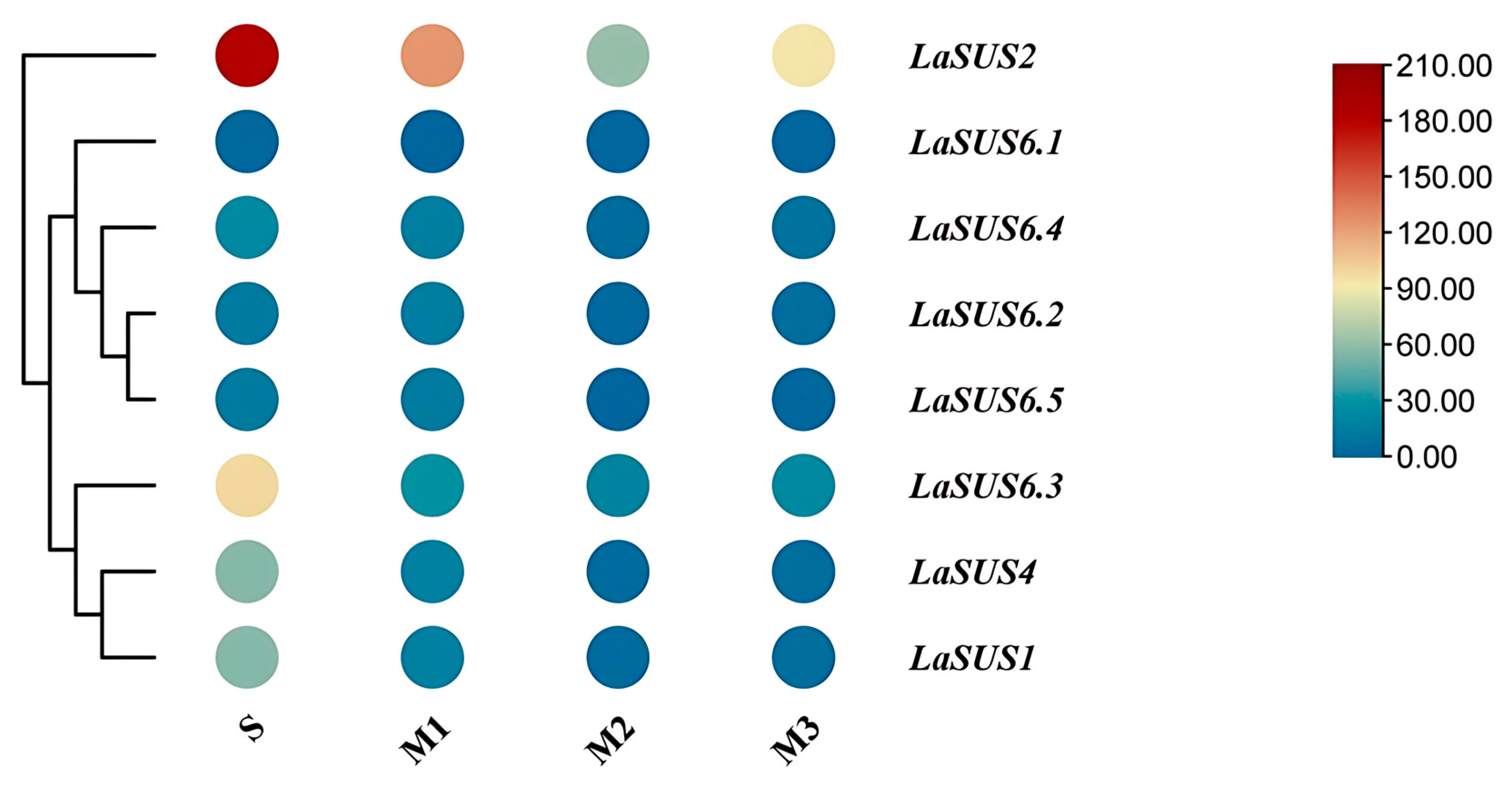
2.9. Expression Patterns of LaSUS in Luffa Different Tissues
2.10. Correlation of LaSUS Expression with Sugar and Cellulose Content
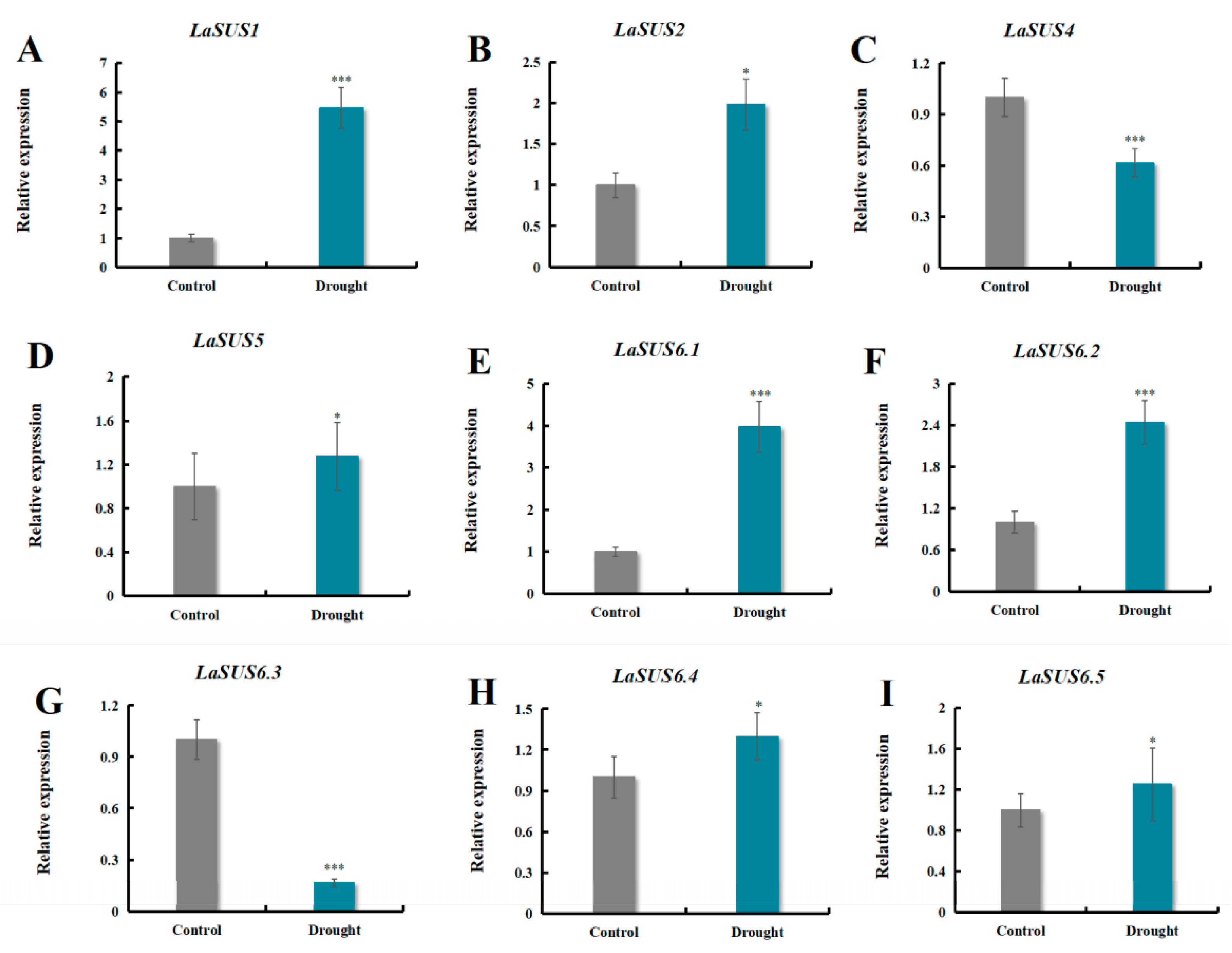
2.11. LaSUS Gene Expression Profiles Under Drought Stress
3. Discussion
4. Materials and Methods
4.1. Identification of LaSUS and Phylogenetic Analysis
4.2. Chromosomal Mapping and Syntenic Analysis of LaSUS
4.3. Gene Structure, Conserved Motifs, and Subcellular Localization Analysis of LaSUS
4.4. Cis Element Screening in the Promoters
4.5. Selective Pressure Analysis of LaSUS
4.6. Molecular Docking
4.7. The Expression Level Measurement of LaSUS
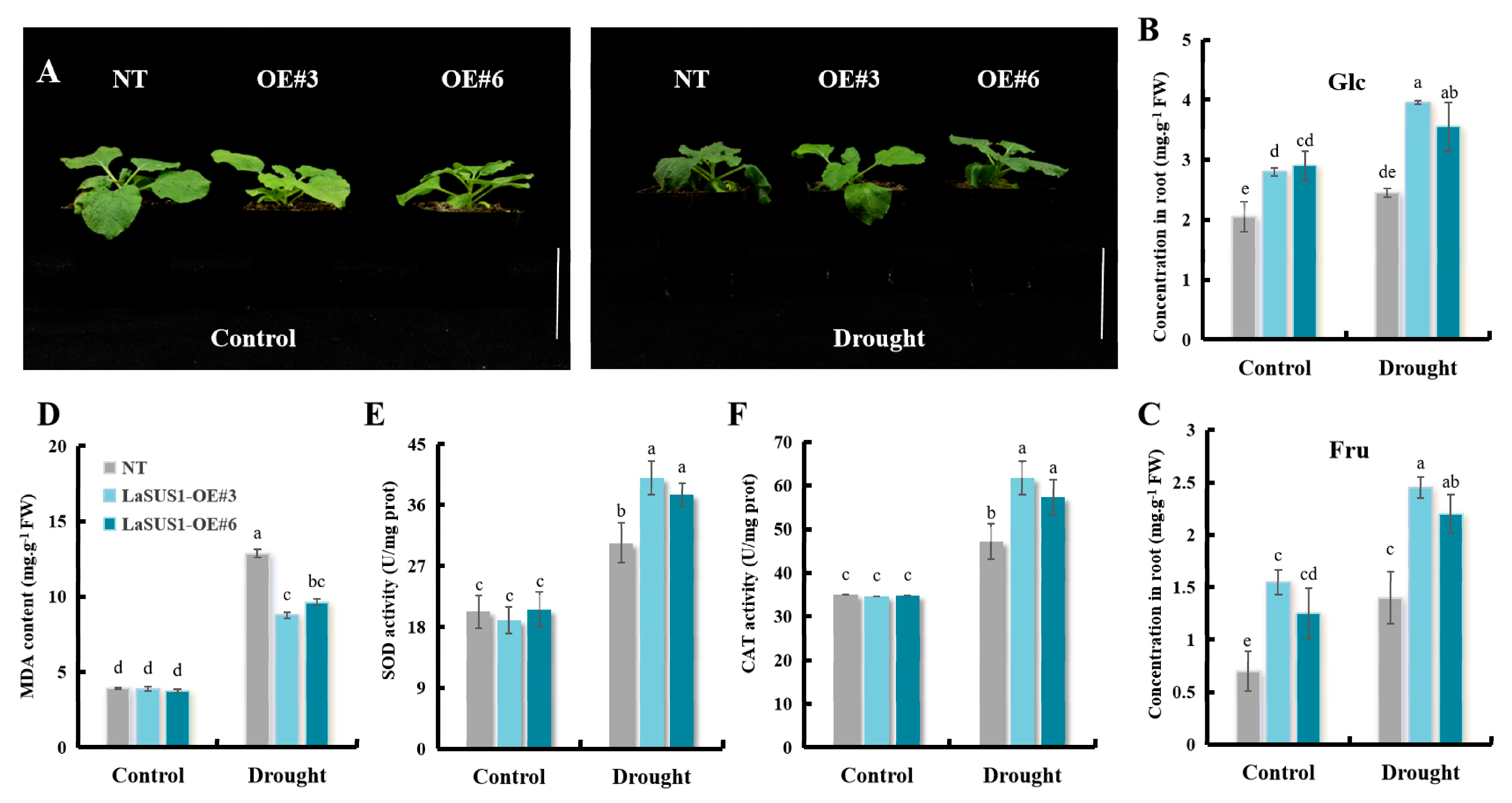
4.8. Construction of LaSUS Transgenic Tobacco Plants
4.9. Drought Stress Treatments and Physiological Indicators Measurements
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, X.; Shen, W.; Huang, J.; Zhang, T.; Zhang, T.; Zhang, X.; Cui, Y.; Sang, X.; Ling, Y.; Li, Y.; et al. Mutation of the OsSAC1 Gene, which Encodes an Endoplasmic Reticulum Protein with an Unknown Function, Causes Sugar Accumulation in Rice Leaves. Plant Cell Physiol. 2018, 59, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P.; Stitt, M. Diurnal changes in sucrose, nucleotides, starch synthesis and AGPS transcript in growing potato tubers that are suppressed by decreased expression of sucrose phosphate synthase. Plant J. 2000, 23, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.M. Phloem Loading and Unloading of Sucrose: What a Long, Strange Trip from Source to Sink. Annu. Rev. Plant Biol. 2022, 73, 553–584. [Google Scholar] [CrossRef] [PubMed]
- Eveland, A.L.; Jackson, D.P. Sugars, signalling, and plant development. J. Exp. Bot. 2012, 63, 3367–3377. [Google Scholar] [CrossRef]
- Jia, Z.; Cheng, X.; Dou, W. Research on the Synthesis of Puerarin Based on the Cascade Catalysis of Glycosyltransferase and Sucrose Synthase. Biotechnol. Adv. 2023, 13, 247–256. [Google Scholar]
