The Role of Methylation Modification in Neural Injury and Repair
Abstract
1. Introduction
2. Overview of Methylation in DNA, Histones and RNA
2.1. DNA Methylation
2.2. Histone Methylation Modification
2.3. RNA Methylation Modification
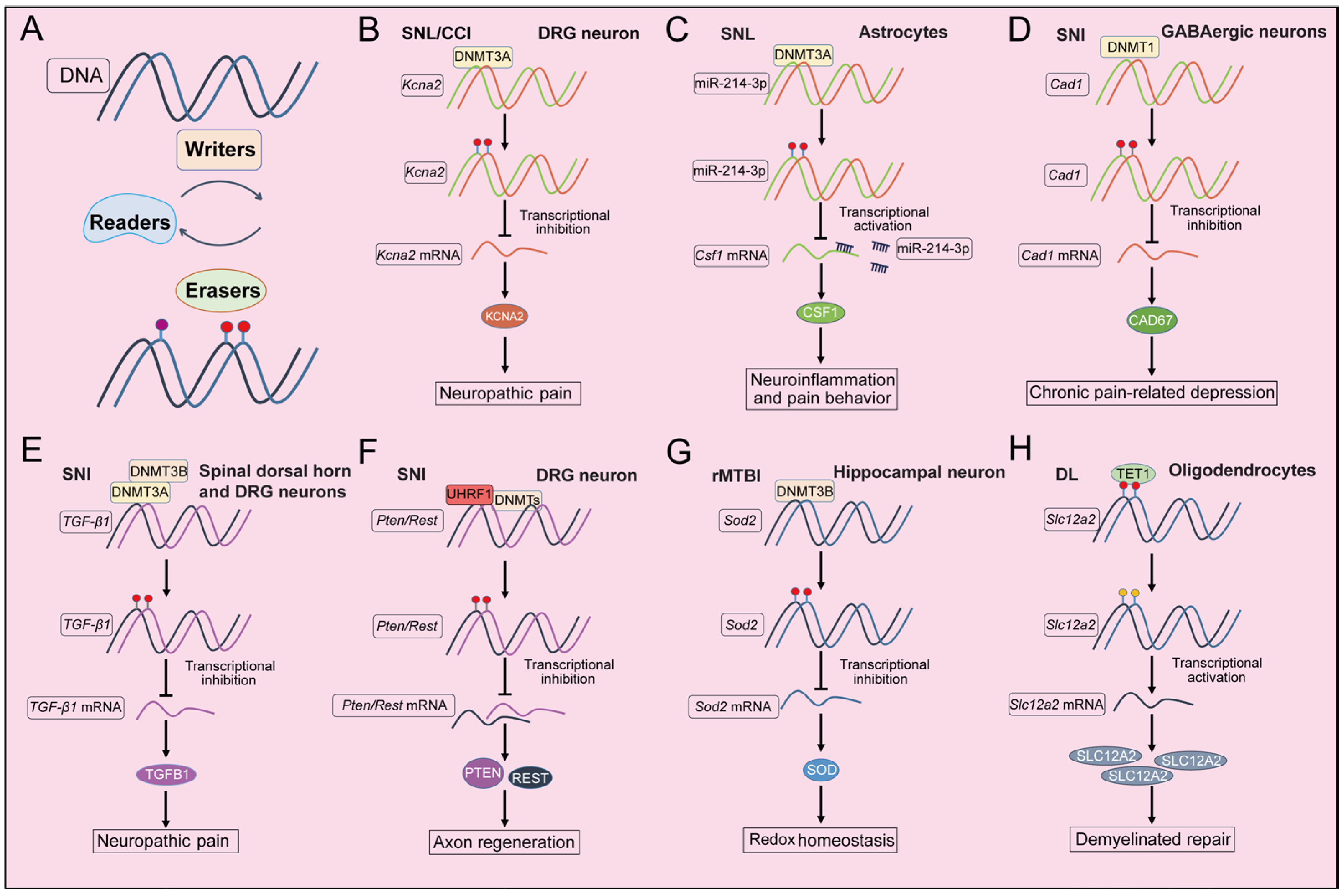
3. DNA Methylation Changes Following Nerve Injury
3.1. Brain Trauma
3.2. Spinal Cord Injury
3.3. Optic Nerve Injury
3.4. Sciatic Nerve Injury
| Animal | Injury Model | Timeline | Methods | Methylation Type | Sample | Ref. |
|---|---|---|---|---|---|---|
| Mice | Sciatic nerve crush | 0, 3 days | Dot blot Assay | 6mA | DRG | [23] |
| Rat | T10–T11 spinal cord transection/ Bilateral sciatic nerves ligation | 0, 14 days | WGBS/MeDIP-Seq | 5mC/5hmC | Spinal cord/Sciatic nerve | [118] |
| Mice | T8 spinal cord transection | 24 h | 5hmC enriched ChIP-Seq, | 5hmC | L4–L6 DRGs | [119] |
| Rat | T8–T10 spinal cord transection | Postnatal day 10 | Bisulfite sequencing PCR (BSP) | 5mC/5hmC | Spinal cord | [120] |
| Rat | C3 dorsal column spinal cord injury | N/A | 5hmC enriched ChIP-Seq, WGBS | 5mC/5hmC | Spinal cord | [124] |
| Mice | lysolecithin-induced demyelinating injury | N/A | RRHP | 5hmC | OPCs/OLs | [127] |
| Mice/rat | T9 dorsal hemisection | 0, 2 h, 0.5 h, 1 day, 7 days | Bisulfite sequencing | 5mC/5hmC | L4-L6 DRG | [135] |
| Rat | Sciatic nerve transection | 0, 1 day, 7 days | MeDIP-Seq | 5mC | L4–L6 DRGs | [136] |
| Mice | Sciatic nerve transection | 0, 7 days | WGBS | 5mC/5hmC | Sciatic nerve | [138] |
| Mice | Diabetes-induced myelin sheaths neuropathy | 6 months | RRBS | 5mC/5hmC | Sciatic nerve | [139] |
| Rat | Spinal nerve ligation | 0, 24 h | RRBS | 5mC/5hmC | L5 DRG | [141] |
| Rat | T9 moderate contusion injury | 0, 1 h, 1 week, 1 month, 3 months | DNA Dot blot Assay | 5mC/5hmC | Brain motor cortex | [125] |
| Rat | T9 moderate contusion injury | 0, 12 weeks | DNA Dot blot Assay | 5mC/5hmC | Brain motor cortex | [126] |
| Rat | C3 dorsal column spinal cord injury | N/A | MeDIP microarrays | 5mC/5hmC | Spinal cord | [155] |
| Mice | T10 dorsal hemisection | 0,1 day,3 days, 7 days | MeDIP-chip | 5mC | L4–L6 DRGs | [156] |
| Rat | Spinal nerve ligation | 0, 3 days, 3 weeks | DREAM/RRBS | 5mC/5hmC | Brain, spinal cord, and L5-6 DRG | [143] |
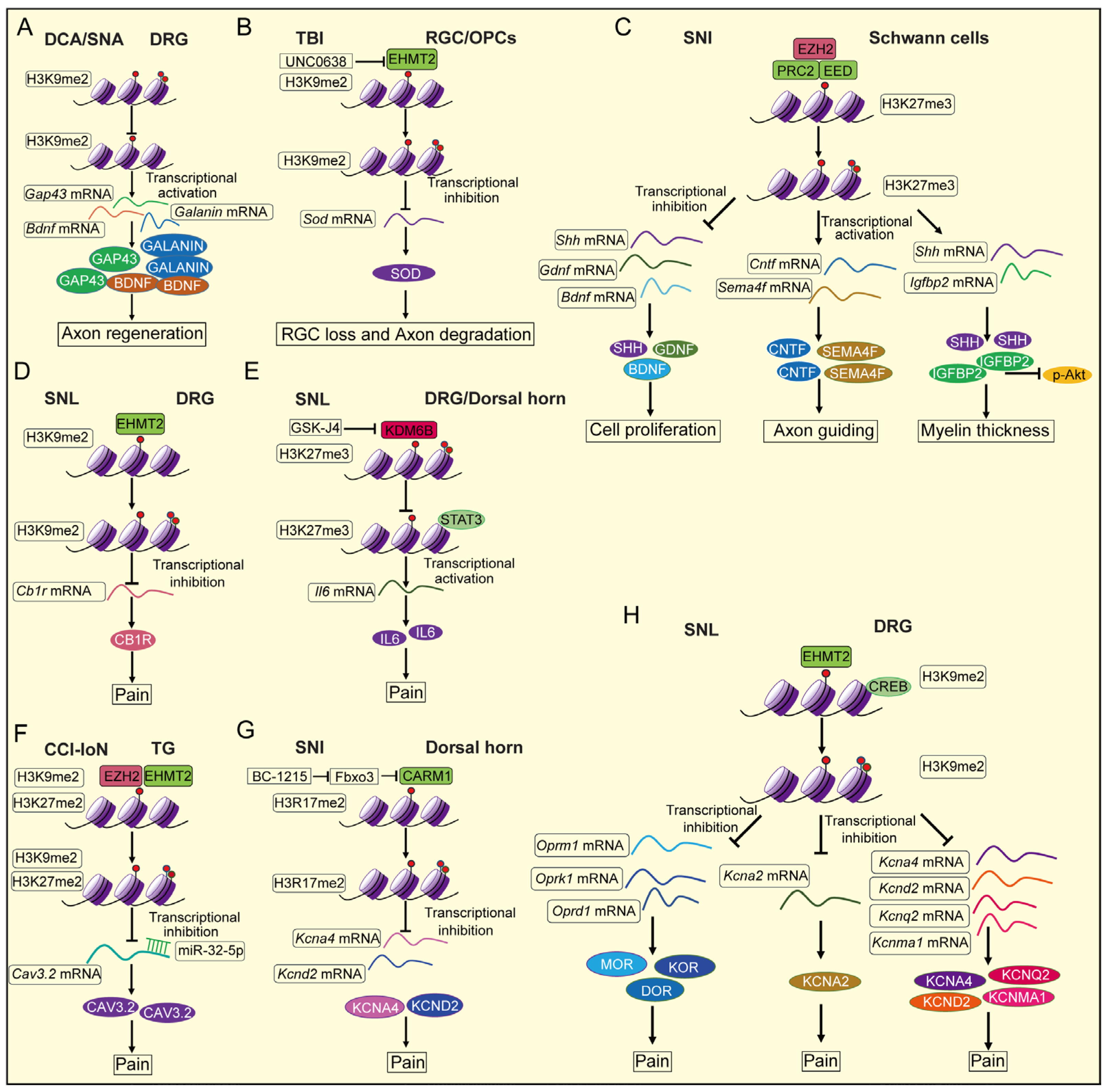
4. The Effect of Histone Methylation in Nerve Injury
4.1. Brain Trauma
4.2. Spinal Cord Injury
4.3. Optic Nerve Injury
4.4. Sciatic Nerve Injury
| Animal | Injury Model | Timeline | Methods | Methylation Type | Sample | Ref. |
|---|---|---|---|---|---|---|
| Rat | SNL | 0, 3 weeks | WB | H3K9me2, H3K27me3 | DRG | [24] |
| Mice | SNT | 0, 1 day | ChIP-Seq, ChIP-PCR | H3K27me3 | Sciatic nerve | [26] |
| Rat | CCI | 0, 6, 24, 72 h | IHC | H3K9me2, H3K4me2 | Hippocampus | [157] |
| Rat | CCI | 0, 3, 14 days | ChIP | H3K9me2/3, H3K27me3, H3K4me 3, H3K36me3 | Hippocampus | [158] |
| Mice | DCA/SNA | 0, 1, 3, 7 days | ChIP | H3K9me2, H3K27me3 | DRG | [159] |
| Mice | CCI | 0, 7 days | WB, IF | H3K9me2 | RGC | [161] |
| Rat | SNL | 0, 3, 10 days | WB | H3K27me3 | Spinal dorsal horn | [176] |
| Rat | CCI | 0, 1, 3, 5, 7, 10, 14 days | WB, IF | H3K27me3 | Spinal dorsal cord | [177] |
| Rat | SNL | 0, 1, 3, 7, 10, 14 days | WB | H3K27me3 | DRG, dorsal horn | [178] |
| Rat | SNL | 0, 7 days | WB, IF | H3R17me2 | Spinal dorsal horn | [183] |
5. The Role of RNA Methylation in Nerve Injury
5.1. Brain Trauma
5.2. Spinal Cord Injury
5.3. Optic Nerve Injury
5.4. Sciatic Nerve Injury
| Animal | Injury Model | TimeLine | Methods | Sample | Ref. |
|---|---|---|---|---|---|
| Mice | Sciatic nerve crush | 0, 1 day | m6A-SMART-seq, m6A-CLIP-SMART-seq | L4-L5 DRG | [27] |
| Rat | Controlled cortical impact | 0, 1 day | MeRIP-seq | Cerebral cortex | [185] |
| Rat | T10 spinal cord contusion | 0, 4 weeks | MeRIP-Seq | Spinal cord | [189] |
| Rat | Sciatic nerve injury | 0, 7 days | MeRIP-seq | Sciatic nerve | [197] |
| Rat | Middle cerebral artery occlusion and reperfusion model | 0, 1 day | MeRIP-seq | Brain cortex | [203] |
6. Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mar, F.M.; Bonni, A.; Sousa, M.M. Cell intrinsic control of axon regeneration. EMBO Rep. 2014, 15, 254–263. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Jin, Y. Intrinsic Control of Axon Regeneration. Neuron 2016, 90, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Scheib, J.; Höke, A. Advances in peripheral nerve regeneration. Nat. Rev. Neurol. 2013, 9, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Faroni, A.; Mobasseri, S.A.; Kingham, P.J.; Reid, A.J. Peripheral nerve regeneration: Experimental strategies and future perspectives. Adv. Drug Deliv. Rev. 2015, 82–83, 160–167. [Google Scholar] [CrossRef]
- Arthur-Farraj, P.J.; Latouche, M.; Wilton, D.K.; Quintes, S.; Chabrol, E.; Banerjee, A.; Woodhoo, A.; Jenkins, B.; Rahman, M.; Turmaine, M.; et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron 2012, 75, 633–647. [Google Scholar] [CrossRef]
- Seijffers, R.; Mills, C.D.; Woolf, C.J. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 7911–7920. [Google Scholar] [CrossRef]
- Shin, J.E.; Cho, Y.; Beirowski, B.; Milbrandt, J.; Cavalli, V.; DiAntonio, A. Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron 2012, 74, 1015–1022. [Google Scholar] [CrossRef]
- Park, K.K.; Liu, K.; Hu, Y.; Smith, P.D.; Wang, C.; Cai, B.; Xu, B.; Connolly, L.; Kramvis, I.; Sahin, M.; et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 2008, 322, 963–966. [Google Scholar] [CrossRef]
- Zhang, Y.; Williams, P.R.; Jacobi, A.; Wang, C.; Goel, A.; Hirano, A.A.; Brecha, N.C.; Kerschensteiner, D.; He, Z. Elevating Growth Factor Responsiveness and Axon Regeneration by Modulating Presynaptic Inputs. Neuron 2019, 103, 39–51.e35. [Google Scholar] [CrossRef]
- Wang, X.W.; Yang, S.G.; Zhang, C.; Hu, M.W.; Qian, J.; Ma, J.J.; Zhang, Y.; Yang, B.B.; Weng, Y.L.; Ming, G.L.; et al. Knocking Out Non-muscle Myosin II in Retinal Ganglion Cells Promotes Long-Distance Optic Nerve Regeneration. Cell Rep. 2020, 31, 107537. [Google Scholar] [CrossRef]
