Genome-Wide Identification and Expression Analysis of Zona Pellucida (ZP) Gene Family in Cynoglossus semilaevis
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of ZP Genes
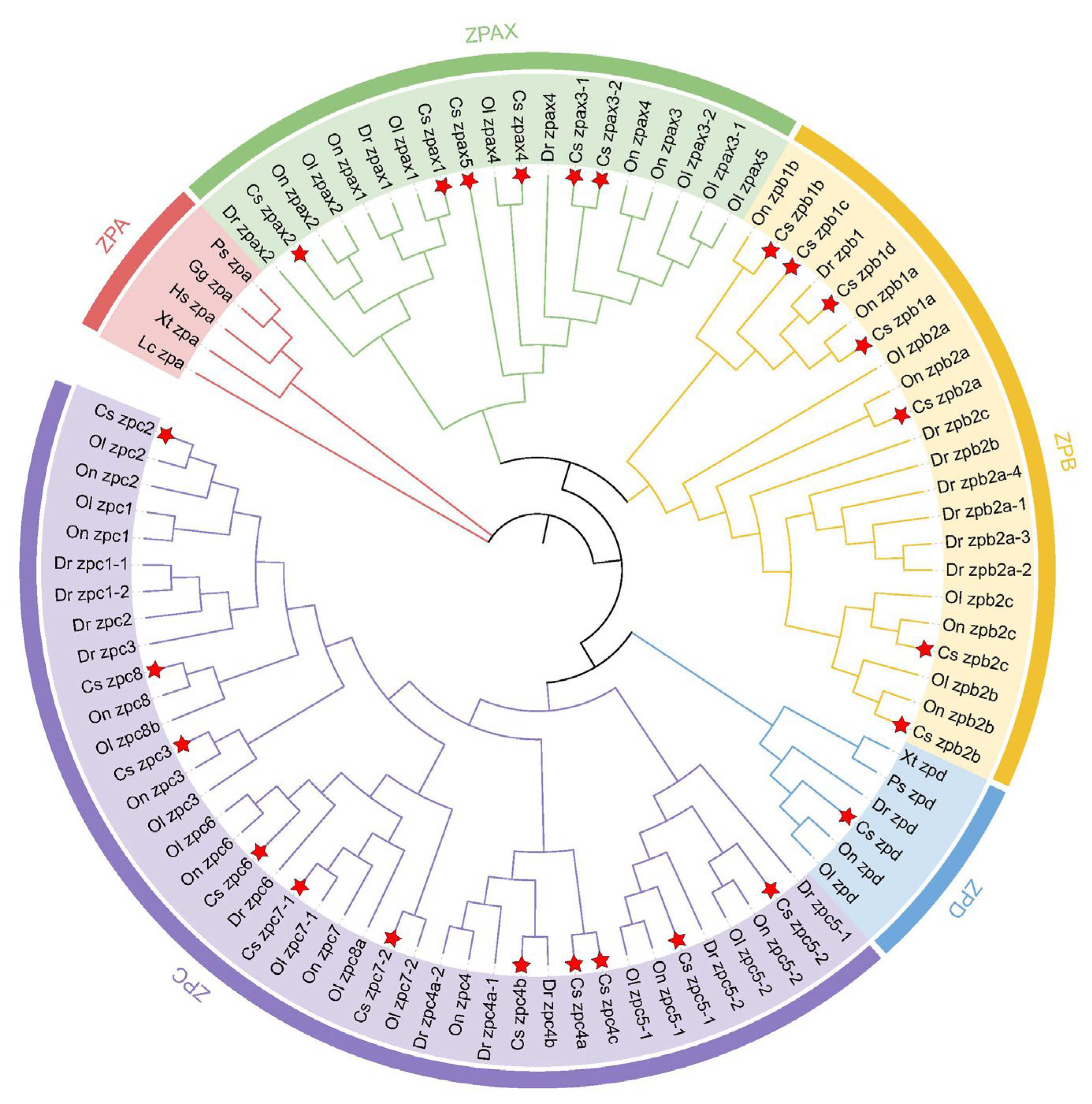
2.2. Phylogenetic Analysis of ZP Family
2.3. Gene Structural Features of ZP Family
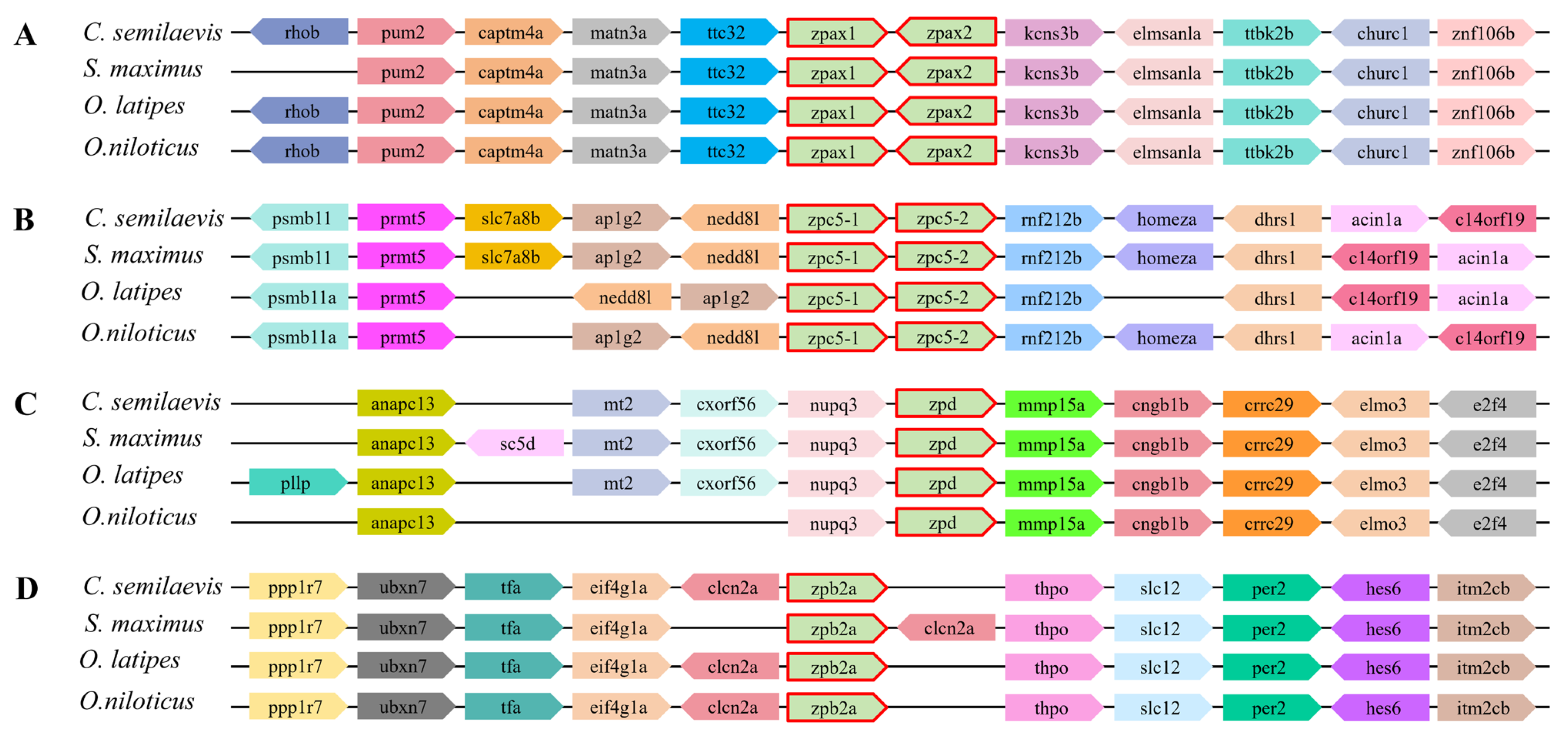
2.4. Syntenic Analysis of ZP Genes in Teleost
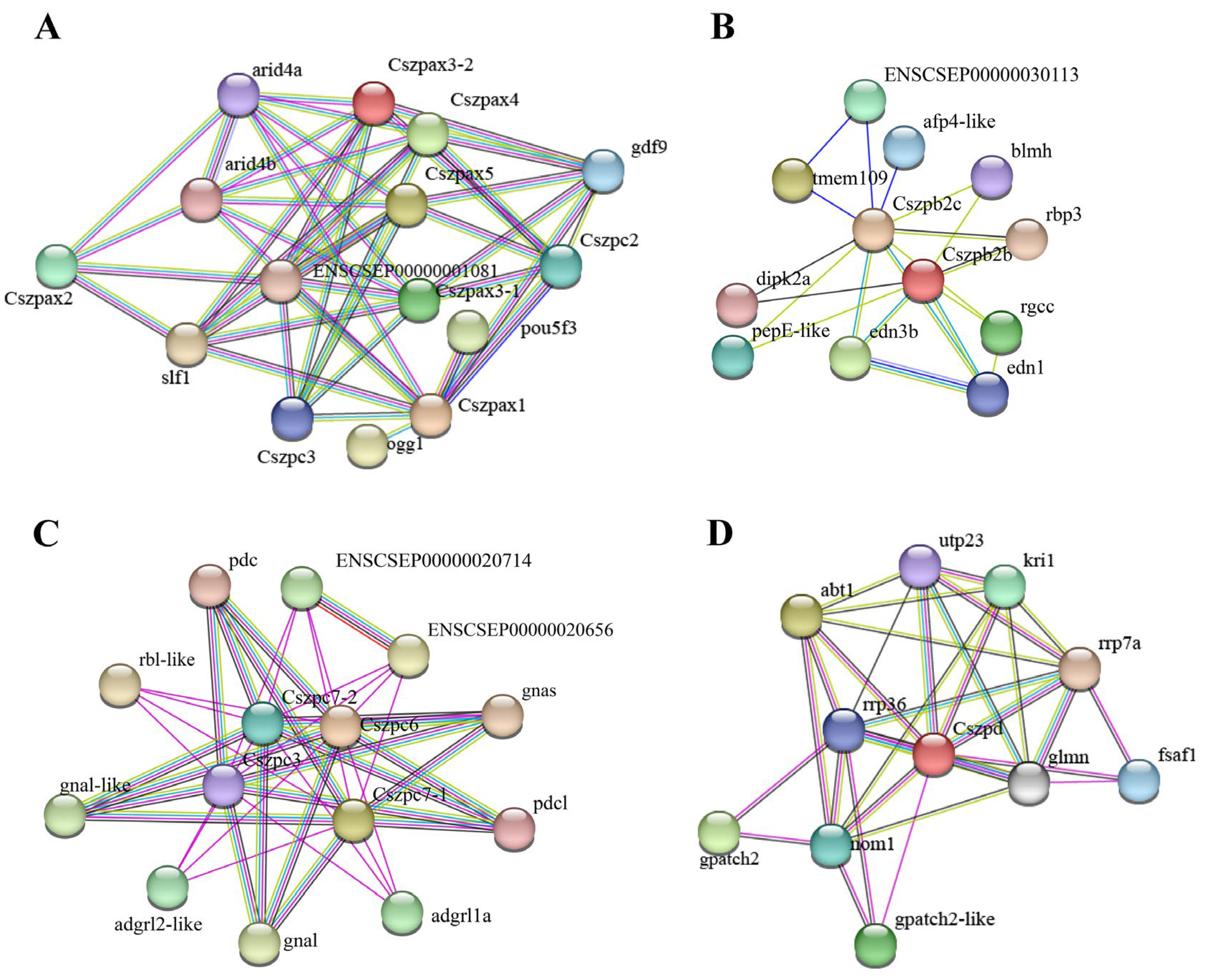
2.5. Protein–Protein Interaction (PPI) Network Analysis
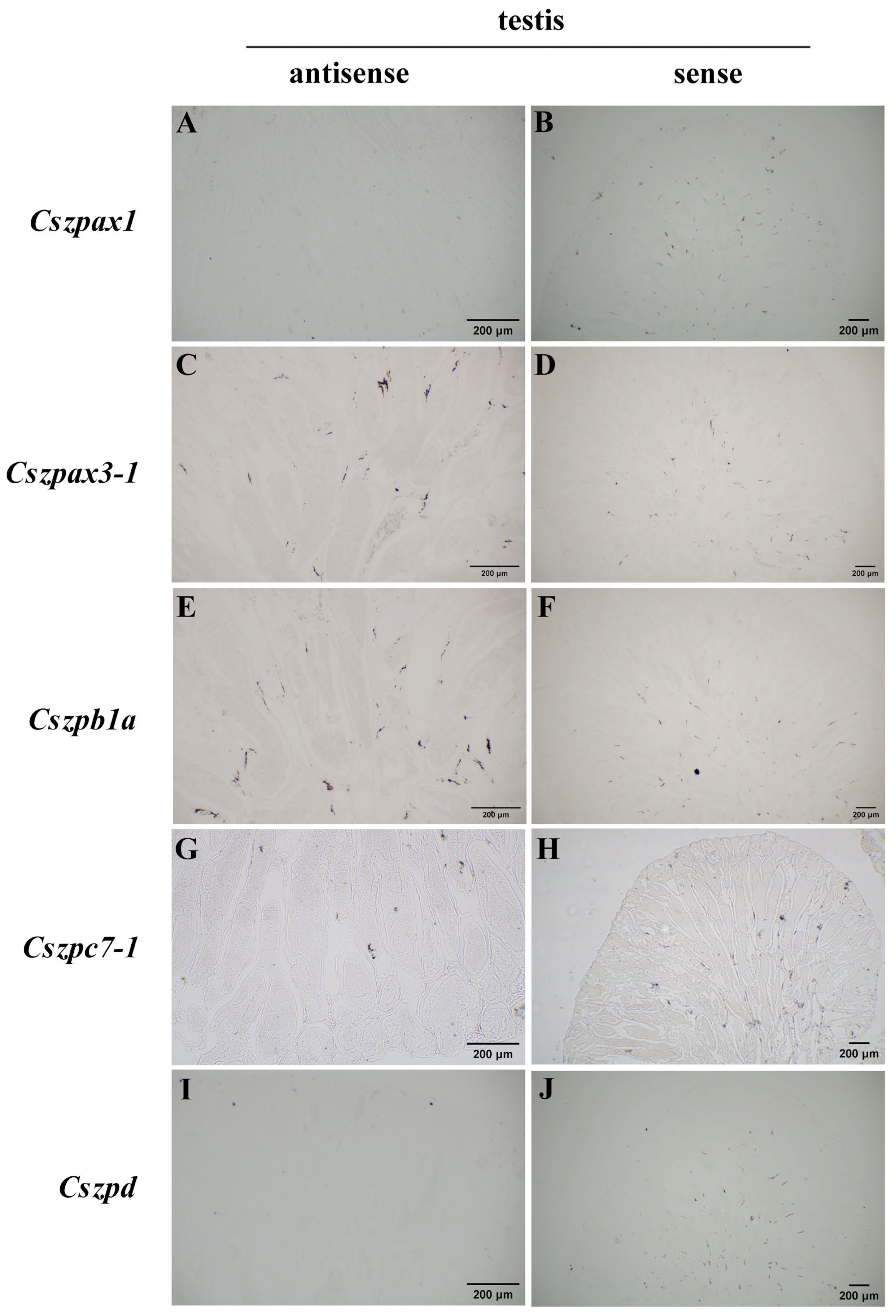
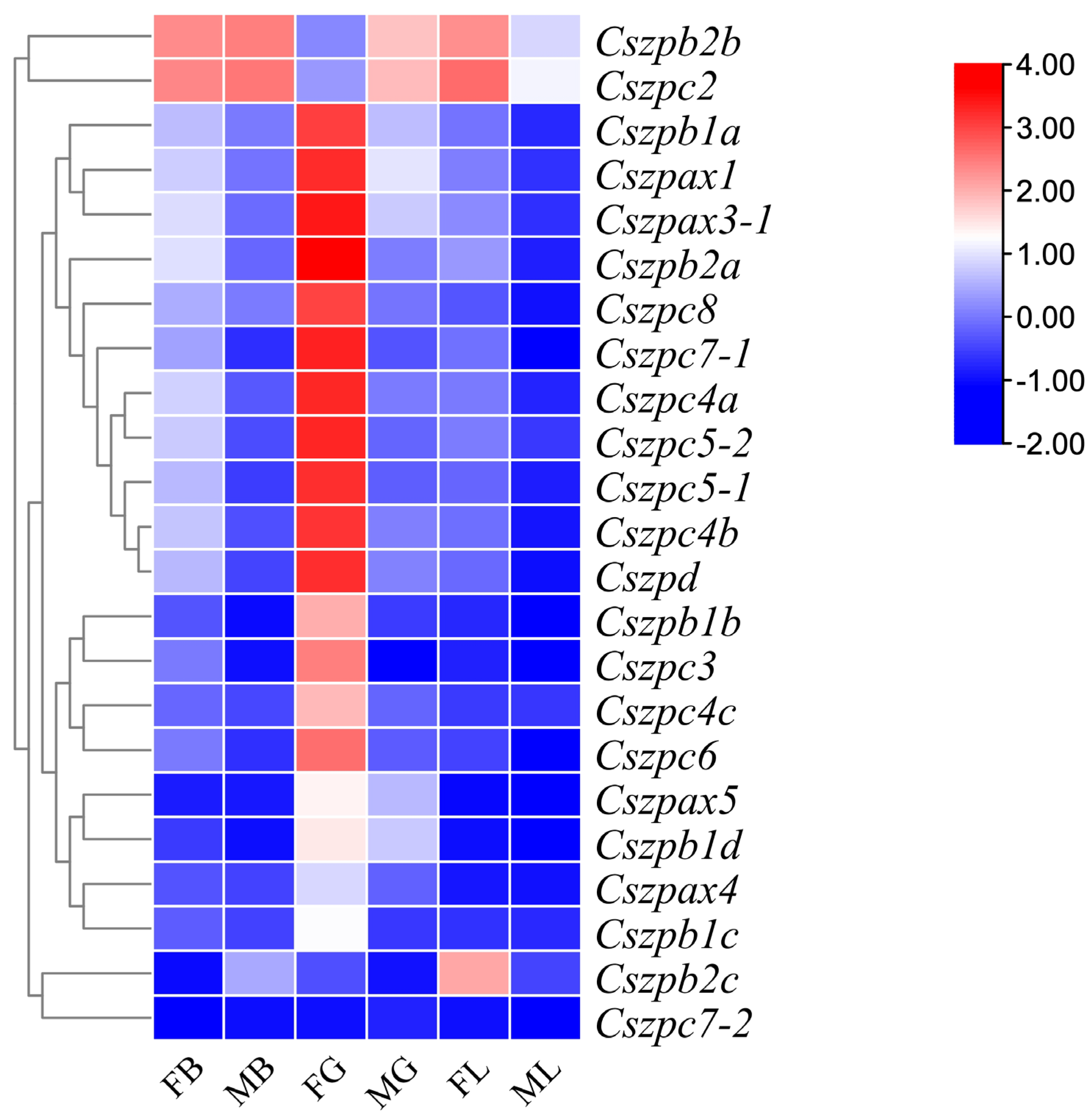
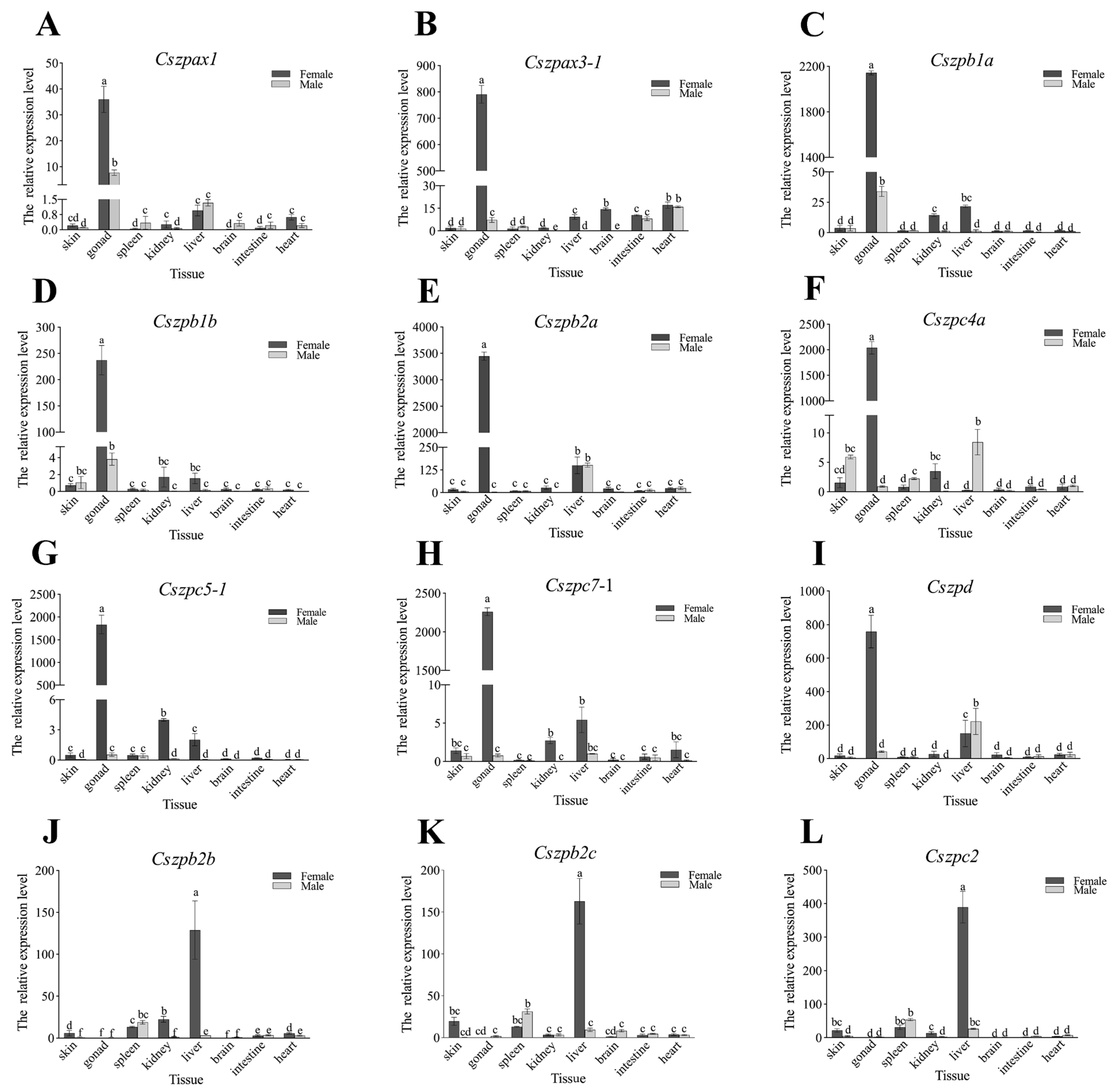
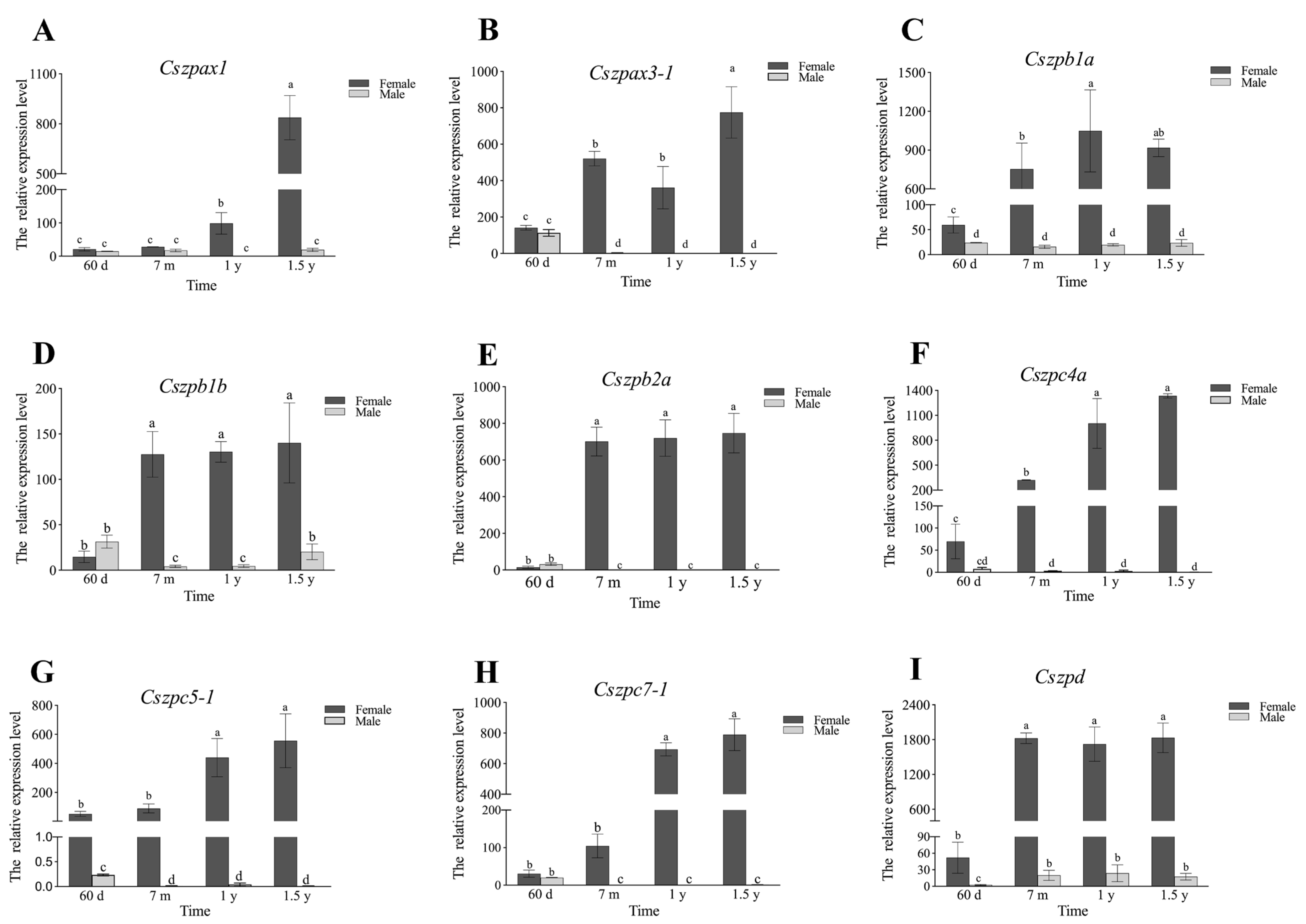
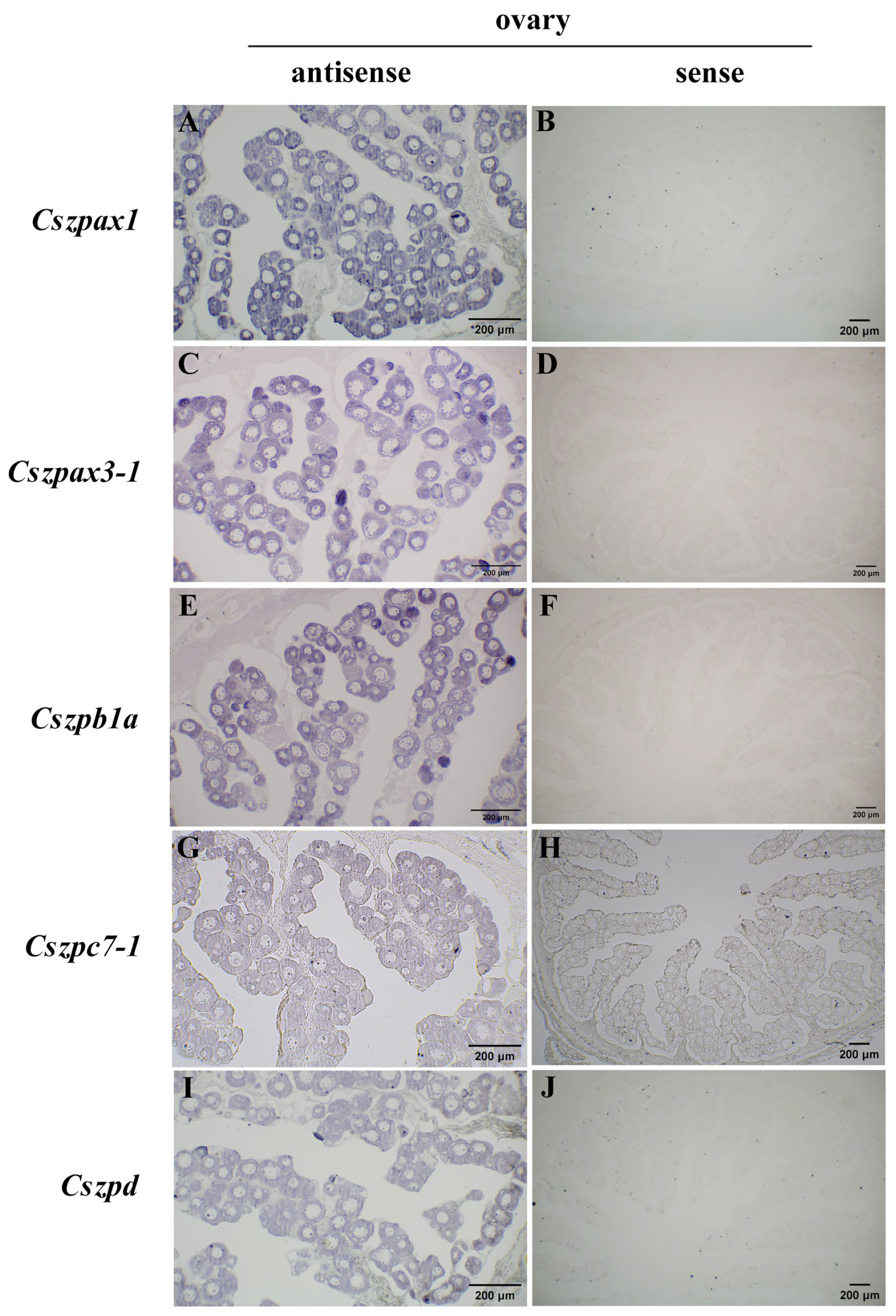
2.6. Expression Patterns of ZP Genes in C. semilaevis
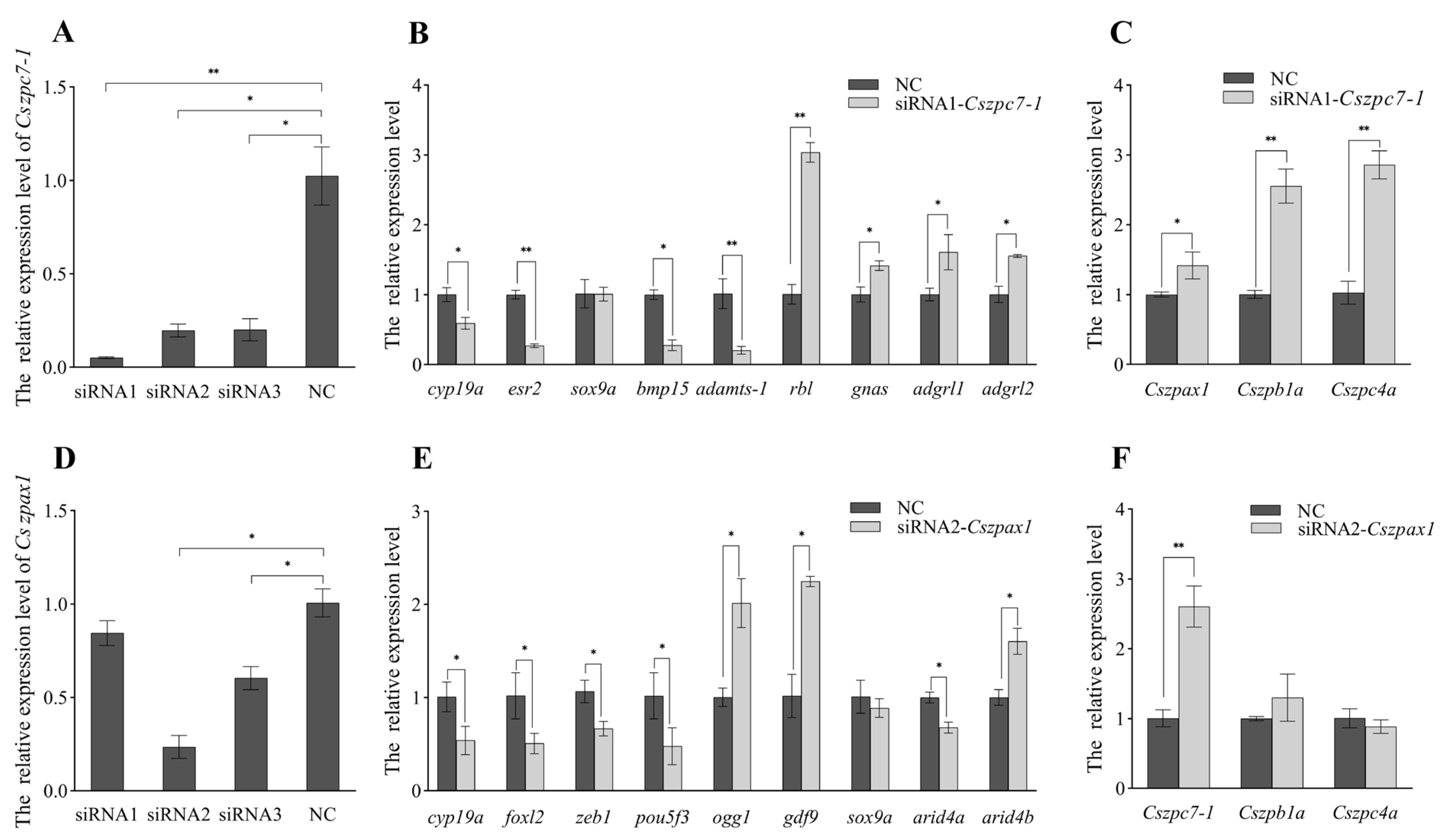
2.7. Knockdown Effects of Cszpax1, Cszpc7-1 on CO
3. Discussion
4. Materials and Methods
4.1. Animal Euthanasia and Ethics Statement
4.2. Fish and Sample Collection
4.3. Identification of ZP Genes in C. semilaevis Genome
4.4. Sequence and Evolutionary Analysis
4.5. Synteny Analysis
4.6. Protein–Protein Interaction Prediction
4.7. Heatmap Plotting with RNA-Seq Data
4.8. Quantitative Real-Time (qPCR) Analysis
4.9. In Situ Hybridization Analysis
4.10. Cell Culture and siRNA Transfection Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| cyp19a | cytochrome p450 19a |
| foxl2 | forkhead box L2 |
| esr2 | estrogen receptor 2 |
| bmp15 | bone morphogenetic protein 15 |
| adamts-1 | ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| sox9a | SRY-box transcription factor 9a |
| zeb1 | zinc finger E-box bingding homeobox 1 |
| rbl | rhamnose-binding lectin |
| gnas | guanine nucleotide binding protein, alpha stimulating |
| gdf9 | growth differentiation factor 9 |
| pou5f3 | POU class 5 homeobox 3 |
| adgrl1 | adhesion G protein-coupled receptor L1 |
| adgrl2 | adhesion G protein-coupled receptor L2 |
| gnal | guanine nucleotide-binding protein G(s) subunit alpha-like |
| ogg1 | 8-oxoguanine DNA glycosylase 1 |
| arid4a | AT-rich interaction domain 4A |
| arid4b | AT-rich interaction domain 4B |
Appendix A

| Primer | Purpose | Sequences (5′→3′) |
|---|---|---|
| zpb2a-q-F | qPCR | GCACCAGGGATGGACAGTTT |
| zpb2a-q-R | qPCR | AGGAGGGGTCATTAGGGTCC |
| zpb1b-q-F | qPCR | CTCGGTGTGGTTGCTTCTCT |
| zpb1b-q-R | qPCR | CTTGGGAGTGGGAAGTGGTC |
| zpb1a-q-F | qPCR | CCCAAAAGCAACACTTCGGG |
| zpb1a-q-R | qPCR | GGGTCTAAAGGGTTGGCACA |
| zpax1-q-F | qPCR | TGTCCAGGAGCACACGTTTT |
| zpax1-q-R | qPCR | AGAGGCAGGAAGTAGACCGT |
| zpd-q-F | qPCR | ATTGCCATCAGAGCCGTTCA |
| zpd-q-R | qPCR | CTTGGAGACGCTGGACATGT |
| zpax3-1-q-F | qPCR | CCGTATGGACAGCCTGACTC |
| zpax3-1-q-R | qPCR | CCATGATGACCACCCAGCTT |
| zpc4a-q-F | qPCR | TCAGTACCGGCCCTTTTGTC |
| zpc4a-q-R | qPCR | CCCTCATTACAGCAGCACCA |
| zpc5-1-q-F | qPCR | AATGGAGGAGTGTGTGGCAG |
| zpc5-1-q-R | qPCR | ATGGCAGATGAGTGGTAGCG |
| zpc7-1-q-F | qPCR | AACCGGAACAAACCCACAGT |
| zpc7-1-q-R | qPCR | ACAACTGGTCCCACTCTTGC |
| β-actin-F | qPCR | TCAATTCTCCACGAACCA |
| β-actin-R | qPCR | CTTACACAGCGAGCAACC |
| cyp19a-q-F | qPCR | GGTGAGGATGTGACCCAGTGT |
| cyp19a-q-R | qPCR | ACGGGCTGAAATCGCAAG |
| foxl2-q-F | qPCR | GAGAGGAAGGGCAACTACTGGA |
| foxl2-q-R | qPCR | TGGTTGGAAGTGCGTGGG |
| sox9a-q-F | qPCR | AAGAACCACACAGATCAAGACAGA |
| sox9a-q-R | qPCR | TAGTCATACTGTGCTCTGGTGATG |
| bmp15-q-F | qPCR | CGGACGGAAGAACTAAACA |
| bmp15-q-R | qPCR | TCATACTGGACCTGGAACG |
| esr2-q-F | qPCR | GATTAGGAGAAGGTGGAGAAGG |
| esr2-q-R | qPCR | GGTAACCAGAGGCATAGTCGTG |
| zeb1-q-F | qPCR | GCTCCTCTTCAAGGCACAGT |
| zeb1-q-R | qPCR | GACGTTACCATCCACTGCCA |
| adamts-1-q-F | qPCR | CTGGCCTCAAACCCTACCTG |
| adamts-1-q-R | qPCR | GGCCTCGCTCCTCTTCATAC |
