Tripeptide-Loaded Liposomes as Multifunctional Components in Topical Formulations
Abstract
1. Introduction
2. Results and Discussion
2.1. Peptide Synthesis and Characterization
2.2. Elastase Inhibition Assay: In Silico and In Vitro Studies
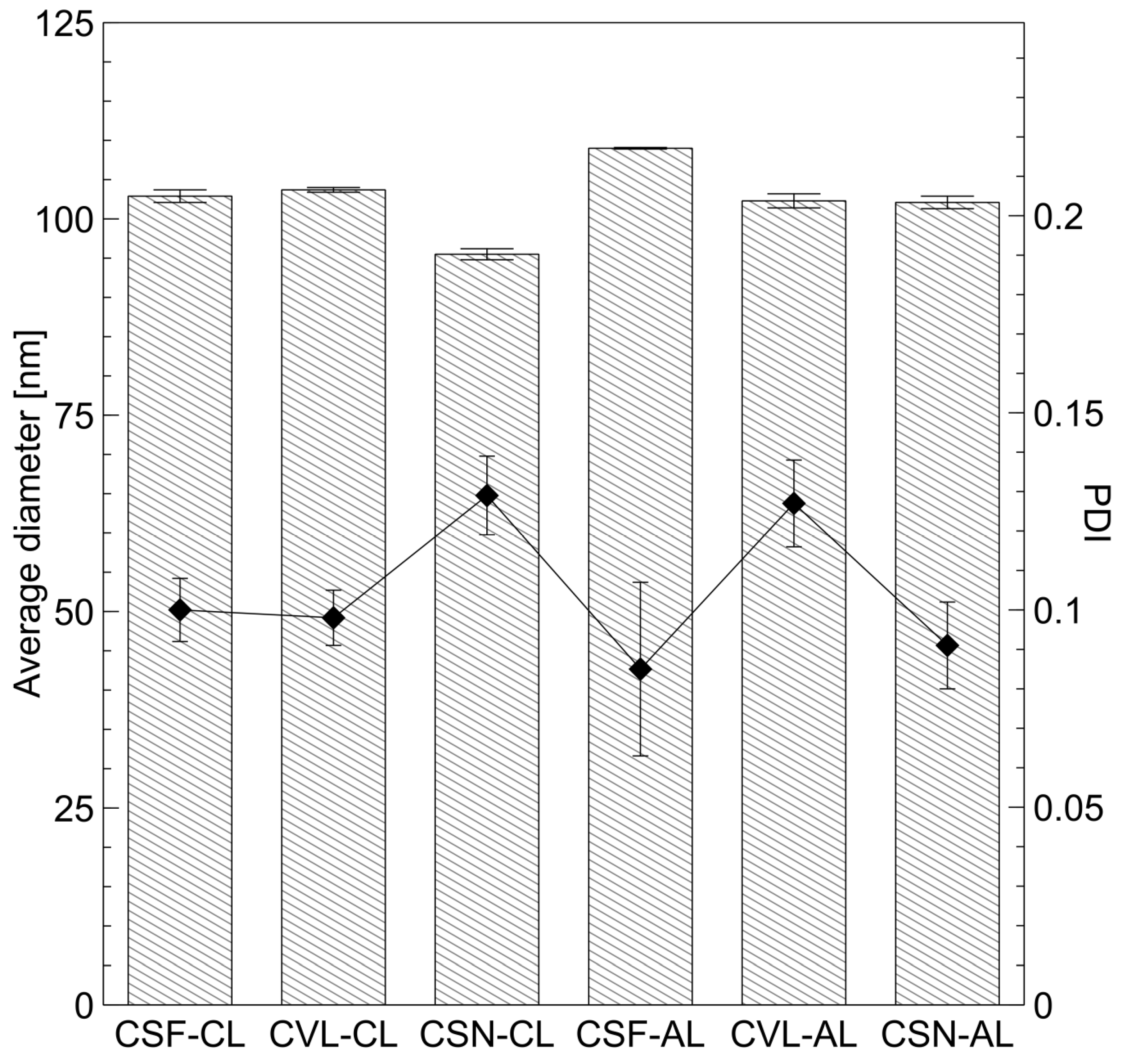
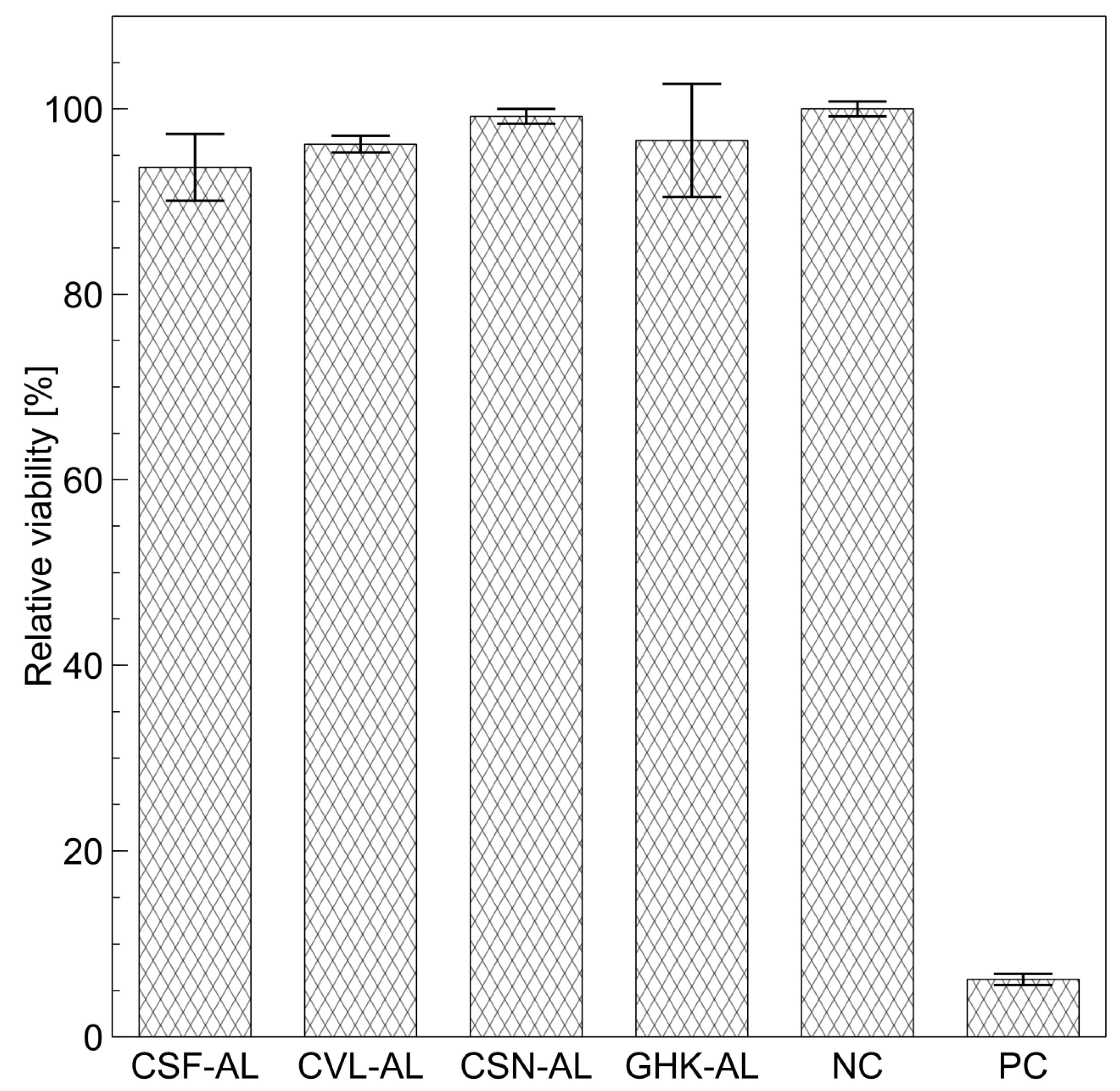
2.3. Physicochemical Properties and Cytotoxicity of Peptide-Loaded Liposomes
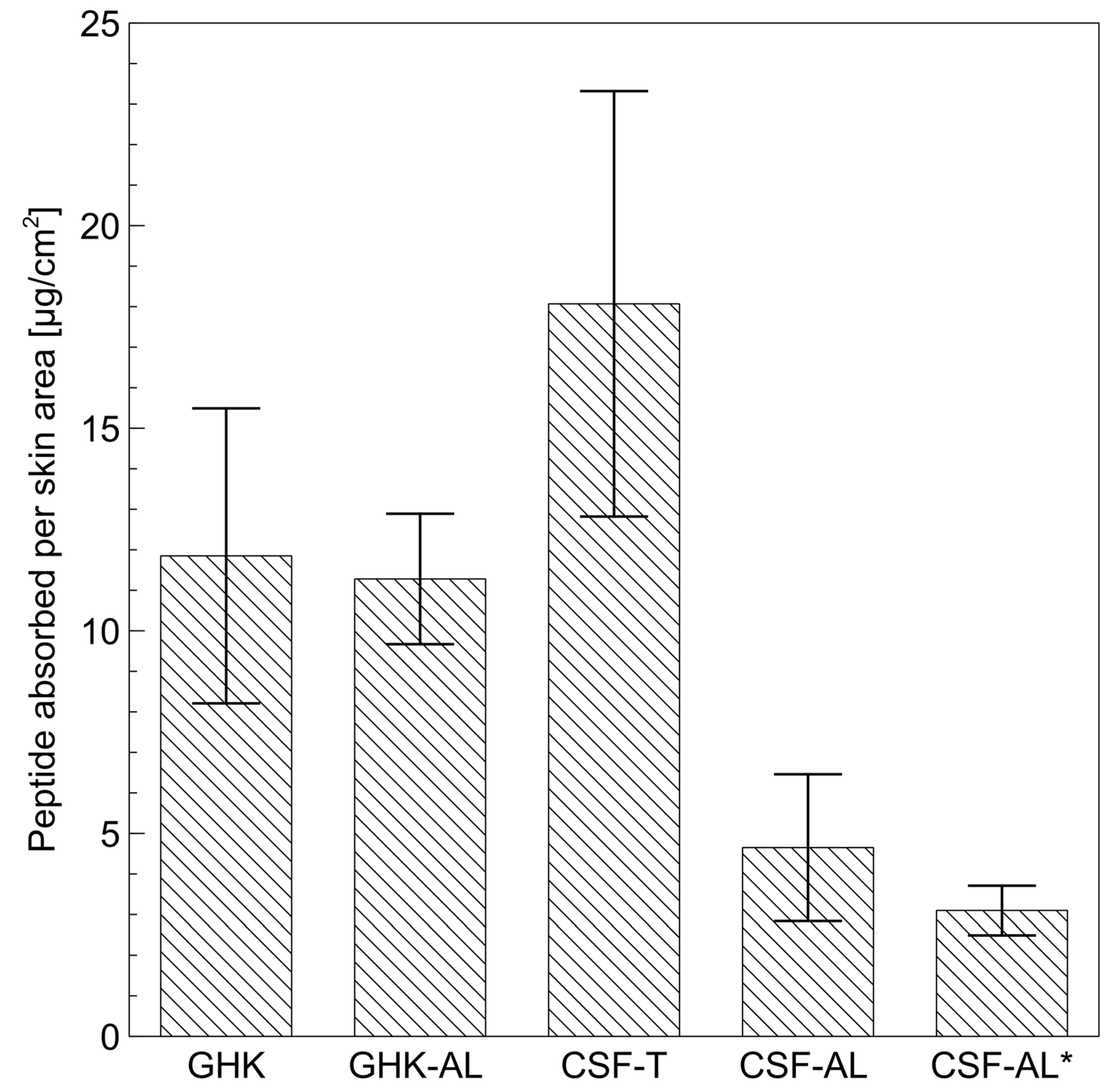
2.4. Permeation Studies
3. Materials and Methods
3.1. Materials
3.2. Peptide Synthesis and Characterization
3.2.1. Peptide Solid Phase Synthesis (PSPS)
3.2.2. Structural Characterization of Peptides
3.3. In Silico Elastase Inhibition Study
3.4. Elastase Inhibition Assay
3.5. Preparation of Empty Liposomes and Peptide-Loaded Liposomes (PLLs)
3.6. Characterization of PLLs
3.6.1. Size Analysis and Zeta Potential
3.6.2. Transmission Electron Microscopy (TEM)
3.6.3. Encapsulation Efficiency (EE) and Loading Capacity (LC)
3.6.4. Cytotoxicity
3.7. Permeation Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Dreno, B.; Araviiskaia, E.; Berardesca, E.; Bieber, T.; Hawk, J.; Sanchez-Viera, M.; Wolkenstein, P. The Science of Dermocosmetics and Its Role in Dermatology. J. Eur. Acad. Dermatol. Venereol. 2014, 28, 1409–1417. [Google Scholar] [CrossRef] [PubMed]
- Park, K.Y.; López Gehrke, I. Combined Multilevel Anti-Aging Strategies and Practical Applications of Dermocosmetics in Aesthetic Procedures. J. Eur. Acad. Dermatol. Venereol. 