- Sas, A.R.; Carbajal, K.S.; Jerome, A.D.; Menon, R. A new neutrophil subset promotes CNS neuron survival and axon regeneration. Nat. Immunol. 2020, 21, 1496–1505. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Sloutsky, R.; Naegle, K.M.; Cavalli, V. Injury-induced HDAC5 nuclear export is essential for axon regeneration. Cell 2013, 155, 894–908. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.L.; An, R.; Cassin, J.; Joseph, J.; Mi, R.; Wang, C.; Zhong, C.; Jin, S.G.; Pfeifer, G.P.; Bellacosa, A.; et al. An Intrinsic Epigenetic Barrier for Functional Axon Regeneration. Neuron 2017, 94, 337–346.e336. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, Y.; Liu, M.; Cao, Q.; Wang, Q.; Zhou, S.; Wang, Y.; Mao, S. The long noncoding RNA Arrl1 inhibits neurite outgrowth by functioning as a competing endogenous RNA during neuronal regeneration in rats. J. Biol. Chem. 2020, 295, 8374–8386. [Google Scholar] [CrossRef]
- Yao, C.; Wang, Y. lncRNA TNXA-PS1 Modulates Schwann Cells by Functioning As a Competing Endogenous RNA Following Nerve Injury. J. Neurosci. Off. J. Soc. Neurosci. 2018, 38, 6574–6585. [Google Scholar] [CrossRef]
- Michalak, E.M.; Burr, M.L.; Bannister, A.J. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 573–589. [Google Scholar] [CrossRef]
- Guo, J.U.; Su, Y.; Shin, J.H.; Shin, J.; Li, H.; Xie, B.; Zhong, C.; Hu, S.; Le, T.; Fan, G. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014, 17, 215–222. [Google Scholar] [CrossRef]
- Bergman, Y.; Cedar, H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013, 20, 274–281. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Frye, M.; Harada, B.T. RNA modifications modulate gene expression during development. Science 2018, 361, 1346–1349. [Google Scholar] [CrossRef] [PubMed]
- Gontier, G.; Iyer, M.; Shea, J.M.; Bieri, G.; Wheatley, E.G.; Ramalho-Santos, M.; Villeda, S.A. Tet2 Rescues Age-Related Regenerative Decline and Enhances Cognitive Function in the Adult Mouse Brain. Cell Rep. 2018, 22, 1974–1981. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Qian, C.; Feng, H.; Lin, T.; Zhu, Q.; Huang, Y.; Zhou, F.Q. N6-methyladenine DNA Demethylase ALKBH1 Regulates Mammalian Axon Regeneration. Neurosci. Bull. 2021, 37, 809–814. [Google Scholar] [CrossRef] [PubMed]
- Laumet, G.; Garriga, J.; Chen, S.R.; Zhang, Y.; Li, D.P.; Smith, T.M.; Dong, Y.; Jelinek, J.; Cesaroni, M.; Issa, J.P.; et al. G9a is essential for epigenetic silencing of K(+) channel genes in acute-to-chronic pain transition. Nat. Neurosci. 2015, 18, 1746–1755. [Google Scholar] [CrossRef]
- Duong, P.; Ma, K.H.; Ramesh, R.; Moran, J.J.; Won, S.; Svaren, J. H3K27 demethylases are dispensable for activation of Polycomb-regulated injury response genes in peripheral nerve. J. Biol. Chem. 2021, 297, 100852. [Google Scholar] [CrossRef]
- Ma, K.H.; Hung, H.A. Epigenomic Regulation of Schwann Cell Reprogramming in Peripheral Nerve Injury. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 9135–9147. [Google Scholar] [CrossRef]
- Weng, Y.L.; Wang, X.; An, R.; Cassin, J.; Vissers, C.; Liu, Y.; Liu, Y.; Xu, T.; Wang, X.; Wong, S.Z.H.; et al. Epitranscriptomic m(6)A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 2018, 97, 313–325.e316. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, Y. FOXD3-mediated transactivation of ALKBH5 promotes neuropathic pain via m(6)A-dependent stabilization of 5-HT3A mRNA in sensory neurons. Proc. Natl. Acad. Sci. USA 2024, 121, e2312861121. [Google Scholar] [CrossRef]
- Wang, D.; Zheng, T.; Zhou, S.; Liu, M.; Liu, Y. Promoting axon regeneration by inhibiting RNA N6-methyladenosine demethylase ALKBH5. ELife 2023, 12, e85309. [Google Scholar] [CrossRef]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef]
- Kaluscha, S.; Domcke, S.; Wirbelauer, C.; Stadler, M.B. Evidence that direct inhibition of transcription factor binding is the prevailing mode of gene and repeat repression by DNA methylation. Nat. Genet. 2022, 54, 1895–1906. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Hajkova, P. Dynamic DNA methylation: In the right place at the right time. Science 2018, 361, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Rottach, A.; Leonhardt, H.; Spada, F. DNA methylation-mediated epigenetic control. J. Cell. Biochem. 2009, 108, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Spruijt, C.G.; Gnerlich, F.; Smits, A.H.; Pfaffeneder, T.; Jansen, P.W.; Bauer, C.; Münzel, M.; Wagner, M.; Müller, M.; Khan, F.; et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 2013, 152, 1146–1159. [Google Scholar] [CrossRef]
- Pecori, R.; Di Giorgio, S. Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination. Nat. Rev. Genet. 2022, 23, 505–518. [Google Scholar] [CrossRef]
- Nabel, C.S.; Jia, H.; Ye, Y.; Shen, L.; Goldschmidt, H.L.; Stivers, J.T.; Zhang, Y.; Kohli, R.M. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 2012, 8, 751–758. [Google Scholar] [CrossRef]
- Jones, P.A.; Takai, D. The role of DNA methylation in mammalian epigenetics. Science 2001, 293, 1068–1070. [Google Scholar] [CrossRef]
- Hu, S.; Wan, J.; Su, Y.; Song, Q.; Zeng, Y.; Nguyen, H.N.; Shin, J.; Cox, E.; Rho, H.S.; Woodard, C.; et al. DNA methylation presents distinct binding sites for human transcription factors. ELife 2013, 2, e00726. [Google Scholar] [CrossRef]
- Buck-Koehntop, B.A.; Defossez, P.A. On how mammalian transcription factors recognize methylated DNA. Epigenetics 2013, 8, 131–137. [Google Scholar] [CrossRef]
- Song, J.; Pfeifer, G.P. Are there specific readers of oxidized 5-methylcytosine bases? BioEssays News Rev. Mol. Cell. Dev. biology 2016, 38, 1038–1047. [Google Scholar] [CrossRef]
- Kubo, N.; Ishii, H.; Xiong, X.; Bianco, S. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 2021, 28, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Maurano, M.T.; Wang, H.; John, S.; Shafer, A.; Canfield, T.; Lee, K.; Stamatoyannopoulos, J.A. Role of DNA Methylation in Modulating Transcription Factor Occupancy. Cell Rep. 2015, 12, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, G.P.; Szabó, P.E.; Song, J. Protein Interactions at Oxidized 5-Methylcytosine Bases. J. Mol. Biol. 2020, 432, 1718–1730. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Zhang, Z.; Chen, J.; Huang, H.; Xu, Y.; Ding, X.; Zheng, Y.; Nishinakamura, R.; Xu, G.L.; Wang, H.; et al. Cooperative Action between SALL4A and TET Proteins in Stepwise Oxidation of 5-Methylcytosine. Mol. Cell 2016, 64, 913–925. [Google Scholar] [CrossRef]