| zpax1-siRNA-1 | siRNA | CCAGAGUCCUUCAGAUAUA |
| zpax1-siRNA-2 | siRNA | CGACUGUGCCGGCAAUCUA |
| zpax1-siRNA-3 | siRNA | CAGACAAGCAGUGGUUUAA |
| zpc7-1-siRNA-1 | siRNA | GGCUGUGUAGCUACGUUAA |
| zpc7-1-siRNA-2 | siRNA | CGCUGGAGAUCGUCAUCAA |
| zpc7-1-siRNA-3 | siRNA | CGCGAUUGUUGUCGGGCUA |
| zpb1a-SP6 | ISH | ATTTAGGTGACACTATAGAATCGAGGCTACGAGGTGGATT |
| zpb1a-T7 | ISH | TAATACGACTCACTATAGGGGCTCATAAACACGGGCTCCA |
| zpd-SP6 | ISH | ATTTAGGTGACACTATAGAATGTGGCAGGTGTAGGACTCT |
| zpd-T7 | ISH | TAATACGACTCACTATAGGGATCTGAATGGTGGCGTCGAG |
| zpax3-1-SP6 | ISH | ATTTAGGTGACACTATAGAAGAGCGTCTGACACCAGAACT |
| zpax3-1-T7 | ISH | TAATACGACTCACTATAGGGAGCTTGAGTCTTCGTGCTGAA |
| zpc7-1-SP6 | ISH | ATTTAGGTGACACTATAGAACCTTGAAAATGGGTGCCTGC |
| zpc7-1-T7 | ISH | TAATACGACTCACTATAGGGCCTCAACCTGGTCTTGAGGTC |
| zpax1-SP6 | ISH | ATTTAGGTGACACTATAGAATCTGTTGAATATACAGTCCCAGAGG |
| zpax1-T7 | ISH | TAATACGACTCACTATAGGGAGCTGAAGTGTGTGCGGTTA |
| sex-F | Gender detection | CCTAAATGATGGATGTAGATTCTGTC |
| sex-R | Gender detection | GATCCAGAGAAAATAAACCCAGG |
| gdf9-q-F | qPCR | ACGCTCACCTTCTGAGTTCC |
| gdf9-q-R | qPCR | AAGATCATGACGCTCAGGGG |
| adgrl1-q-F | qPCR | GTGGAAAGGCCGTGTCCTAA |
| adgrl1-q-R | qPCR | CTATTTGGCTGACCCACGGT |
| rbl-q-F | qPCR | GACGCCGTGATCAGACTACC |
| rbl-q-R | qPCR | ACATACGTAGGCCACCTCCA |
| gnas-q-F | qPCR | TGCACGCTACACTACACCTG |
| gnas-q-R | qPCR | ATGTTCTCTGTGTCGACCGC |
| pou5f3-q-F | qPCR | GCCAGAGTCGGCCATATGAT |
| pou5f3-q-R | qPCR | TAGAGGATGCTCGGGTCTGG |
| adgrl1-q-F | qPCR | CGGATGCACGAGCTTATCCT |
| adgrl1-q-R | qPCR | AGATGGGACAGGCAATGTGG |
| adgrl2-q-F | qPCR | AACGCACTCGCAACATTGTC |
| adgrl2-q-R | qPCR | TCACCCATAAGCCCCTCTCA |
| ogg1-q-F | qPCR | GGCCAAACATGCGGTACTTT |
| ogg1-q-R | qPCR | CGTGCCACATTTCCTTTCGT |
| arid4b-q-F | qPCR | CTGTGGCTGGAACGTCAGAT |
| arid4b-q-R | qPCR | GAGCGATGGACACGGTTAGT |
| arid4a-q-F | qPCR | CCTCCTCCATGTCATCGTCG |
| arid4a-q-R | qPCR | CGAGGCTTCTGGGACTTTGT |
| Species | Gene | Accession No |
|---|---|---|
| Cynoglossus semilaevis | Cszpax1 | XP_024912764.1 |
| Cszpax2 | XP_024912783.1 | |
| Cszpax3-1 | XP_008319235.1 | |
| Cszpax3-2 | XP_024915769.1 | |
| Cszpax4 | XP_008310879.2 | |
| Cszpax5 | XP_008319357.1 | |
| Cszpb1a | XP_008329736.1 | |
| Cszpb1b | XP_008328541.1 | |
| Cszpb1c | XP_024920423.1 | |
| Cszpb1d | XP_008315308.1 | |
| Cszpb2a | XP_008332246.1 | |
| Cszpb2b | XP_024912115.1 | |
| Cszpb2c | XP_008310180.2 | |
| Cszpc2 | XP_016888570.1 | |
| Cszpc3 | XP_008323734.1 | |
| Cszpc4a | XP_008314339.1 | |
| Cszpc4b | XP_024914509.1 | |
| Cszpc4c | XP_008325694.1 | |
| Cszpc5-1 | XP_008332968.3 | |
| Cszpc5-2 | NP_001284511.2 | |
| Cszpc6 | XP_008332190.1 | |
| Cszpc7 | XP_008327912.1 | |
| Cszpc8 | XP_008306734.1 | |
| Cszpc9 | XP_008325416.3 | |
| Cszpd | XP_008309362.1 | |
| Danio rerio | Drzpax1 | ENSDARG00000069251 |
| Drzpax2 | ENSDARG00000017188 | |
| Drzpax4 | ENSDARG00000079034 | |
| Drzpb2a-1 | ENSDARG00000090237 | |
| Drzpb2a-2 | ENSDARG00000091409 | |
| Drzpb2a-3 | ENSDARG00000086352 | |
| Drzpb2a-4 | ENSDARG00000086522 | |
| Drzpb2b | ENSDARG00000055415 | |
| Drzpb2c | ENSDARG00000004898 | |
| Drzpd | ENSDARG00000051959 | |
| Drzpc1-1 | ENSDARG00000042129 | |
| Drzpc1-2 | ENSDARG00000042130 | |
| Drzpc2 | ENSDARG00000090768 | |
| Drzpc3 | ENSDARG00000016908 | |
| Drzpc4a-1 | ENSDARG00000040081 | |
| Drzpc4a-2 | ENSDARG00000070178 | |
| Drzpc4b | ENSDARG00000092919 | |
| Drzpc5-1 | ENSDARG00000038720 | |
| Drzpc5-2 | ENSDARG00000054313 | |
| Drzpc6 | ENSDARG00000039828 | |
| Oreochromis niloticus | Onzpax1 | ENSONIP00000007688 |
| Onzpax2 | ENSONIP00000007675 | |
| Onzpax3 | ENSONIP00000008459 | |
| Onzpax4 | ENSONIP00000006775 | |
| Onzpb1a | XP_003452556.1 | |
| Onzpb1b | ENSONIP00000010516 | |