2024, 38, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Souto, E.B.; Jäger, E.; Jäger, A.; Štěpánek, P.; Cano, A.; Viseras, C.; Barbosa, R.d.M.; Chorilli, M.; Zielińska, A.; Severino, P.; et al. Lipid Nanomaterials for Targeted Delivery of Dermocosmetic Ingredients: Advances in Photoprotection and Skin Anti-Aging. Nanomaterials 2022, 12, 377. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.M.; Gonçalves, M.C.; Marques, M.S.; Veiga, F.; Paiva-Santos, A.C.; Pires, P.C. In Vitro Models for Anti-Aging Efficacy Assessment: A Critical Update in Dermocosmetic Research. Cosmetics 2023, 10, 66. [Google Scholar] [CrossRef]
- Draelos, Z.; Bogdanowicz, P.; Saurat, J.H. Top Weapons in Skin Aging and Actives to Target the Consequences of Skin Cell Senescence. J. Eur. Acad. Dermatol. Venereol. 2024, 38, 15–22. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Blume-Peytavi, U.; Kosmadaki, M.; Roó, E.; Vexiau-Robert, D.; Kerob, D.; Goldstein, S.R. Skin, Hair and beyond: The Impact of Menopause. Climacteric 2022, 25, 434–442. [Google Scholar] [CrossRef]
- Masum, M.N.; Yamauchi, K.; Mitsunaga, T. Tyrosinase Inhibitors from Natural and Synthetic Sources as Skin-Lightening Agents. Rev. Agric. Sci. 2019, 7, 41–58. [Google Scholar] [CrossRef]
- Thirion, L.; Piérard-Franchimont, C.; Piérard, G.E. Whitening Effect of a Dermocosmetic Formulation: A Randomized Double-Blind Controlled Study on Melasma. Int. J. Cosmet. Sci. 2006, 28, 263–267. [Google Scholar] [CrossRef]
- Guerrero, D. Dermocosmetic Management of Hyperpigmentations. Ann. Dermatol. Venereol. 2012, 139, S166–S169. [Google Scholar] [CrossRef]
- Hubert, J.; Angelis, A.; Aligiannis, N.; Rosalia, M.; Abedini, A.; Bakiri, A.; Reynaud, R.; Nuzillard, J.M.; Gangloff, S.C.; Skaltsounis, A.L.; et al. In Vitro Dermo-Cosmetic Evaluation of Bark Extracts from Common Temperate Trees. Planta Med. 2016, 82, 1351–1358. [Google Scholar] [CrossRef]