- Xiao, C.L.; Zhu, S.; He, M.; Chen, D.; Zhang, Q.; Chen, Y.; Yu, G.; Liu, J.; Xie, S.Q.; Luo, F.; et al. N(6)-Methyladenine DNA Modification in the Human Genome. Mol. Cell 2018, 71, 306–318.e307. [Google Scholar] [CrossRef]
- Xie, Q.; Wu, T.P.; Gimple, R.C.; Li, Z.; Prager, B.C.; Wu, Q.; Yu, Y.; Wang, P.; Wang, Y.; Gorkin, D.U.; et al. N(6)-methyladenine DNA Modification in Glioblastoma. Cell 2018, 175, 1228–1243.e1220. [Google Scholar] [CrossRef]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizábal-Corrales, D.; Hsu, C.H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. Elegans Cell 2015, 161, 868–878. [Google Scholar] [CrossRef]
- Fernandes, S.B.; Grova, N.; Roth, S.; Duca, R.C.; Godderis, L.; Guebels, P.; Mériaux, S.B.; Lumley, A.I.; Bouillaud-Kremarik, P.; Ernens, I.; et al. N(6)-Methyladenine in Eukaryotic DNA: Tissue Distribution, Early Embryo Development, and Neuronal Toxicity. Front. Genet. 2021, 12, 657171. [Google Scholar] [CrossRef]
- Kweon, S.M.; Chen, Y.; Moon, E.; Kvederaviciutė, K.; Klimasauskas, S.; Feldman, D.E. An Adversarial DNA N(6)-Methyladenine-Sensor Network Preserves Polycomb Silencing. Mol. Cell 2019, 74, 1138–1147.e1136. [Google Scholar] [CrossRef]
- Yu, D.; Horton, J.R.; Yang, J.; Hajian, T.; Vedadi, M.; Sagum, C.A.; Bedford, M.T.; Blumenthal, R.M.; Zhang, X.; Cheng, X. Human MettL3-MettL14 RNA adenine methyltransferase complex is active on double-stranded DNA containing lesions. Nucleic Acids Res. 2021, 49, 11629–11642. [Google Scholar] [CrossRef]
- Musheev, M.U.; Baumgärtner, A. The origin of genomic N(6)-methyl-deoxyadenosine in mammalian cells. Nat. Chem. Biol. 2020, 16, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Liu, Y. DNA N(6)-methyladenine demethylase ALKBH1 enhances osteogenic differentiation of human MSCs. Bone Res. 2016, 4, 16033. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, C.B.; Horton, J.R.; Zhou, J.; Bedford, M.T.; Blumenthal, R.M.; Zhang, X.; Cheng, X. Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA. Nucleic Acids Res. 2020, 48, 10329–10341. [Google Scholar] [CrossRef] [PubMed]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef]
- Guccione, E.; Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 642–657. [Google Scholar] [CrossRef]
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; Van Calcar, S.; Qu, C.; Ching, K.A.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007, 39, 311–318. [Google Scholar] [CrossRef]
- Kim, T.; Buratowski, S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5’ transcribed regions. Cell 2009, 137, 259–272. [Google Scholar] [CrossRef]
- Lauberth, S.M.; Nakayama, T.; Wu, X.; Ferris, A.L.; Tang, Z.; Hughes, S.H.; Roeder, R.G. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 2013, 152, 1021–1036. [Google Scholar] [CrossRef]
- Weinberg, D.N.; Papillon-Cavanagh, S.; Chen, H.; Yue, Y.; Chen, X.; Rajagopalan, K.N.; Horth, C.; McGuire, J.T.; Xu, X.; Nikbakht, H.; et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 2019, 573, 281–286. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Zhang, Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011, 25, 1345–1358. [Google Scholar] [CrossRef]
- Rice, J.C.; Briggs, S.D.; Ueberheide, B.; Barber, C.M.; Shabanowitz, J.; Hunt, D.F.; Shinkai, Y.; Allis, C.D. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 2003, 12, 1591–1598. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Zegerman, P.; Partridge, J.F.; Miska, E.A.; Thomas, J.O.; Allshire, R.C.; Kouzarides, T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001, 410, 120–124. [Google Scholar] [CrossRef]
- Vavouri, T.; Lehner, B. Human genes with CpG island promoters have a distinct transcription-associated chromatin organization. Genome Biol. 2012, 13, R110. [Google Scholar] [CrossRef]
- Laugesen, A.; Højfeldt, J.W.; Helin, K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol. Cell 2019, 74, 8–18. [Google Scholar] [CrossRef]
- Sato, Y.; Kujirai, T.; Arai, R.; Asakawa, H.; Ohtsuki, C.; Horikoshi, N.; Yamagata, K.; Ueda, J.; Nagase, T.; Haraguchi, T.; et al. A Genetically Encoded Probe for Live-Cell Imaging of H4K20 Monomethylation. J. Mol. Biol. 2016, 428, 3885–3902. [Google Scholar] [CrossRef]
- Jørgensen, S.; Schotta, G.; Sørensen, C.S. Histone H4 lysine 20 methylation: Key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013, 41, 2797–2806. [Google Scholar] [CrossRef]
- Shinkai, Y.; Tachibana, M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011, 25, 781–788. [Google Scholar] [CrossRef]
- Tachibana, M.; Matsumura, Y.; Fukuda, M.; Kimura, H.; Shinkai, Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008, 27, 2681–2690. [Google Scholar] [CrossRef]
- Jiao, L.; Liu, X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 2015, 350, aac4383. [Google Scholar] [CrossRef]
- Weirich, S.; Khella, M.S. Structure, Activity and Function of the Suv39h1 and Suv39h2 Protein Lysine Methyltransferases. Life 2021, 11, 703. [Google Scholar] [CrossRef]
- Montavon, T.; Shukeir, N.; Erikson, G.; Engist, B.; Onishi-Seebacher, M.; Ryan, D.; Musa, Y.; Mittler, G.; Meyer, A.G.; Genoud, C.; et al. Complete loss of H3K9 methylation dissolves mouse heterochromatin organization. Nat. Commun. 2021, 12, 4359. [Google Scholar] [CrossRef]
- Park, K.; Kim, J.A.; Kim, J. Transcriptional regulation by the KMT2 histone H3K4 methyltransferases. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194545. [Google Scholar] [CrossRef]
- Xiang, Y.; Zhu, Z.; Han, G.; Lin, H.; Xu, L.; Chen, C.D. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007, 17, 850–857. [Google Scholar] [CrossRef]
- Swigut, T.; Wysocka, J. H3K27 demethylases, at long last. Cell 2007, 131, 29–32. [Google Scholar] [CrossRef]
- Wang, H.; Fan, Z. H3K4me3 regulates RNA polymerase II promoter-proximal pause-release. Nature 2023, 615, 339–348. [Google Scholar] [CrossRef]
- Rose, N.R.; Klose, R.J. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim. Biophys. Acta 2014, 1839, 1362–1372. [Google Scholar] [CrossRef]