| Onzpb2a | ENSONIP00000012346 | |
| Onzpb2b | ENSONIP00000018582 | |
| Onzpb2c | ENSONIP00000018726 | |
| Onzpd | ENSONIP00000012896 | |
| Onzpc1 | ENSONIP00000004983 | |
| Onzpc2 | ENSONIP00000018727 | |
| Onzpc3 | ENSONIP00000015344 | |
| Onzpc4 | ENSONIP00000002777 | |
| Onzpc5-1 | ENSONIP00000019557 | |
| Onzpc5-2 | ENSONIP00000019559 | |
| Onzpc6 | ENSONIP00000012513 | |
| Onzpc7 | ENSONIP00000002749 | |
| Onzpc8 | ENSONIP00000013206 | |
| Oryzias latipes | Olzpax1 | ENSORLG00000012471 |
| Olzpax2 | ENSORLG00000012527 | |
| Olzpax3-1 | ENSORLG00000006604 | |
| Olzpax3-2 | ENSORLG00000006621 | |
| Olzpax4 | ENSORLG00000018174 | |
| Olzpax5 | ENSORLG00000006629 | |
| Olzpb2a | ENSORLG00000016580 | |
| Olzpb2b | ENSORLG00000010880 | |
| Olzpb2c | ENSORLG00000010086 | |
| Olzpd | ENSORLG00000015608 | |
| Olzpc1 | ENSORLG00000020441 | |
| Olzpc2 | ENSORLG00000010134 | |
| Olzpc3 | ENSORLG00000005414 | |
| Olzpc5-1 | ENSORLG00000012273 | |
| Olzpc5-2 | ENSORLG00000012249 | |
| Olzpc6 | ENSORLG00000016064 | |
| Olzpc7-1 | ENSORLG00000009853 | |
| Olzpc7-2 | ENSORLG00000009867 | |
| Olzpc8a | ENSORLG00000006875 | |
| Olzpc8b | ENSORLG00000015033 | |
| Pelodiscus sinensis | Pszpa | ENSPSIG00000004165 |
| Pszpd | ENSPSIG00000004205 | |
| Pszpb1 | ENSPSIG00000005870 | |
| Pszpb2a-1 | ENSPSIG00000005091 | |
| Pszpb2a-2 | ENSPSIG00000014055 | |
| Pszpb2b | ENSPSIG00000006328 | |
| Pszpc1 | ENSPSIG00000010174 | |
| Pszpc1 | ENSPSIG00000010174 | |
| Pszpc2 | ENSPSIG00000013197 | |
| Pszpc3 | ENSPSIG00000016070 | |
| Pszpax1 | ENSPSIG00000011815 | |
| Pszpax2 | ENSPSIG00000011721 | |
| Xenopus tropicalis | Xtzpa | ENSXETG00000016704 |
| Xtzpd | ENSXETG00000013767 | |
| Xtzpb1 | ENSXETG00000015911 | |
| Xtzpb2 | ENSXETG00000011855 | |
| Xtzpc1 | ENSXETG00000019465 | |
| Xtzpc2 | ENSXETG00000025433 | |
| Xtzpax1 | ENSXETG00000015572 | |
| Xtzpax2 | ENSXETG00000014953 | |
| Gallus gallus | Ggzpa | ENSGALP00000038302 |
| Ggzpb1 | CAC16087.1 | |
| Ggzpb2 | ENSGALP00000032884 | |
| Ggzpc1 | ENSGALP00000002636 | |
| Ggzpc2-1 | ENSGALP00000002356 | |
| Ggzpc2-2 | ENSGALP00000002368 | |
| Ggzpax1 | ENSGALP00000026515 | |
| Ggzpax2 | ENSGALP00000026528 | |
| Ggzpd | BAD13713.2 | |
| Homo sapiens | Hszpa | ENSG00000103310 |
| Hszpb1 | ENSG00000149506 | |
| Hszpb2 | ENSG00000116996 | |
| Hszpc | ENSG00000188372 | |
| Latimeria chalumnae | Lczpa | ENSLACG00000005253 |
| Lczpb1 | ENSLACG00000005027 | |
| Lczpb2a-1 | ENSLACG00000008179 | |
| Lczpb2a-2 | ENSLACG00000007163 | |
| Lczpb2a-3 | ENSLACG00000005832 | |
| Lczpb2a-4 | ENSLACG00000004264 | |
| Lczpb2b | ENSLACG00000011054 | |
| Lczpb2c | ENSLACG00000007972 | |
| Lczpc1 | ENSLACG00000001864 | |
| Lczpc2-1 | ENSLACG00000000590 | |
| Lczpc2-2 | ENSLACG00000001956 | |
| Lczpc2-3 | ENSLACG00000002494 | |
| Lczpc2-4 | ENSLACG00000004624 | |
| Lczpc5 | ENSLACG00000007941 | |
| Lczpd | ENSLACG00000002854 | |
| Lczpax1 | ENSLACG00000015100 | |
| Lczpax2 | ENSLACG00000006540 |
References
- Xu, S.; Tian, Z.; Zhu, H. Comparison of Induced and Natural Spawning in Southern Flounder Paralichthys lethostigma. Fish. Sci. 2009, 28, 3. [Google Scholar]
- Huang, L.; Liang, M.; Zhang, H.; Qu, J.; Wang, X.; Zheng, K. The effect of dietary vitamin A level on reproductive performance of broodstock Scophthamus maximus. Prog. Fish. Sci. 2013, 34, 62–70. [Google Scholar]
- Bleil, J.D.; Wassarman, P.M. Structure and function of the zona pellucida: Identification and characterization of the proteins of the mouse oocyte’s zona pellucida. Dev. Biol. 1980, 76, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Bleil, J.D.; Wassarman, P.M. Mammalian sperm-egg interaction: Identification of a glycoprotein in mouse egg zonae pellucidae possessing receptor activity for sperm. Cell 1980, 20, 873–882. [Google Scholar] [CrossRef]