- Coleman, H.D.; Yan, J.; Mansfield, S.D. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc. Natl. Acad. Sci. USA 2009, 106, 13118. [Google Scholar] [CrossRef]
- Li, J.; Baroja-Fernández, E.; Bahaji, A.; Muñoz, F.J.; Ovecka, M.; Montero, M.; Sesma, M.T.; Alonso-Casajús, N.; Almagro, G.; Sánchez-López, A.M.; et al. Enhancing sucrose synthase activity results in increased levels of starch and ADP-glucose in maize (Zea mays L.) seed endosperms. Plant Cell Physiol. 2013, 54, 282–294. [Google Scholar] [CrossRef]
- Zhang, Q.; Dai, W.T.; Li, J.X.; Jin, X.W. Effects of sugar metabolism on post-ripening softening of melon fruits. Food Ferment. Ind. 2020, 46, 112–117. [Google Scholar]
- Quinet, M.; Angosto, T.; Yuste-Lisbona, F.J.; Blanchard-Gros, R.; Bigot, S.; Martinez, J.P.; Lutts, S. Tomato Fruit Development and Metabolism. Front. Plant Sci. 2019, 10, 1554. [Google Scholar] [CrossRef]
- Jia, H.; Wang, Y.; Sun, M.; Li, B.; Han, Y.; Zhao, Y.; Li, X.; Ding, N.; Li, C.; Ji, W.; et al. Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytol. 2013, 198, 453–465. [Google Scholar] [CrossRef]
- Geigenberger, P.; Reimholz, R.; Deiting, U.; Sonnewald, U.; Stitt, M. Decreased expression of sucrose phosphate synthase strongly inhibits the water stress–induced synthesis of sucrose in growing potato tubers. Plant J. Cell Mol. Biol. 1999, 19, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L.; Chourey, P.S. A fiberless seed mutation in cotton is associated with lack of fiber cell initiation in ovule epidermis and alterations in sucrose synthase expression and carbon partitioning in developing seeds. Plant Physiol. 1998, 118, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.M.; Brill, E.; Llewellyn, D.J.; Furbank, R.T.; Ruan, Y.L. Overexpression of a potato sucrose synthase gene in cotton accelerates leaf expansion, reduces seed abortion, and enhances fiber production. Mol. Plant 2012, 5, 430–441. [Google Scholar] [CrossRef]