- Hashimoto, H.; Vertino, P.M.; Cheng, X. Molecular coupling of DNA methylation and histone methylation. Epigenomics 2010, 2, 657–669. [Google Scholar] [CrossRef]
- Ooi, S.K.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.P.; Allis, C.D.; et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef]
- Harris, R.A.; Wang, T.; Coarfa, C.; Nagarajan, R.P.; Hong, C.; Downey, S.L.; Johnson, B.E.; Fouse, S.D.; Delaney, A.; Zhao, Y.; et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010, 28, 1097–1105. [Google Scholar] [CrossRef]
- Lewis, C.J.; Pan, T.; Kalsotra, A. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2017, 18, 202–210. [Google Scholar] [CrossRef]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Wiener, D.; Schwartz, S. The epitranscriptome beyond m(6)A. Nat. Rev. Genet. 2021, 22, 119–131. [Google Scholar] [CrossRef]
- Xu, W.; Li, J.; He, C.; Wen, J.; Ma, H.; Rong, B. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature 2021, 591, 317–321. [Google Scholar] [CrossRef]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835.e814. [Google Scholar] [CrossRef]
- Mendel, M.; Chen, K.M.; Homolka, D.; Gos, P.; Pandey, R.R.; McCarthy, A.A.; Pillai, R.S. Methylation of Structured RNA by the m(6)A Writer METTL16 Is Essential for Mouse Embryonic Development. Mol. Cell 2018, 71, 986–1000.e1011. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015, 29, 1343–1355. [Google Scholar] [CrossRef]
- Ontiveros, R.J.; Stoute, J.; Liu, K.F. The chemical diversity of RNA modifications. Biochem. J. 2019, 476, 1227–1245. [Google Scholar] [CrossRef]
- He, P.C.; He, C. m(6) A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977. [Google Scholar] [CrossRef]
- He, Y.; Yu, X.; Li, J.; Zhang, Q.; Zheng, Q.; Guo, W. Role of m(5)C-related regulatory genes in the diagnosis and prognosis of hepatocellular carcinoma. Am. J. Transl. Res. 2020, 12, 912–922. [Google Scholar]
- Mei, L.; Shen, C.; Miao, R.; Wang, J.Z.; Cao, M.D.; Zhang, Y.S.; Shi, L.H.; Zhao, G.H.; Wang, M.H.; Wu, L.S. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57(Kip2) by an m(5)C-dependent manner. Cell Death Dis. 2020, 11, 270. [Google Scholar] [CrossRef]
- Huang, T.; Chen, W.; Liu, J.; Gu, N.; Zhang, R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol. 2019, 26, 380–388. [Google Scholar] [CrossRef]
- Dai, X.; Gonzalez, G.; Li, L.; Li, J.; You, C.; Miao, W.; Hu, J.; Fu, L.; Zhao, Y.; Li, R.; et al. YTHDF2 Binds to 5-Methylcytosine in RNA and Modulates the Maturation of Ribosomal RNA. Anal. Chem. 2020, 92, 1346–1354. [Google Scholar] [CrossRef]
- Li, X.; Xiong, X.; Zhang, M.; Wang, K.; Chen, Y.; Zhou, J.; Mao, Y.; Lv, J.; Yi, D.; Chen, X.W.; et al. Base-Resolution Mapping Reveals Distinct m(1)A Methylome in Nuclear- and Mitochondrial-Encoded Transcripts. Mol. Cell 2017, 68, 993–1005.e1009. [Google Scholar] [CrossRef]
- Safra, M.; Sas-Chen, A.; Nir, R.; Winkler, R.; Nachshon, A.; Bar-Yaacov, D.; Erlacher, M.; Rossmanith, W.; Stern-Ginossar, N.; Schwartz, S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 2017, 551, 251–255. [Google Scholar] [CrossRef]
- Zhang, H.; Shi, X.; Huang, T.; Zhao, X.; Chen, W.; Gu, N.; Zhang, R. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020, 48, 6251–6264. [Google Scholar] [CrossRef]
- Li, G.; Ma, L.; He, S.; Luo, R.; Wang, B.; Zhang, W.; Song, Y.; Liao, Z.; Ke, W.; Xiang, Q.; et al. WTAP-mediated m(6)A modification of lncRNA NORAD promotes intervertebral disc degeneration. Nat. Commun. 2022, 13, 1469. [Google Scholar] [CrossRef]
- Peng, W.; Li, J.; Chen, R.; Gu, Q.; Yang, P.; Qian, W.; Ji, D.; Wang, Q.; Zhang, Z.; Tang, J.; et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 393. [Google Scholar] [CrossRef]
- Deng, X.; Qing, Y. The roles and implications of RNA m(6)A modification in cancer. Nat. Rev. Clin. Oncol. 2023, 20, 507–526. [Google Scholar] [CrossRef]
- Flamand, M.N.; Tegowski, M.; Meyer, K.D. The Proteins of mRNA Modification: Writers, Readers, and Erasers. Annu. Rev. Biochem. 2023, 92, 145–173. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. ELife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Weng, H.; Huang, F.; Yu, Z.; Chen, Z.; Prince, E.; Kang, Y.; Zhou, K.; Li, W.; Hu, J.; Fu, C.; et al. The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell 2022, 40, 1566–1582.e1510. [Google Scholar] [CrossRef]
- Zhao, G.; Liu, R.; Ge, L.; Qi, D.; Wu, Q.; Lin, Z.; Song, H.; Zhong, L.; Cui, H. NONO regulates m(5)C modification and alternative splicing of PTEN mRNAs to drive gastric cancer progression. J. Exp. Clin. Cancer Res. 2025, 44, 81. [Google Scholar] [CrossRef]
- Haghighi, F.; Ge, Y.; Chen, S.; Xin, Y.; Umali, M.U.; De Gasperi, R.; Gama Sosa, M.A.; Ahlers, S.T.; Elder, G.A. Neuronal DNA Methylation Profiling of Blast-Related Traumatic Brain Injury. J. Neurotrauma 2015, 32, 1200–1209. [Google Scholar] [CrossRef]
- Treble-Barna, A.; Heinsberg, L.W.; Puccio, A.M.; Shaffer, J.R.; Okonkwo, D.O.; Beers, S.R.; Weeks, D.E.; Conley, Y.P. Acute Brain-Derived Neurotrophic Factor DNA Methylation Trajectories in Cerebrospinal Fluid and Associations with Outcomes Following Severe Traumatic Brain Injury in Adults. Neurorehabilit. Neural Repair 2021, 35, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Zhang, Z.; Fauser, U.; Schluesener, H.J. Global hypomethylation defines a sub-population of reactive microglia/macrophages in experimental traumatic brain injury. Neurosci. Lett. 2007, 429, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Cuautle, D.G.; Donna, S.; Cieri, M.B.; Villarreal, A.; Ramos, A.J. Pathological remodeling of reactive astrocytes: Involvement of DNA methylation and downregulation of homeostatic genes. J. Neurochem. 2024, 168, 2935–2955. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, J.; Karimi, M.; von Gertten, C.; Holmin, S.; Ekström, T.J.; Sandberg-Nordqvist, A.C. Traumatic brain injury induces relocalization of DNA-methyltransferase 1. Neurosci. Lett. 2009, 457, 8–11. [Google Scholar] [CrossRef]
- Balasubramanian, N.; Jadhav, G.; Sakharkar, A.J. Repeated mild traumatic brain injuries perturb the mitochondrial biogenesis via DNA methylation in the hippocampus of rat. Mitochondrion 2021, 61, 11–24. [Google Scholar] [CrossRef]