- Goh, C.L.; Noppakun, N.; Micali, G.; Azizan, N.; Boonchai, W.; Chan, Y.; Cheong, W.; Chiu, P.; Etnawati, K.; Gulmatico-Flores, Z.; et al. Meeting the Challenges of Acne Treatment in Asian Patients: A Review of the Role of Dermocosmetics as Adjunctive Therapy. J. Cutan. Aesthet. Surg. 2016, 9, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, D.; Wang, L.; Zhang, R.; Wei, J.; Gao, L.; Zhang, X.; Kerob, D.; Zhang, Z. Efficacy Evaluation of a Dermocosmetic with Skin Repair Properties after Fractional Laser Surgery in Acne Scars. Ski. J. Cutan. Med. 2022, 6, s87. [Google Scholar] [CrossRef]
- Conforti, C.; Giuffrida, R.; Fadda, S.; Fai, A.; Romita, P.; Zalaudek, I.; Dianzani, C. Topical Dermocosmetics and Acne Vulgaris. Dermatol. Ther. 2021, 34, e14436. [Google Scholar] [CrossRef]
- Araviiskaia, E.; Lopez Estebaranz, J.L.; Pincelli, C. Dermocosmetics: Beneficial Adjuncts in the Treatment of Acne Vulgaris. J. Dermatolog. Treat. 2021, 32, 3–10. [Google Scholar] [CrossRef]
- Vanessa, V.V.; Kammal, W.S.L.W.A.; Lai, Z.W.; How, K.N. A Review of Moisturizing Additives for Atopic Dermatitis. Cosmetics 2022, 9, 75. [Google Scholar] [CrossRef]
- Gayraud, F.; Sayag, M.; Jourdan, E. Efficacy and Tolerance Assessment of a New Type of Dermocosmetic in Infants and Children with Moderate Atopic Dermatitis. J. Cosmet. Dermatol. 2015, 14, 107–112. [Google Scholar] [CrossRef]
- Maintz, L.; Bieber, T.; Simpson, H.D.; Demessant-Flavigny, A.L. From Skin Barrier Dysfunction to Systemic Impact of Atopic Dermatitis: Implications for a Precision Approach in Dermocosmetics and Medicine. J. Pers. Med. 2022, 12, 893. [Google Scholar] [CrossRef]