- Lefièvre, L.; Conner, S.J.; Salpekar, A.; Olufowobi, O.; Ashton, P.; Pavlovic, B.; Lenton, W.; Afnan, M.; Brewis, I.A.; Monk, M.; et al. Four zona pellucida glycoproteins are expressed in the human. Hum. Reprod. 2004, 19, 1580–1586. [Google Scholar] [CrossRef]
- Smith, J.; Paton, I.R.; Hughes, D.C.; Burt, D.W. Isolation and mapping the chicken zona pellucida genes: An insight into the evolution of orthologous genes in different species. Mol. Reprod. Dev. 2005, 70, 133–145. [Google Scholar] [CrossRef]
- Murata, K.; Sasaki, T.; Yasumasu, S.; Iuchi, I.; Enami, J.; Yasumasu, I.; Yamagami, K. Cloning of cDNAs for the precursor protein of a low-molecular-weight subunit of the inner layer of the egg envelope (chorion) of the fish Oryzias latipes. Dev. Biol. 1995, 167, 9–17. [Google Scholar] [CrossRef]
- Wu, T.; Cheng, Y.; Liu, Z.; Tao, W.; Zheng, S.; Wang, D. Bioinformatic analyses of zona pellucida genes in vertebrates and their expression in Nile tilapia. Fish. Physiol. Biochem. 2018, 44, 435–449. [Google Scholar] [CrossRef]
- Qi, H.; Williams, Z.; Wassarman, P.M. Secretion and assembly of zona pellucida glycoproteins by growing mouse oocytes microinjected with epitope-tagged cDNAs for mZP2 and mZP3. Mol. Biol. Cell 2002, 13, 530–541. [Google Scholar] [CrossRef]
- Greve, J.M.; Wassarman, P.M. Mouse egg extracellular coat is a matrix of interconnected filaments possessing a structural repeat. J. Mol. Biol. 1985, 181, 253–264. [Google Scholar] [CrossRef]
- Bleil, J.D.; Greve, J.M.; Wassarman, P.M. Identification of a secondary sperm receptor in the mouse egg zona pellucida: Role in maintenance of binding of acrosome-reacted sperm to eggs. Dev. Biol. 1988, 128, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.L.; Zhou, C.X.; Zhang, X.L.; Liu, P.; Jin, Z.; Zhu, G.Y.; Ma, Y.; Li, J.; Yang, Z.X.; Zhang, D. ZP3 is Required for Germinal Vesicle Breakdown in Mouse Oocyte Meiosis. Sci. Rep. 2017, 7, 41272. [Google Scholar] [CrossRef] [PubMed]
- Yokokawa, R.; Watanabe, K.; Kanda, S.; Nishino, Y.; Yasumasu, S.; Sano, K. Egg envelope formation of medaka Oryzias latipes requires ZP proteins originating from both the liver and ovary. J. Biol. Chem. 2023, 299, 104600. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.-Q.; Wang, Y.-X.; Zhao, Y.; Zhang, J.; He, X.; Xie, P.; Chen, J.; Sun, Y.-H. Transfer of the zp3a gene results in changes in egg adhesiveness and buoyancy in transgenic zebrafish. Zool. Res. 2023, 44, 259–268. [Google Scholar] [CrossRef]
- Cao, L.; Huang, Q.; Wu, Z.; Cao, D.-d.; Ma, Z.; Xu, Q.; Hu, P.; Fu, Y.; Shen, Y.; Chan, J.; et al. Neofunctionalization of zona pellucida proteins enhances freeze-prevention in the eggs of Antarctic notothenioids. Nat. Commun. 2016, 7, 12987. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Fan, C.; Wang, T.; Chen, C.; Li, Z.; Li, Q.; Chen, S. Study on the method of quick cultivation of female parental half-smooth tongue sole (Cynoglossus semilaevis) with nereid. J. Shanghai Ocean Univ. 2013, 22, 7. [Google Scholar]
- Wang, N.; Yang, Q.; Wang, J.; Shi, R.; Li, M.; Gao, J.; Xu, W.; Yang, Y.; Chen, Y.; Chen, S. Integration of Transcriptome and Methylome Highlights the Roles of Cell Cycle and Hippo Signaling Pathway in Flatfish Sexual Size Dimorphism. Front. Cell Dev. Biol. 2021, 9, 743722. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.-M.; Brunet, F.; Petit, J.-L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; Naruse, K.; Sasaki, S.; Nakatani, Y.; Qu, W.; Ahsan, B.; Yamada, T.; Nagayasu, Y.; Doi, K.; Kasai, Y.; et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 2007, 447, 714–719. [Google Scholar] [CrossRef]