- Balasubramanian, N.; Sagarkar, S.; Choudhary, A.G.; Kokare, D.M.; Sakharkar, A.J. Epigenetic Blockade of Hippocampal SOD2 Via DNMT3b-Mediated DNA Methylation: Implications in Mild Traumatic Brain Injury-Induced Persistent Oxidative Damage. Mol. Neurobiol. 2021, 58, 1162–1184. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, J.; Zhang, K.; Lu, G.; Liu, Y.; Ren, K.; Wang, W. Ten-eleven translocation 1 mediated-DNA hydroxymethylation is required for myelination and remyelination in the mouse brain. Nat. Commun. 2021, 12, 5091. [Google Scholar] [CrossRef]
- Shi, G.; Zhou, X.; Wang, X.; Zhang, X.; Zhang, P.; Feng, S. Signatures of altered DNA methylation gene expression after central and peripheral nerve injury. J. Cell. Physiol. 2020, 235, 5171–5181. [Google Scholar] [CrossRef]
- Loh, Y.E.; Koemeter-Cox, A.; Finelli, M.J.; Shen, L.; Friedel, R.H.; Zou, H. Comprehensive mapping of 5-hydroxymethylcytosine epigenetic dynamics in axon regeneration. Epigenetics 2017, 12, 77–92. [Google Scholar] [CrossRef]
- Doherty, T.S.; Bozeman, A.L.; Roth, T.L.; Brumley, M.R. DNA methylation and behavioral changes induced by neonatal spinal transection. Infant Behav. Dev. 2019, 57, 101381. [Google Scholar] [CrossRef]
- Wu, Z.; Li, C.; Zhu, R.; Cao, Y.; Chen, T.C.; Cheng, L. Reduced non-CpG methylation is a potential epigenetic target after spinal cord injury. Neural Regen. Res. 2023, 18, 2489–2496. [Google Scholar] [CrossRef] [PubMed]
- Iskandar, B.J.; Rizk, E.; Meier, B.; Hariharan, N.; Bottiglieri, T.; Finnell, R.H.; Jarrard, D.F.; Banerjee, R.V.; Skene, J.H.; Nelson, A.; et al. Folate regulation of axonal regeneration in the rodent central nervous system through DNA methylation. J. Clin. Investig. 2010, 120, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Madrid, A.; Alisch, R.S.; Rizk, E.; Papale, L.A.; Hogan, K.J.; Iskandar, B.J. Transgenerational epigenetic inheritance of axonal regeneration after spinal cord injury. Environ. Epigenetics 2023, 9, dvad002. [Google Scholar] [CrossRef] [PubMed]
- Madrid, A.; Borth, L.E. DNA methylation and hydroxymethylation have distinct genome-wide profiles related to axonal regeneration. Epigenetics 2021, 16, 64–78. [Google Scholar] [CrossRef]
- Hong, J.Y.; Davaa, G.; Yoo, H.; Hong, K.; Hyun, J.K. Ascorbic Acid Promotes Functional Restoration after Spinal Cord Injury Partly by Epigenetic Modulation. Cells 2020, 9, 1310. [Google Scholar] [CrossRef]
- Davaa, G.; Hong, J.Y.; Kim, T.U.; Lee, S.J. Exercise Ameliorates Spinal Cord Injury by Changing DNA Methylation. Cells 2021, 10, 143. [Google Scholar] [CrossRef]
- Moyon, S.; Frawley, R.; Marechal, D.; Huang, D.; Marshall-Phelps, K.L.H.; Kegel, L.; Bøstrand, S.M.K. TET1-mediated DNA hydroxymethylation regulates adult remyelination in mice. Nat. Commun. 2021, 12, 3359. [Google Scholar] [CrossRef]
- Lu, Y.; Brommer, B.; Tian, X. Reprogramming to recover youthful epigenetic information and restore vision. Nature 2020, 588, 124–129. [Google Scholar] [CrossRef]
- Rizk, E.; Madrid, A.; Koueik, J.; Sun, D. Purified regenerating retinal neurons reveal regulatory role of DNA methylation-mediated Na+/K+-ATPase in murine axon regeneration. Commun. Biol. 2023, 6, 120. [Google Scholar] [CrossRef]
- Hu, X.; Xu, W.; Ren, Y.; Wang, Z.; He, X.; Huang, R.; Ma, B.; Zhao, J.; Zhu, R.; Cheng, L. Spinal cord injury: Molecular mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2023, 8, 245. [Google Scholar] [CrossRef]
- Zheng, B.; Tuszynski, M.H. Regulation of axonal regeneration after mammalian spinal cord injury. Nat. Rev. Mol. Cell Biol. 2023, 24, 396–413. [Google Scholar] [CrossRef] [PubMed]
- Renthal, W.; Tochitsky, I.; Yang, L.; Cheng, Y.C.; Li, E.; Kawaguchi, R.; Geschwind, D.H.; Woolf, C.J. Transcriptional Reprogramming of Distinct Peripheral Sensory Neuron Subtypes after Axonal Injury. Neuron 2020, 108, 128–144.e129. [Google Scholar] [CrossRef] [PubMed]
- Jessen, K.R.; Mirsky, R. The repair Schwann cell and its function in regenerating nerves. J. Physiol. 2016, 594, 3521–3531. [Google Scholar] [CrossRef] [PubMed]
- Van Emmenis, L.; Mòdol-Caballero, G.; Harford-Wright, E.; Power, A.; Cattin, A.L.; White, I.J.; Casal, G.; Boal-Carvalho, I.; Bennett, C.L.; Lloyd, A.C. Identification of CCL3 as a Schwann cell chemotactic factor essential for nerve regeneration. Cell Rep. 2025, 44, 115322. [Google Scholar] [CrossRef]
- Shin, H.Y.; Kwon, M.J.; Lee, E.M.; Kim, K. Role of Myc Proto-Oncogene as a Transcriptional Hub to Regulate the Expression of Regeneration-Associated Genes following Preconditioning Peripheral Nerve Injury. J. Neurosci. Off. J. Soc. Neurosci. 2021, 41, 446–460. [Google Scholar] [CrossRef]
- Shin, H.Y.; Kim, K.; Kwon, M.J.; Oh, Y.J.; Kim, E.H.; Kim, H.S.; Hong, C.P.; Lee, J.H.; Lee, K.; Kim, B.G. Alteration in global DNA methylation status following preconditioning injury influences axon growth competence of the sensory neurons. Exp. Neurol. 2020, 326, 113177. [Google Scholar] [CrossRef]
- Oh, Y.M.; Mahar, M.; Ewan, E.E.; Leahy, K.M.; Zhao, G. Epigenetic regulator UHRF1 inactivates REST and growth suppressor gene expression via DNA methylation to promote axon regeneration. Proc. Natl. Acad. Sci. USA 2018, 115, E12417–E12426. [Google Scholar] [CrossRef]
- Arthur-Farraj, P.J.; Morgan, C.C.; Adamowicz, M.; Gomez-Sanchez, J.A.; Fazal, S.V.; Beucher, A.; Razzaghi, B.; Mirsky, R.; Jessen, K.R.; Aitman, T.J. Changes in the Coding and Non-coding Transcriptome and DNA Methylome that Define the Schwann Cell Repair Phenotype after Nerve Injury. Cell Rep. 2017, 20, 2719–2734. [Google Scholar] [CrossRef]
- Varela-Rey, M.; Iruarrizaga-Lejarreta, M.; Lozano, J.J.; Aransay, A.M.; Fernandez, A.F.; Lavin, J.L.; Mósen-Ansorena, D.; Berdasco, M.; Turmaine, M.; Luka, Z.; et al. S-adenosylmethionine levels regulate the schwann cell DNA methylome. Neuron 2014, 81, 1024–1039. [Google Scholar] [CrossRef]
- Wu, Q.; Xie, J.; Zhu, X.; He, J. Runt-related transcription factor 3, mediated by DNA-methyltransferase 1, regulated Schwann cell proliferation and myelination during peripheral nerve regeneration via JAK/STAT signaling pathway. Neurosci. Res. 2023, 192, 1–10. [Google Scholar] [CrossRef]