- Mourelle, M.L.; Gómez, C.P.; Legido, J.L. Hydrobiome of Thermal Waters: Potential Use in Dermocosmetics. Cosmetics 2023, 10, 94. [Google Scholar] [CrossRef]
- Olszanecka-Glinianowicz, M. The Subjective Assessment of the Effect and Satisfaction with Dermocosmetics Use by Patients with Skin Disturbances. Postep. Dermatol. I Alergol. 2015, 32, 195–203. [Google Scholar] [CrossRef]
- Kerob, D.; Czermanska, A.; Karamon, E.M.; Moga, A.; Lecerf, G.; Nioré, M.; Le Dantec, G.; Le Floc’H, C.; Tan, J. Dermocosmetic Significantly Reduces the Frequency and Intensity of Facial Skin Intolerability and Sensitivity in Subjects with Skin Intolerant to Skin Care Products and Sensitive Skin. Clin. Cosmet. Investig. Dermatol. 2023, 16, 1787–1794. [Google Scholar] [CrossRef]
- Casetti, F.; Wölfle, U.; Gehring, W.; Schempp, C.M. Dermocosmetics for Dry Skin: A New Role for Botanical Extracts. Ski. Pharmacol. Physiol. 2011, 24, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.F.; Magalhães, W.V.; Di Stasi, L.C. Recent Advances in Herbal-Derived Products with Skin Anti-Aging Properties and Cosmetic Applications. Molecules 2022, 27, 7518. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, L.T.N.; Moon, J.Y.; Lee, Y.C. Insights into Bioactive Peptides in Cosmetics. Cosmetics 2023, 10, 111. [Google Scholar] [CrossRef]
- Waszkielewicz, A.M.; Mirosław, K. Peptides and Their Mechanisms of Action in the Skin. Appl. Sci. 2024, 14, 11495. [Google Scholar] [CrossRef]
- Skibska, A.; Perlikowska, R. Signal Peptides—Promising Ingredients in Cosmetics. Curr. Protein Pept. Sci. 2021, 22, 716–728. [Google Scholar] [CrossRef]