- Wei, Z.; Qu, Z.; Zhang, L.; Zhao, S.; Bi, Z.; Ji, X.; Wang, X.; Wei, H. Overexpression of poplar xylem sucrose synthase in tobacco leads to a thickened cell wall and increased height. PLoS ONE 2015, 10, e0120669. [Google Scholar] [CrossRef]
- Zhao, C.; Hua, L.N.; Liu, X.F.; Li, Y.Z.; Shen, Y.Y. Sucrose synthase FaSS1 plays an important role in the regulation of strawberry fruit ripening. Plant Growth Regul. 2017, 81, 175–181. [Google Scholar] [CrossRef]
- Zhu, Q.; Tan, Q.; Gao, Q.; Zheng, S.; Chen, W.; Galaud, J.P.; Li, X.; Zhu, X. Calmodulin-like protein CML15 interacts with PP2C46/65 to regulate papaya fruit ripening via integrating calcium, ABA and ethylene signals. Plant Biotechnol. 2024, 22, 1703–1723. [Google Scholar] [CrossRef]
- Lindsay, E.W.; Susan, R.S.; Lukas, A.M.; Michael, M. Comparative analysis of Cucurbita pepo metabolism throughout fruit development in acorn squash and oilseed pumpkin. Hortic. Res. 2016, 3, 16045. [Google Scholar]
- Gonzalez, E.M.; Gordon, A.J.; James, C.L.; Arrese-lgor, C. The role of sucrose synthase in the response of soybean nodules to drought. J. Exp. Bot. 1995, 46, 1515–1523. [Google Scholar] [CrossRef]
- Verma, E.S.; Singal, B.; Munjal, R. Purification of sucrose synthase from thermotolerant wheat grains and its characterization. J. Environ. Biol. 2018, 39, 459–466. [Google Scholar] [CrossRef]
- Salvi, P.; Agarrwal, R.; Kajal; Gandass, N.; Manna, M.; Kaur, H.; Deshmukh, R. Sugar transporters and their molecular tradeoffs during abiotic stress responses in plants. Physiol. Plant 2022, 3, e13652. [Google Scholar] [CrossRef]
- Gujjar, R.S.; Roytrakul, S.; Chuekong, W.; Supaibulwatana, K. A synthetic cytokinin influences the accumulation of leaf soluble sugars and sugar transporters, and enhances the drought adaptability in rice. 3 Biotech. 2021, 11, 369. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sui, X.; Guo, J.; Wang, Z.; Cheng, J.; Ma, S.; Li, X.; Zhang, Z. Antisense suppression of cucumber (Cucumis sativus L.) sucrose synthase 3 (CsSUS3) reduces hypoxic stress tolerance. Plant Cell Environ. 2014, 37, 795–810. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Lin, Y.; Meng, Y.; Si, C. Genome-Wide Identification and Analysis of SUS and AGPase Family Members in Sweet Potato: Response to Excessive Nitrogen Stress during Storage Root Formation. Int. J. Mol. Sci. 2024, 25, 8236. [Google Scholar] [CrossRef]
- Arise, R.O.; Idi, J.J.; Mic-Braimoh, I.M.; Korode, E.; Ahmed, R.N.; Osemwegie, O. In vitro Angiotesin-1-converting enzyme, α-amylase and α-glucosidase inhibitory and antioxidant activities of Luffa cylindrical (L.) M. Roem seed protein hydrolysate. Heliyon 2019, 5, e01634. [Google Scholar] [CrossRef]
- Felix, A.O.; Chigozie, I.O.; Chidimma, C.; Blessing, U.; Peter, E. Evaluation of Antioxidant, Anti-inflammatory and Antimicrobial Activities of the Leaf Extracts of Luffa cylindrica. J. Health Sci. 2018, 8, 101–109. [Google Scholar]
- Nemati, F.; Ghanati, F.; Ahmadi Gavlighi, H.; Sharifi, M. Comparison of sucrose metabolism in wheat seedlings during drought stress and subsequent recovery. Biol Plant 2018, 62, 595–599. [Google Scholar] [CrossRef]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Schmölzer, K.; Gutmann, A.; Diricks, M.; Desmet, T.; Nidetzky, B. Sucrose synthase: A unique glycosyltransferase for biocatalytic glycosylation process development. Biotechnol. Adv. 2016, 34, 88–111. [Google Scholar] [CrossRef]
- Zhang, J.; Arro, J.; Chen, Y.; Ming, R. Haplotype analysis of sucrose synthase gene family in three Saccharum species. BMC Genom. 2013, 14, 314. [Google Scholar] [CrossRef]
- Tong, X.L.; Wang, Z.Y.; Ma, B.Q.; Zhang, C.X.; Zhu, L.C.; Ma, F.W.; Li, M.J. Structure and expression analysis of the sucrose synthase gene family in apple. J. Integr. Agric. 2018, 17, 847–856. [Google Scholar] [CrossRef]