- Gölzenleuchter, M.; Kanwar, R.; Zaibak, M.; Al Saiegh, F.; Hartung, T.; Klukas, J.; Smalley, R.L.; Cunningham, J.M.; Figueroa, M.E.; Schroth, G.P.; et al. Plasticity of DNA methylation in a nerve injury model of pain. Epigenetics 2015, 10, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.; Monteiro, C.; Cardoso-Cruz, H. Altered Brain Expression of DNA Methylation and Hydroxymethylation Epigenetic Enzymes in a Rat Model of Neuropathic Pain. Int. J. Mol. Sci. 2023, 24, 7305. [Google Scholar] [CrossRef] [PubMed]
- Garriga, J.; Laumet, G. Nerve Injury-Induced Chronic Pain Is Associated with Persistent DNA Methylation Reprogramming in Dorsal Root Ganglion. J. Neurosci. 2018, 38, 6090–6101. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, D.; Dai, Z.; You, Y.; Chen, Y.; Lei, C.; Lv, Y.; Wang, Y. Intraperitoneal 5-Azacytidine Alleviates Nerve Injury-Induced Pain in Rats by Modulating DNA Methylation. Mol. Neurobiol. 2023, 60, 2186–2199. [Google Scholar] [CrossRef]
- Jiang, B.C.; He, L.N.; Wu, X.B.; Shi, H.; Zhang, W.W.; Zhang, Z.J.; Cao, D.L.; Li, C.H.; Gu, J. Promoted Interaction of C/EBPα with Demethylated Cxcr3 Gene Promoter Contributes to Neuropathic Pain in Mice. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 685–700. [Google Scholar] [CrossRef]
- Imai, S.; Saeki, M.; Yanase, M.; Horiuchi, H.; Abe, M.; Narita, M.; Kuzumaki, N.; Suzuki, T.; Narita, M. Change in microRNAs associated with neuronal adaptive responses in the nucleus accumbens under neuropathic pain. J. Neurosci. Off. J. Soc. Neurosci. 2011, 31, 15294–15299. [Google Scholar] [CrossRef]
- Zhao, J.Y.; Liang, L.; Gu, X.; Li, Z.; Wu, S.; Sun, L.; Atianjoh, F.E.; Feng, J.; Mo, K.; Jia, S.; et al. DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat. Commun. 2017, 8, 14712. [Google Scholar] [CrossRef]
- Liu, L.; Xu, D.; Wang, T.; Zhang, Y.; Yang, X.; Wang, X.; Tang, Y. Epigenetic reduction of miR-214-3p upregulates astrocytic colony-stimulating factor-1 and contributes to neuropathic pain induced by nerve injury. Pain 2020, 161, 96–108. [Google Scholar] [CrossRef]
- Tochiki, K.K.; Cunningham, J.; Hunt, S.P.; Géranton, S.M. The expression of spinal methyl-CpG-binding protein 2, DNA methyltransferases and histone deacetylases is modulated in persistent pain states. Mol. Pain 2012, 8, 1477–8069. [Google Scholar] [CrossRef]
- Francés, R.; Mata-Garrido, J.; de la Fuente, R.; Carcelén, M.; Lafarga, M. Identification of Epigenetic Interactions between MicroRNA-30c-5p and DNA Methyltransferases in Neuropathic Pain. Int. J. Mol. Sci. 2022, 23, 13994. [Google Scholar] [CrossRef]
- Ding, X.; Lin, Y.; Chen, C.; Yan, B.; Liu, Q.; Zheng, H.; Wu, Y.; Zhou, C. DNMT1 Mediates Chronic Pain-Related Depression by Inhibiting GABAergic Neuronal Activation in the Central Amygdala. Biol. Psychiatry 2023, 94, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lin, Z.P.; Zheng, H.Z.; Zhang, S.; Zhang, Z.L.; Chen, Y.; You, Y.S.; Yang, M.H. Abnormal DNA methylation in the lumbar spinal cord following chronic constriction injury in rats. Neurosci. Lett. 2016, 610, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, C.; Guo, Q.L.; Yan, J.Q.; Zhu, X.Y.; Huang, C.S.; Zou, W.Y. Intrathecal 5-azacytidine inhibits global DNA methylation and methyl- CpG-binding protein 2 expression and alleviates neuropathic pain in rats following chronic constriction injury. Brain Res. 2011, 1418, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Yu, L.; Gao, Y.; Ma, L.; Ren, J.; Liu, Y.; Gao, D.S.; Xie, C.; Wu, Y.; Wang, L.; et al. MeCP2 Epigenetic Silencing of Oprm1 Gene in Primary Sensory Neurons Under Neuropathic Pain Conditions. Frontiers in Neuroscience 2021, 15, 743207. [Google Scholar] [CrossRef]
- Patel, N.J.; Hogan, K.J.; Rizk, E.; Stewart, K.; Madrid, A.; Vadakkadath Meethal, S.; Alisch, R.; Borth, L.; Papale, L.A.; Ondoma, S.; et al. Ancestral Folate Promotes Neuronal Regeneration in Serial Generations of Progeny. Mol. Neurobiol. 2020, 57, 2048–2071. [Google Scholar] [CrossRef]
- Lindner, R.; Puttagunta, R.; Nguyen, T.; Di Giovanni, S. DNA methylation temporal profiling following peripheral versus central nervous system axotomy. Sci. Data 2014, 1, 140038. [Google Scholar] [CrossRef]
- Gao, W.M.; Chadha, M.S.; Kline, A.E.; Clark, R.S.; Kochanek, P.M.; Dixon, C.E.; Jenkins, L.W. Immunohistochemical analysis of histone H3 acetylation and methylation--evidence for altered epigenetic signaling following traumatic brain injury in immature rats. Brain Res. 2006, 1070, 31–34. [Google Scholar] [CrossRef]
- Schober, M.E.; Ke, X.; Xing, B.; Block, B.P.; Requena, D.F.; McKnight, R.; Lane, R.H. Traumatic brain injury increased IGF-1B mRNA and altered IGF-1 exon 5 and promoter region epigenetic characteristics in the rat pup hippocampus. J. Neurotrauma 2012, 29, 2075–2085. [Google Scholar] [CrossRef]
- Puttagunta, R.; Tedeschi, A.; Sória, M.G.; Hervera, A.; Lindner, R.; Rathore, K.I.; Gaub, P.; Joshi, Y.; Nguyen, T.; Schmandke, A.; et al. PCAF-dependent epigenetic changes promote axonal regeneration in the central nervous system. Nat. Commun. 2014, 5, 3527. [Google Scholar] [CrossRef]
- Hervera, A.; Zhou, L.; Palmisano, I.; McLachlan, E.; Kong, G.; Hutson, T.H.; Danzi, M.C.; Lemmon, V.P. PP4-dependent HDAC3 dephosphorylation discriminates between axonal regeneration and regenerative failure. EMBO J. 2019, 38, e101032. [Google Scholar] [CrossRef]
- Gupta, R.; Saha, P.; Sen, T.; Sen, N. An augmentation in histone dimethylation at lysine nine residues elicits vision impairment following traumatic brain injury. Free Radic. Biol. Med. 2019, 134, 630–643. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Yang, S.G.; Hu, M.W.; Wang, R.Y.; Zhang, C.; Kosanam, A.R.; Ochuba, A.J.; Jiang, J.J.; Luo, X.; Guan, Y.; et al. Histone methyltransferase Ezh2 coordinates mammalian axon regeneration via regulation of key regenerative pathways. J. Clin. Investig. 2024, 134, e163145. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.G.; Li, C.P.; Wang, X.W.; Huang, T.; Qian, C.; Li, Q.; Zhao, L.R.; Zhou, S.Y.; Ding, C.Y.; Nie, R.; et al. Roles of Kdm6a and Kdm6b in Regulation of Mammalian Neural Regeneration. Adv. Sci. 2025, 12, e2405537. [Google Scholar] [CrossRef] [PubMed]