- Lu, W.; Dong, C. Research Progress of Metal Chelating Peptides. Food Health 2022, 4, 19. [Google Scholar] [CrossRef]
- Echavarría, J.A.C.; El Hajj, S.; Irankunda, R.; Selmeczi, K.; Paris, C.; Udenigwe, C.C.; Canabady-Rochelle, L. Screening, Separation and Identification of Metal-Chelating Peptides for Nutritional, Cosmetics and Pharmaceutical Applications. Food Funct. 2024, 15, 3300–3326. [Google Scholar] [CrossRef]
- Veiga, E.; Ferreira, L.; Correia, M.; Pires, P.C.; Hameed, H.; Araújo, A.R.T.S.; Cefali, L.C.; Mazzola, P.G.; Hamishehkar, H.; Veiga, F.; et al. Anti-Aging Peptides for Advanced Skincare: Focus on Nanodelivery Systems. J. Drug Deliv. Sci. Technol. 2023, 89, 105087. [Google Scholar] [CrossRef]
- Ledwoń, P.; Papini, A.M.; Rovero, P.; Latajka, R. Peptides and Peptidomimetics as Inhibitors of Enzymes Involved in Fibrillar Collagen Degradation. Materials 2021, 14, 3217. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Liceaga, A.M. Identification of Chia Seed (Salvia hispanica L.) Peptides with Enzyme Inhibition Activity towards Skin-Aging Enzymes. Amino Acids 2020, 52, 1149–1159. [Google Scholar] [CrossRef]
- Maiman, R. Neurotransmitter-Affecting Peptides. In Cosmeceutical Science in Clinical Practice; CRC Press: Boca Raton, FL, USA, 2023; pp. 83–88. ISBN 9781315165905. [Google Scholar]
- Suthiram, K.T.; Ghazi, T.; Abdul, N.S.; Chuturgoon, A.A. Kojic Acid Induces Oxidative Stress and Anti-Inflammatory Effects in Human Hepatocellular Carcinoma (HepG2) Cells. Toxicon 2023, 232, 107221. [Google Scholar] [CrossRef] [PubMed]