- Wang, Z.; Wei, P.; Wu, M.; Xu, Y.; Li, F.; Luo, Z.; Zhang, J.; Chen, A.; Xie, X.; Cao, P.; et al. Analysis of the sucrose synthase gene family in tobacco: Structure, phylogeny, and expression patterns. Planta 2015, 242, 153–166. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Chen, Z.; Wang, J.; Ye, M.; Ji, L.; Wang, J.; Liao, W.; Ma, H. Identification and characterization of the Populus sucrose synthase gene family. Gene 2014, 539, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.; Cao, Y.; Cheng, X.; Meng, D.; Chen, Y.; Shakoor, A.; Gao, J.; Cai, Y. The Sucrose Synthase Gene Family in Chinese Pear (Pyrus bretschneideri Rehd.): Structure, Expression, and Evolution. Molecules 2018, 23, 1144. [Google Scholar] [CrossRef]
- Huang, T.W.; Luo, X.L.; Fan, Z.P.; Yang, Y.N.; Wan, W. Genome-wide identification and analysis of the sucrose synthase gene family in cassava (Manihot esculenta Crantz). Gene 2021, 769, 145278. [Google Scholar] [CrossRef]
- Jacob, A.G.; Smith, C.W.J. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef]
- Dai, X.; Li, R.; Li, X.; Liang, Y.; Gao, Y.; Xu, Y.; Shi, L.; Zhou, Y.; Wang, H. Gene duplication and subsequent functional diversification of sucrose hydrolase in Papilio xuthus. Insect Mol. Biol. 2019, 28, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhou, H.; Ma, B.; Owiti, A.; Korban, S.S.; Han, Y. Divergent Evolutionary Pattern of Sugar Transporter Genes is Associated with the Difference in Sugar Accumulation between Grasses and Eudicots. Sci. Rep. 2016, 6, 29153. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, J.; Zhang, Z.; Wu, Y. Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize. Plant Biotechnol. J. 2020, 18, 1897–1907. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, M.; Wang, M.; Zhang, W.; Qiao, C.; Luo, Q.; Lu, X. Freshwater Cyanobacterium Synechococcus elongatus PCC 7942 Adapts to an Environment with Salt Stress via Ion-Induced Enzymatic Balance of Compatible Solutes. Appl. Environ. Microbiol. 2020, 86, e02904-19. [Google Scholar] [CrossRef]