- Berry, K.P.; Lu, Q.R. Chromatin modification and epigenetic control in functional nerve regeneration. Semin. Cell Dev. Biol. 2020, 97, 74–83. [Google Scholar] [CrossRef]
- Ghosh, K.; Pan, H.L. Epigenetic Mechanisms of Neural Plasticity in Chronic Neuropathic Pain. ACS Chem. Neurosci. 2022, 13, 432–441. [Google Scholar] [CrossRef]
- Descalzi, G.; Ikegami, D.; Ushijima, T.; Nestler, E.J.; Zachariou, V.; Narita, M. Epigenetic mechanisms of chronic pain. Trends Neurosci. 2015, 38, 237–246. [Google Scholar] [CrossRef]
- Penas, C.; Navarro, X. Epigenetic Modifications Associated to Neuroinflammation and Neuropathic Pain After Neural Trauma. Front. Cell. Neurosci. 2018, 12, 158. [Google Scholar] [CrossRef]
- Fuhrmann, D.; Elsässer, H.P. Schwann cell Myc-interacting zinc-finger protein 1 without pox virus and zinc finger: Epigenetic implications in a peripheral neuropathy. Neural Regen. Res. 2018, 13, 1534–1537. [Google Scholar] [CrossRef]
- Guo, T.T.; Zhao, Y.; Huang, W.X.; Zhang, T.; Zhao, L.L.; Gu, X.S.; Zhou, S.L. Silencing the enhancer of zeste homologue 2, Ezh2, represses axon regeneration of dorsal root ganglion neurons. Neural Regen. Res. 2022, 17, 1518–1525. [Google Scholar] [CrossRef]
- Ma, K.H.; Duong, P.; Moran, J.J.; Junaidi, N.; Svaren, J. Polycomb repression regulates Schwann cell proliferation and axon regeneration after nerve injury. Glia 2018, 66, 2487–2502. [Google Scholar] [CrossRef]
- Ma, K.H.; Hung, H.A.; Srinivasan, R.; Xie, H.; Orkin, S.H.; Svaren, J. Regulation of Peripheral Nerve Myelin Maintenance by Gene Repression through Polycomb Repressive Complex 2. J. Neurosci. Off. J. Soc. Neurosci. 2015, 35, 8640–8652. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Zhao, J.Y.; Gu, X.; Wu, S.; Mo, K.; Xiong, M.; Marie Lutz, B.; Bekker, A.; Tao, Y.X. G9a inhibits CREB-triggered expression of mu opioid receptor in primary sensory neurons following peripheral nerve injury. Mol. Pain 2016, 12, 1744806916682242. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Gu, X.; Zhao, J.Y.; Wu, S.; Miao, X.; Xiao, J.; Mo, K.; Zhang, J.; Lutz, B.M.; Bekker, A.; et al. G9a participates in nerve injury-induced Kcna2 downregulation in primary sensory neurons. Sci. Rep. 2016, 6, 37704. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhang, J.; Chen, L.; Chen, S.R.; Chen, H.; Zhang, G.; Pan, H.L. Histone methyltransferase G9a diminishes expression of cannabinoid CB(1) receptors in primary sensory neurons in neuropathic pain. J. Biol. Chem. 2020, 295, 3553–3562. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, L.; Miao, X.; Wu, S.; Cao, J.; Tao, B.; Mao, Q.; Mo, K.; Xiong, M.; Lutz, B.M.; et al. Contribution of the Suppressor of Variegation 3-9 Homolog 1 in Dorsal Root Ganglia and Spinal Cord Dorsal Horn to Nerve Injury-induced Nociceptive Hypersensitivity. Anesthesiology 2016, 125, 765–778. [Google Scholar] [CrossRef]
- Yadav, R.; Weng, H.R. EZH2 regulates spinal neuroinflammation in rats with neuropathic pain. Neuroscience 2017, 349, 106–117. [Google Scholar] [CrossRef]
- An, Q.; Sun, C.; Li, R.; Chen, S.; Gu, X.; An, S.; Wang, Z. Calcitonin gene-related peptide regulates spinal microglial activation through the histone H3 lysine 27 trimethylation via enhancer of zeste homolog-2 in rats with neuropathic pain. J. Neuroinflammation 2021, 18, 117. [Google Scholar] [CrossRef]
- Li, L.; Bai, L.; Yang, K.; Zhang, J.; Gao, Y.; Jiang, M.; Yang, Y.; Zhang, X.; Wang, L.; Wang, X.; et al. KDM6B epigenetically regulated-interleukin-6 expression in the dorsal root ganglia and spinal dorsal horn contributes to the development and maintenance of neuropathic pain following peripheral nerve injury in male rats. Brain Behav. Immun. 2021, 98, 265–282. [Google Scholar] [CrossRef]
- Qi, R.; Cao, J. Histone methylation-mediated microRNA-32-5p down-regulation in sensory neurons regulates pain behaviors via targeting Cav3.2 channels. Proc. Natl. Acad. Sci. USA 2022, 119, e2117209119. [Google Scholar] [CrossRef]
- Mo, K.; Xu, H.; Gong, H.; Lei, H.; Wang, Y.; Guo, W.; Xu, S.; Tu, W. Dorsal Root Ganglia Coactivator-associated Arginine Methyltransferase 1 Contributes to Peripheral Nerve Injury-induced Pain Hypersensitivities. Neuroscience 2018, 394, 232–242. [Google Scholar] [CrossRef]
- Penney, J.; Seo, J.; Kritskiy, O.; Elmsaouri, S. Loss of Protein Arginine Methyltransferase 8 Alters Synapse Composition and Function, Resulting in Behavioral Defects. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 8655–8666. [Google Scholar] [CrossRef] [PubMed]
- Lo, L.H.; Dong, R.; Lyu, Q.; Lai, K.O. The Protein Arginine Methyltransferase PRMT8 and Substrate G3BP1 Control Rac1-PAK1 Signaling and Actin Cytoskeleton for Dendritic Spine Maturation. Cell Rep. 2020, 31, 107744. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.C.; Ho, Y.C.; Lai, C.Y.; Wang, H.H.; Yang, P.S.; Cheng, J.K.; Chen, G.D.; Ng, S.C.; Lee, A.S.; Tseng, K.W.; et al. Blocking the Spinal Fbxo3/CARM1/K(+) Channel Epigenetic Silencing Pathway as a Strategy for Neuropathic Pain Relief. Neurotherapeutics 2021, 18, 1295–1315. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mao, J.; Wang, X.; Lin, Y.; Hou, G.; Zhu, J.; Xie, B. Genome-wide screening of altered m6A-tagged transcript profiles in the hippocampus after traumatic brain injury in mice. Epigenomics 2019, 11, 805–819. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, Y.; Ma, H.; Zeng, R.; Liu, R.; Wang, P.; Jin, X.; Zhao, Y. Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury. Mol. Brain 2020, 13, 11. [Google Scholar] [CrossRef]
- Xiang, Z.; Luo, Y.; Yu, J.; Ma, H.; Zhao, Y. Comprehensive Transcriptome-Wide Profiling of 5-Methylcytosine Modifications in Long Non-Coding RNAs in a Rat Model of Traumatic Brain Injury. Curr. Issues Mol. Biol. 2024, 46, 14497–14513. [Google Scholar] [CrossRef]
- Liu, D.; Fan, B.; Li, J.; Sun, T.; Ma, J.; Zhou, X.; Feng, S. N6-methyladenosine modification: A potential regulatory mechanism in spinal cord injury. Front. Cell. Neurosci. 2022, 16, 989637. [Google Scholar] [CrossRef]
- Wang, S.; Xie, X.; Li, C.; Jia, J.; Chen, C. Integrative network analysis of N(6) methylation-related genes reveal potential therapeutic targets for spinal cord injury. Math. Biosci. Eng. MBE 2021, 18, 8174–8187. [Google Scholar] [CrossRef]