- Conner, S.J.; Hughes, D.C. Analysis of fish ZP1/ZPB homologous genes—Evidence for both genome duplication and species-specific amplification models of evolution. Reproduction 2003, 126, 347–352. [Google Scholar] [CrossRef]
- Goudet, G.; Mugnier, S.; Callebaut, I.; Monget, P. Phylogenetic analysis and identification of pseudogenes reveal a progressive loss of zona pellucida genes during evolution of vertebrates. Biol. Reprod. 2008, 78, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Sano, K.; Kawaguchi, M.; Yoshikawa, M.; Iuchi, I.; Yasumasu, S. Evolution of the teleostean zona pellucida gene inferred from the egg envelope protein genes of the Japanese eel, Anguilla japonica. FEBS J. 2010, 277, 4674–4684. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yu, H.; Zhang, Q.; Qi, J.; Zhong, Q.; Chen, Y.; Li, C. Molecular characterization and expression pattern of two zona pellucida genes in half-smooth tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010, 155, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Braun, B.C.; Ringleb, J.; Waurich, R.; Viertel, D.; Jewgenow, K. Functional role of feline zona pellucida protein 4 trefoil domain: A sperm receptor or structural component of the domestic cat zona pellucida? Reprod. Domest. Anim. 2009, 44 (Suppl. 2), 234–238. [Google Scholar] [CrossRef]
- Sano, K.; Kawaguchi, M.; Katano, K.; Tomita, K.; Inokuchi, M.; Nagasawa, T.; Hiroi, J.; Kaneko, T.; Kitagawa, T.; Fujimoto, T.; et al. Comparison of Egg Envelope Thickness in Teleosts and its Relationship to the Sites of ZP Protein Synthesis. J. Exp. Zool. B Mol. Dev. Evol. 2017, 328, 240–258. [Google Scholar] [CrossRef]
- Yamagami, K.; Hamazaki, T.S.; Yasumasu, S.; Masuda, K.; Iuchi, I. Molecular and cellular basis of formation, hardening, and breakdown of the egg envelope in fish. Int. Rev. Cytol. 1992, 136, 51–92. [Google Scholar]
- Kudo, S. Enzymes responsible for the bactericidal effect in extracts of vitelline and fertilisation envelopes of rainbow trout eggs. Zygote 2000, 8, 257–265. [Google Scholar] [CrossRef]
- Murata, K.; Sugiyama, H.; Yasumasu, S.; Iuchi, I.; Yasumasu, I.; Yamagami, K. Cloning of cDNA and estrogen-induced hepatic gene expression for choriogenin H, a precursor protein of the fish egg envelope (chorion). Proc. Natl. Acad. Sci. USA 1997, 94, 2050–2055. [Google Scholar] [CrossRef]
- Hyllner, S.J.; Westerlund, L.; Olsson, P.E.; Schopen, A. Cloning of rainbow trout egg envelope proteins: Members of a unique group of structural proteins. Biol. Reprod. 2001, 64, 805–811. [Google Scholar] [CrossRef]
- Wang, H.; Gong, Z. Characterization of two zebrafish cDNA clones encoding egg envelope proteins ZP2 and ZP3. Biochim. Biophys. Acta 1999, 1446, 156–160. [Google Scholar] [CrossRef]
- Hasuwa, H.; Ueda, J.; Ikawa, M.; Okabe, M. miR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science 2013, 341, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, Y.; Tan, F.; Wang, H.Y.; Chen, J.Y.; Zhang, X.; Zhao, X.; Liu, K.; Wang, Q.; Liu, S.; et al. Single-Cell Atlas of the Chinese Tongue Sole (Cynoglossus semilaevis) Ovary Reveals Transcriptional Programs of Oogenesis in Fish. Front. Cell Dev. Biol. 2022, 10, 828124. [Google Scholar] [CrossRef] [PubMed]
- Nosek, J.; Krajhanzl, A.; Kocourek, J. Studies on lectins. LVII. Immunofluorescence localization of lectins present in fish ovaries. Histochemistry 1983, 79, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.-H.; Yang, S.-T.; Yang, Z.-A.; Zhang, L.; Gui, J.-F. A C-type lectin associated and translocated with cortical granules during oocyte maturation and egg fertilization in fish. Dev. Biol. 2004, 265, 341–354. [Google Scholar] [CrossRef]