- Ge, S.J.; Zhao, D.; Zhou, X.; Sun, Q.; Liu, L. In situ and intravesicular degradation of lipid droplets during cucumber leaf senescence. J. Hortic. 2016, 43, 2021–2028. [Google Scholar]
- Hussain, M.; Farooq, S.; Hasan, W.; Ul-Allah, T.M.; Farooq, M.; Nawaz, A. Drought stress in sunflower: Physiological effects and its management through breeding and agronomic alternatives. Agric. Water Manag. 2018, 201, 152–166. [Google Scholar] [CrossRef]
- Chalivendra, C.S.; Martin, M.S. Altered Patterns of Sucrose Synthase Phosphorylation and Localization Precede Callose Induction and Root Tip Death in Anoxic Maize Seedlings. Plant Physiol. 2001, 125, 585–594. [Google Scholar]
- Wang, J.; Ji, L.X.; Chen, Z.; Ye, M.X.; Li, Y.; An, X.M. Cloning and expression analysis of the sugarcane sucrase gene PtSS2 in Populus tomentosa. Acta Bot. Boreali-Occident. Sin. 2013, 33, 1501–1507. [Google Scholar]
- Bao, Y.Y.; Peng, X.R.; Pan, R.Y.; Ren, R.; Fang, T. Identification and expression analysis of the SUS gene family in passion fruit. Jiangsu Agric. Sci. 2024, 52, 45–51. [Google Scholar]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Large-Scale Plant Protein Subcellular Location Prediction. J. Cell Biochem. 2007, 100, 665–678. [Google Scholar] [CrossRef]
- Chou, K.C. Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 2005, 21, 10–19. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Ensemble classifier for protein folding pattern recognition. Bioinformatics 2006, 22, 1717–1722. [Google Scholar]
- Tian, X.; Li, Y.; Wang, S.; Zou, H.; Xiao, Q.; Ma, B.; Ma, F.; Li, M. Glucose uptake from the rhizosphere mediated by MdDOF3-MdHT1.2 regulates drought resistance in apple. Plant Biotechnol. J. 2024, 22, 1566–1581. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Tang, Y.; Pang, B.; Li, X.; Yang, Y.; Deng, J.; Feng, C.; Li, L.; Ren, G.; Wang, Y.; et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic. Res. 2020, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Horsch, R.B.; Fry, J.; Hoffmann, N.; Neidermeyer, J.; Rogers, S.G.; Fraley, R.T. Leaf disc transformation. In Plant Molecular Biology Manual A5; Gelvin, S.B., Schilperoort, R.A., Verma, D.P.S., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1988; pp. 1–9. [Google Scholar]
- Mathis, N.L.; Hinchee, M.A.W. Agrobacterium inoculation techniques for plant tissues. In Plant Molecular Biology Manual B6; Gelvin, S.B., Schilperoort, R.A., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994; pp. 1–9. [Google Scholar]
- Murugeasn, S.; Amaresan, N.; Narayanan, S. Estimation of Malondialdehyde (MDA) by Thiobarbituric Acid (TBA) Assay. In Plant Stress Physiology; Springer: Cham, Switzerland, 2021; pp. 435–444. [Google Scholar]
- Li, M.J.; Li, P.; Ma, F.W.; Dandekar, A.M.; Cheng, L.L. Sugar metabolism and accumulation in the fruit of transgenic apple trees with decreased sorbitol synthesis. Hortic. Res. 2018, 5, 60. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | CDS | Chromosome Location | Amino Acids | MW (kDa) | PI | GRAVY | Instability Index | Aliphatc Index |
|---|---|---|---|---|---|---|---|---|---|
| LaSUS1 | LC_Maker00039001_R1 | 2392 | Chr3 | 796 | 91.63 | 5.95 | −0.29 | 30.86 | 93.19 |
| LaSUS2 | LC_Maker00039001_R0 | 2436 | Chr11 | 811 | 92.44 | 6.02 | −0.221 | 34.62 | 94.35 |
| LaSUS6.1 | LC_Maker00036130_R0 | 2580 | Chr10 | 859 | 97.28 | 7.57 | −0.341 | 43.17 | 85.72 |
| LaSUS5 | LC_Maker00008755_R0 | 2499 | Chr8 | 832 | 94.45 | 6.24 | −0.292 | 38.16 | 85.32 |
| LaSUS4 | CS_CsGy1G028230.2_R4 | 1131 | Chr12 | 376 | 44.11 | 6.71 | −0.246 | 29.17 | 103.38 |
| LaSUS6.5 | LC_Maker00030741_R0 | 3081 | Chr8 | 1026 | 115.76 | 6.44 | −0.440 | 42.36 | 89.42 |
| LaSUS6.4 | CS_CsGy2G009040.2_R0 | 3192 | Chr12 | 1063 | 119.28 | 6.05 | −0.453 | 45.36 | 85.42 |
| LaSUS6.3 | CS_CsGy1G017180.1_R0 | 3174 | Chr3 | 1057 | 117.29 | 6.04 | −0.382 | 41.66 | 87.38 |