- Liu, X.; Li, Z.; Tong, J.; Wu, F.; Jin, H.; Liu, K. Characterization of the Expressions and m6A Methylation Modification Patterns of mRNAs and lncRNAs in a Spinal Cord Injury Rat Model. Mol. Neurobiol. 2025, 62, 806–818. [Google Scholar] [CrossRef]
- Xing, L.; Cai, Y. Epitranscriptomic m6A regulation following spinal cord injury. J. Neurosci. Res. 2021, 99, 843–857. [Google Scholar] [CrossRef]
- Wang, H.; Yuan, J.; Dang, X.; Shi, Z.; Ban, W.; Ma, D. Mettl14-mediated m6A modification modulates neuron apoptosis during the repair of spinal cord injury by regulating the transformation from pri-mir-375 to miR-375. Cell Biosci. 2021, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Duan, Y.; Chang, F.; Zhang, T.; Huang, X.; Yu, C. METTL14 promotes apoptosis of spinal cord neurons by inducing EEF1A2 m6A methylation in spinal cord injury. Cell Death Discov. 2022, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Cao, J.; Hong, Z.; Lu, Y.; Qin, Z.; Tao, T. Epigenetic combined with transcriptomic analysis of the m6A methylome after spared nerve injury-induced neuropathic pain in mice. Neural Regen. Res. 2023, 18, 2545–2552. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Pi, W.J.; Zhang, Q.; Li, Q. Molecular Characterization and Clinical Characteristics of m5C-Based RNA Methylation in Spinal Cord Injury: Validated by qPCR. Int. J. Genom. 2022, 2022, 5433860. [Google Scholar] [CrossRef]
- Duan, X.; Qiao, M.; Bei, F.; Kim, I.J.; He, Z.; Sanes, J.R. Subtype-specific regeneration of retinal ganglion cells following axotomy: Effects of osteopontin and mTOR signaling. Neuron 2015, 85, 1244–1256. [Google Scholar] [CrossRef]
- Albik, S.; Tao, Y.X. Emerging role of RNA m6A modification in chronic pain. Pain 2021, 162, 1897–1898. [Google Scholar] [CrossRef]
- Zhang, L.; Hao, D.; Ma, P.; Ma, B.; Qin, J.; Tian, G.; Liu, Z.; Zhou, X. Epitranscriptomic Analysis of m6A Methylome After Peripheral Nerve Injury. Front. Genet. 2021, 12, 686000. [Google Scholar] [CrossRef]
- Liu, X.; Lv, J.; Tang, W.; Hu, Y.; Wen, Y.; Shen, H. METTL3-mediated maturation of miR-192-5p targets ATG7 to prevent Schwann cell autophagy in peripheral nerve injury. J. Neuropathol. Exp. Neurol. 2023, 82, 1010–1019. [Google Scholar] [CrossRef]
- Ma, L.; Huang, Y.; Zhang, F.; Gao, D.S.; Sun, N.; Ren, J.; Xia, S.; Li, J.; Peng, X.; Yu, L.; et al. MMP24 Contributes to Neuropathic Pain in an FTO-Dependent Manner in the Spinal Cord Neurons. Front. Pharmacol. 2021, 12, 673831. [Google Scholar] [CrossRef]
- Wu, S.F.; Wang, Y.; Zhao, Q.C. Demethylase FTO promotes neuropathic pain development via regulating the m6A methylation levels of CXCR3. Acta Biochim. Pol. 2022, 69, 819–824. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, X.; Wang, J.; Jin, Y.; Gong, M.; Ye, Y.; Li, P. METTL3 suppresses neuropathic pain via modulating N6-methyladenosine-dependent primary miR-150 processing. Cell Death Discov. 2022, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Han, Y.; Liu, L.; Liu, M.; Lin, J.; Tang, Y.; Guo, S.; He, R.; Wu, Q. N(6)-Methyladenosine methylase METTL3 contributes to neuropathic pain by epigenetic silencing of mu opioid receptor. Behav. Brain Res. 2023, 452, 114592. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.; Wang, Q.; Zhao, Y.; Song, Y.; You, H.; Wang, J.; Liu, R.; Shi, Z.; Chen, X.; Luo, Q. Alteration of N (6) -Methyladenosine mRNA Methylation in a Rat Model of Cerebral Ischemia-Reperfusion Injury. Front. Neurosci. 2021, 15, 605654. [Google Scholar] [CrossRef] [PubMed]
- Kan, R.L.; Chen, J.; Sallam, T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. TIG 2022, 38, 182–193. [Google Scholar] [CrossRef]
- Sendinc, E.; Shi, Y. RNA m6A methylation across the transcriptome. Mol. Cell 2023, 83, 428–441. [Google Scholar] [CrossRef]
- Niederberger, E.; Resch, E.; Parnham, M.J.; Geisslinger, G. Drugging the pain epigenome. Nat. Revi. Neurol. 2017, 13, 434–447. [Google Scholar] [CrossRef]
- Jiang, W.; Tan, X.Y.; Li, J.M.; Yu, P.; Dong, M. DNA Methylation: A Target in Neuropathic Pain. Front. Med. 2022, 9, 879902. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Q.; Zhao, X.; Shao, L.; Liu, G.; Zheng, X.; Xie, L.; Zhang, Y.; Sun, C.; Xu, R. YTHDC1 mitigates ischemic stroke by promoting Akt phosphorylation through destabilizing PTEN mRNA. Cell Death Dis. 2020, 11, 977. [Google Scholar] [CrossRef]
- Xu, G.E.; Zhao, X.; Li, G.; Gokulnath, P.; Wang, L.; Xiao, J. The landscape of epigenetic regulation and therapeutic application of N(6)-methyladenosine modifications in non-coding RNAs. Genes Dis. 2024, 11, 101045. [Google Scholar] [CrossRef]
- Mao, S.; Huang, T.; Chen, Y.; Shen, L.; Zhou, S.; Zhang, S.; Yu, B. Circ-Spidr enhances axon regeneration after peripheral nerve injury. Cell Death Dis. 2019, 10, 787. [Google Scholar] [CrossRef]
- Wang, D.; Zheng, T.; Ge, X.; Xu, J.; Feng, L.; Jiang, C.; Tao, J.; Chen, Y.; Liu, X.; Yu, B.; et al. Unfolded protein response-induced expression of long noncoding RNA Ngrl1 supports peripheral axon regeneration by activating the PI3K-Akt pathway. Exp. Neurol. 2022, 352, 114025. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, S.; Pan, Y.; Zheng, T.; Cao, Q.; Yu, B.; Zhou, F.; Wang, D. The Role of Methylation Modification in Neural Injury and Repair. Int. J. Mol. Sci. 2025, 26, 5349. https://doi.org/10.3390/ijms26115349
Lv S, Pan Y, Zheng T, Cao Q, Yu B, Zhou F, Wang D. The Role of Methylation Modification in Neural Injury and Repair. International Journal of Molecular Sciences. 2025; 26(11):5349. https://doi.org/10.3390/ijms26115349
Chicago/Turabian StyleLv, Saizhen, Yanyu Pan, Tiemei Zheng, Qianqian Cao, Bin Yu, Fengquan Zhou, and Dong Wang. 2025. "The Role of Methylation Modification in Neural Injury and Repair" International Journal of Molecular Sciences 26, no. 11: 5349. https://doi.org/10.3390/ijms26115349
APA StyleLv, S., Pan, Y., Zheng, T., Cao, Q., Yu, B., Zhou, F., & Wang, D. (2025). The Role of Methylation Modification in Neural Injury and Repair. International Journal of Molecular Sciences, 26(11), 5349. https://doi.org/10.3390/ijms26115349