- Zhou, J.; Sun, S.; Li, R.; Xu, H.; Li, M.; Li, Z. Transcriptome analysis of Schizothorax oconnori (Cypriniformes: Cyprinidae) oocytes: The role of K(+) in promoting yolk globule fusion and regulating oocyte maturation. Fish. Physiol. Biochem. 2024, 50, 435–448. [Google Scholar] [CrossRef]
- Mukherjee, D.; Majumder, S.; Roy Moulik, S.; Pal, P.; Gupta, S.; Guha, P.; Kumar, D. Membrane receptor cross talk in gonadotropin-, IGF-I-, and insulin-mediated steroidogenesis in fish ovary: An overview. Gen. Comp. Endocrinol. 2017, 240, 10–18. [Google Scholar] [CrossRef]
- França, M.M.; Mendonca, B.B. Genetics of ovarian insufficiency and defects of folliculogenesis. Best. Pract. Res. Clin. Endocrinol. Metab. 2022, 36, 101594. [Google Scholar] [CrossRef]
- Ren, D.; Wei, X.; Lin, L.; Yuan, F.; Bi, Y.; Guo, Z.; Liu, L.; Ji, L.; Yang, X.; Han, K.; et al. A novel heterozygous missense variant of the ARID4A gene identified in Han Chinese families with schizophrenia-diagnosed siblings that interferes with DNA-binding activity. Mol. Psychiatry 2022, 27, 2777–2786. [Google Scholar] [CrossRef]
- Wu, R.C.; Jiang, M.; Beaudet, A.L.; Wu, M.Y. ARID4A and ARID4B regulate male fertility, a functional link to the AR and RB pathways. Proc. Natl. Acad. Sci. USA 2013, 110, 4616–4621. [Google Scholar] [CrossRef]
- Zuccotti, M.; Merico, V.; Sacchi, L.; Bellone, M.; Brink, T.C.; Bellazzi, R.; Stefanelli, M.; Redi, C.A.; Garagna, S.; Adjaye, J. Maternal Oct-4 is a potential key regulator of the developmental competence of mouse oocytes. BMC Dev. Biol. 2008, 8, 97. [Google Scholar] [CrossRef]
- Sánchez-Sánchez, A.V.; Camp, E.; García-España, A.; Leal-Tassias, A.; Mullor, J.L. Medaka Oct4 is expressed during early embryo development, and in primordial germ cells and adult gonads. Dev. Dyn. 2010, 239, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Paulini, F.; Melo, E.O. The role of oocyte-secreted factors GDF9 and BMP15 in follicular development and oogenesis. Reprod. Domest. Anim. 2011, 46, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Li, X.; Cheng, P.; Yang, Q.; Chen, Z.; Chen, S.; Wang, N. Characterization of growth differentiation factor 9 and bone morphogenetic factor 15 in Chinese tongue sole (Cynoglossus semilaevis): Sex-biased expression pattern and promoter regulation. Theriogenology 2022, 182, 119–128. [Google Scholar] [CrossRef]
- Lord, T.; Aitken, R.J. Fertilization stimulates 8-hydroxy-2′-deoxyguanosine repair and antioxidant activity to prevent mutagenesis in the embryo. Dev. Biol. 2015, 406, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Young, M.; McPhaul, M.J. A steroidogenic factor-1-binding site and cyclic adenosine 3′,5′-monophosphate response element-like elements are required for the activity of the rat aromatase promoter in rat Leydig tumor cell lines. Endocrinology 1998, 139, 5082–5093. [Google Scholar] [CrossRef]
- Shao, C.; Liu, G.; Liu, S.; Liu, C.; Chen, S. Characterization of the cyp19a1a gene from a BAC sequence in half-smooth tongue sole (Cynoglossus semilaevis) and analysis of its conservation among teleosts. Acta Oceanol. Sin. 2013, 32, 35–43. [Google Scholar] [CrossRef]
- Zhang, C.; He, Q.; Cheng, H.; Li, L.; Ruan, X.; Duan, X.; Huang, F.; Yang, H.; Zhang, H.; Shi, H.; et al. Transcription factors foxl2 and foxl3 regulate cyp19a1a and cyp11b in orange-spotted grouper (Epinephelus coioides). Aquac. Rep. 2022, 25, 101243. [Google Scholar] [CrossRef]
- Yang, Y.J.; Wang, Y.; Li, Z.; Zhou, L.; Gui, J.F. Sequential, Divergent, and Cooperative Requirements of Foxl2a and Foxl2b in Ovary Development and Maintenance of Zebrafish. Genetics 2017, 205, 1551–1572. [Google Scholar] [CrossRef]