| LaSUS6.2 | LC_Maker00039364_R2 | 3114 | Chr2 | 1037 | 115.73 | 5.93 | −0.375 | 40.31 | 88.36 |
| Fru | Glu | Suc | Cellulose | |
|---|---|---|---|---|
| LaSUS1 | 0.865 | 0.855 | 0.984 * | −0.749 |
| LaSUS2 | 0.886 | 0.808 | 0.945 | −0.376 |
| LaSUS4 | 0.865 | 0.855 | 0.984 * | −0.690 |
| LaSUS6.1 | 0.469 | 0.507 | 0.747 | −0.935 |
| LaSUS6.2 | 0.941 | 0.849 | 0.811 | −0.806 |
| LaSUS6.3 | 0.768 | 0.766 | 0.942 | −0.749 |
| LaSUS6.4 | 0.928 | 0.856 | 0.958 * | −0.857 |
| LaSUS6.5 | 0.969 * | 0.896 | 0.852 | −0.645 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, X.; Wang, H.; Liu, J.; Huang, L.; Zheng, X.; Li, Y.; Sun, S.; Ma, C.; Zhao, H.; Zhao, P. Genome-Wide Identification of Luffa Sucrose Synthase Genes Reveals LaSUS1-Mediated Sugar Metabolism Boosting Drought Tolerance. Int. J. Mol. Sci. 2025, 26, 5675. https://doi.org/10.3390/ijms26125675
Tian X, Wang H, Liu J, Huang L, Zheng X, Li Y, Sun S, Ma C, Zhao H, Zhao P. Genome-Wide Identification of Luffa Sucrose Synthase Genes Reveals LaSUS1-Mediated Sugar Metabolism Boosting Drought Tolerance. International Journal of Molecular Sciences. 2025; 26(12):5675. https://doi.org/10.3390/ijms26125675
Chicago/Turabian StyleTian, Xiaocheng, Hanyi Wang, Jianting Liu, Liujing Huang, Xiaohui Zheng, Yufeng Li, Shaolong Sun, Chongjian Ma, Hongbo Zhao, and Puyan Zhao. 2025. "Genome-Wide Identification of Luffa Sucrose Synthase Genes Reveals LaSUS1-Mediated Sugar Metabolism Boosting Drought Tolerance" International Journal of Molecular Sciences 26, no. 12: 5675. https://doi.org/10.3390/ijms26125675
APA StyleTian, X., Wang, H., Liu, J., Huang, L., Zheng, X., Li, Y., Sun, S., Ma, C., Zhao, H., & Zhao, P. (2025). Genome-Wide Identification of Luffa Sucrose Synthase Genes Reveals LaSUS1-Mediated Sugar Metabolism Boosting Drought Tolerance. International Journal of Molecular Sciences, 26(12), 5675. https://doi.org/10.3390/ijms26125675