- Xu, X.; Wang, X.; Zhou, L.; Liu, Q.; Li, J. Changes of estrogen, aromatase and estrogen receptor during oogenesis and gestation of Sebastes schlegelii. Oceanol. Limnol. Sin. 2021, 52, 1236–1243. [Google Scholar]
- Dong, J.; Albertini, D.F.; Nishimori, K.; Kumar, T.R.; Lu, N.; Matzuk, M.M. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature 1996, 383, 531–535. [Google Scholar] [CrossRef]
- Curry, T.E., Jr. ADAMTS1 and versican: Partners in ovulation and fertilization. Biol. Reprod. 2010, 83, 505–506. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shozu, M.; Minami, N.; Yokoyama, H.; Inoue, M.; Kurihara, H.; Matsushima, K.; Kuno, K. ADAMTS-1 is involved in normal follicular development, ovulatory process and organization of the medullary vascular network in the ovary. J. Mol. Endocrinol. 2005, 35, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.M.; Dunning, K.R.; Robker, R.L.; Pritchard, M.; Russell, D.L. Requirement for ADAMTS-1 in extracellular matrix remodeling during ovarian folliculogenesis and lymphangiogenesis. Dev. Biol. 2006, 300, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Rankin, T.L.; O’Brien, M.; Lee, E.; Wigglesworth, K.; Eppig, J.; Dean, J. Defective zonae pellucidae in Zp2-null mice disrupt folliculogenesis, fertility and development. Development 2001, 128, 1119–1126. [Google Scholar] [CrossRef]
- Liu, W.; Li, K.; Bai, D.; Yin, J.; Tang, Y.; Chi, F.; Zhang, L.; Wang, Y.; Pan, J.; Liang, S.; et al. Dosage effects of ZP2 and ZP3 heterozygous mutations cause human infertility. Hum. Genet. 2017, 136, 975–985. [Google Scholar] [CrossRef]
- Rankin, T.; Talbot, P.; Lee, E.; Dean, J. Abnormal zonae pellucidae in mice lacking ZP1 result in early embryonic loss. Development 1999, 126, 3847–3855. [Google Scholar] [CrossRef]
- Lamas-Toranzo, I.; Fonseca Balvís, N.; Querejeta-Fernández, A.; Izquierdo-Rico, M.J.; González-Brusi, L.; Lorenzo, P.L.; García-Rebollar, P.; Avilés, M.; Bermejo-Álvarez, P. ZP4 confers structural properties to the zona pellucida essential for embryo development. eLife 2019, 8, e48904. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, S.; Gao, F.; Meng, L.; Hu, Q.; Song, W.; Shao, C.; Lv, W. SCAR-transformation of Sex-specific SSR Marker and Its Application inHalf-smooth Tongue Sole (Cynoglossus semiliaevis). J. Agric. Biotechnol. 2014, 22, 787–792. [Google Scholar]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar Gustavo, A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Eddy, S.R. Hidden Markov models. Curr. Opin. Struct. Biol. 1996, 6, 361–365. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT: Iterative Refinement and Additional Methods. In Multiple Sequence Alignment Methods; Russell, D.J., Ed.; Humana Press: Totowa, NJ, USA, 2014; pp. 131–146. [Google Scholar]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Suppl. 2), W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Louis, A.; Muffato, M.; Roest Crollius, H. Genomicus: Five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 2012, 41, D700–D705. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, S.; Su, B.; Xue, T.; Cao, M.; Li, C. Genome-wide identification, characterization, and expression of the Toll-like receptors in Japanese flounder (Paralichthys olivaceus). Aquaculture 2021, 545, 737127. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhu, Y.; Cui, Z.; Yang, Y.; Xu, W.; Shao, C.; Fu, X.; Li, Y.; Chen, S. Expression analysis and characterization of dmrt2 in Chinese tongue sole (Cynoglossus semilaevis). Theriogenology 2019, 138, 1–8. [Google Scholar] [CrossRef]

| Name | Gene ID | Gene Length (bp) | ORF Length (bp) | Amino Acid Length (aa) | MW (kDa) | pI | Chr | Location | No. of Exons | No. of Introns |
|---|---|---|---|---|---|---|---|---|---|---|
| Cszpax1 | 103380918 | 5850 | 2850 | 950 | 103.73 | 4.84 | 7 | 5,983,260–5,989,109 | 21 | 20 |
| Cszpax2 | 112487247 | 3614 | 1662 | 554 | 61.72 | 5.03 | 7 | 5,977,607–5,981,220 | 13 | 12 |
| Cszpax3-1 | 103386630 | 14,745 | 2412 | 804 | 91.44 | 5.19 | 1 | 23,962,096– 23,976,840 | 25 | 24 |
| Cszpax3-2 | 112487726 | 3896 | 2400 | 800 | 89.83 | 5.44 | 1 | 23,966,848–23,970,743 | 19 | 18 |
| Cszpax4 | 103380637 | 8419 | 3282 | 1094 | 123.50 | 6.67 | 7 | 1,154,026–1,162,444 | 20 | 19 |
| Cszpax5 | 103386730 | 5305 | 2118 | 706 | 79.67 | 5.78 | 1 | 23,976,991–23,982,295 | 16 | 15 |
| Cszpb1a | 103394278 | 2910 | 1461 | 487 | 54.12 | 4.67 | 18 | 9,161,710–9,164,619 | 10 | 9 |
| Cszpb1b | 103393374 | 3542 | 1599 | 533 | 59.41 | 8.84 | 17 | 12,633,779–12,637,320 | 12 | 11 |
| Cszpb1c | 103393397 | 13,959 | 3033 | 1011 | 110.60 | 5.32 | 17 | 12813072–12,827,030 | 16 | 15 |
| Cszpb1d | 103383792 | 11,226 | 3243 | 1081 | 120.00 | 6.1 | 9 | 9,156,026–9,167,251 | 18 | 17 |
| Cszpb2a | 103396071 | 2286 | 1278 | 426 | 47.19 | 5.39 | 20 | 4,011,277–4,013,562 | 8 | 7 |
| Cszpb2b | 103380189 | 4402 | 1938 | 646 | 72.52 | 5.79 | 6 | 11,810,105–11,814,506 | 8 | 7 |
| Cszpb2c | 103380137 | 2580 | 1479 | 493 | 52.91 | 7.34 | 6 | 11,208,465–11,211,044 | 8 | 7 |
| Cszpc2 | 103380130 | 2718 | 1344 | 448 | 48.56 | 5.35 | 6 | 11,212,333–11,215,050 | 8 | 7 |
| Cszpc3 | 103389891 | 5953 | 1455 | 485 | 54.01 | 6.11 | 14 | 17,172,577–17,178,529 | 12 | 11 |
| Cszpc4a | 103383075 | 3615 | 1908 | 636 | 70.82 | 6.19 | 1 | 2,597,477–2,601,091 | 9 | 8 |
| Cszpc4b | 103384013 | 2723 | 1281 | 427 | 47.76 | 8.65 | 9 | 13,824,630–13,827,352 | 9 | 8 |
| Cszpc4c | 103391190 | 4981 | 1749 | 583 | 64.97 | 5.79 | 1 | 4,104,869–4,109,849 | 8 | 7 |
| Cszpc5-1 | 103396600 | 2058 | 1068 | 356 | 39.68 | 8.18 | 20 | 11,811,312–11,813,369 | 9 | 8 |
| Cszpc5-2 | 103396601 | 2153 | 939 | 313 | 34.77 | 5.44 | 20 | 11,814,022–11,816,174 | 10 | 9 |
| Cszpc6 | 103396037 | 5493 | 1401 | 467 | 52.55 | 9.09 | 20 | 3,517,755–3,523,247 | 9 | 8 |
| Cszpc7-1 | 103392929 | 2522 | 1557 | 519 | 56.42 | 4.98 | 17 | 6,833,291–6,835,812 | 10 | 9 |
| Cszpc7-2 | 103391061 | 2702 | 948 | 316 | 34.97 | 6.41 | 1 | 32,117,282–32,119,983 | 8 | 7 |
| Cszpc8 | 103377641 | 1096 | 642 | 214 | 23.67 | 6.59 | 4 | 2,417,479–2,418,574 | 2 | 1 |
| Cszpd | 103379554 | 4431 | 1218 | 406 | 45.21 | 6.55 | 6 | 2,033,303–2,037,733 | 10 | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Chen, Z.; Gao, C.; Li, X.; Wang, N.; Zhang, M.; Yan, H.; Sha, Z.; Chen, S. Genome-Wide Identification and Expression Analysis of Zona Pellucida (ZP) Gene Family in Cynoglossus semilaevis. Int. J. Mol. Sci. 2025, 26, 5346. https://doi.org/10.3390/ijms26115346
Zhang K, Chen Z, Gao C, Li X, Wang N, Zhang M, Yan H, Sha Z, Chen S. Genome-Wide Identification and Expression Analysis of Zona Pellucida (ZP) Gene Family in Cynoglossus semilaevis. International Journal of Molecular Sciences. 2025; 26(11):5346. https://doi.org/10.3390/ijms26115346
Chicago/Turabian StyleZhang, Kaili, Zhangfan Chen, Chengbin Gao, Xihong Li, Na Wang, Min Zhang, Haipeng Yan, Zhenxia Sha, and Songlin Chen. 2025. "Genome-Wide Identification and Expression Analysis of Zona Pellucida (ZP) Gene Family in Cynoglossus semilaevis" International Journal of Molecular Sciences 26, no. 11: 5346. https://doi.org/10.3390/ijms26115346
APA StyleZhang, K., Chen, Z., Gao, C., Li, X., Wang, N., Zhang, M., Yan, H., Sha, Z., & Chen, S. (2025). Genome-Wide Identification and Expression Analysis of Zona Pellucida (ZP) Gene Family in Cynoglossus semilaevis. International Journal of Molecular Sciences, 26(11), 5346. https://doi.org/10.3390/ijms26115346