- Phasha, V.; Senabe, J.; Ndzotoyi, P.; Okole, B.; Fouche, G.; Chuturgoon, A. Review on the Use of Kojic Acid—A Skin-Lightening Ingredient. Cosmetics 2022, 9, 64. [Google Scholar] [CrossRef]
- Irfan, M.; Shafeeq, A.; Siddiq, U.; Bashir, F.; Ahmad, T.; Athar, M.; Butt, M.T.; Ullah, S.; Mukhtar, A.; Hussien, M.; et al. A Mechanistic Approach for Toxicity and Risk Assessment of Heavy Metals, Hydroquinone and Microorganisms in Cosmetic Creams. J. Hazard. Mater. 2022, 433, 128806. [Google Scholar] [CrossRef]
- Dymek, M.; Warszycki, D.; Podlewska, S.; Sikora, E. Novel Tripeptides as Tyrosinase Inhibitors: In Silico and In Vitro Approaches. Int. J. Mol. Sci. 2024, 25, 13509. [Google Scholar] [CrossRef]
- Dymek, M.; Sikora, E. Liposomes as Biocompatible and Smart Delivery Systems—The Current State. Adv. Colloid Interface Sci. 2022, 309, 102757. [Google Scholar] [CrossRef]
- Jin, W.; Stehbens, S.J.; Barnard, R.T.; Blaskovich, M.A.T.; Ziora, Z.M. Dysregulation of Tyrosinase Activity: A Potential Link between Skin Disorders and Neurodegeneration. J. Pharm. Pharmacol. 2024, 76, 13–22. [Google Scholar] [CrossRef]
- Imokawa, G. Recent Advances in Characterizing Biological Mechanisms Underlying UV-Induced Wrinkles: A Pivotal Role of Fibrobrast-Derived Elastase. Arch. Dermatol. Res. 2008, 300, 7–20. [Google Scholar] [CrossRef]
- Bayrak, B.B.; Ertik, O.; Onul, N.; Mermer, N.S.; Yanardag, R. Synthesis Novel N,S-Substituted Nitrobutadiene Derivatives: Some Metabolic Enzyme Inhibition Properties and Antioxidant Activities and in Silico ADMET and Molecular Docking Studies. J. Iran. Chem. Soc. 2024, 21, 1299–1315. [Google Scholar] [CrossRef]
- Eun Lee, K.; Bharadwaj, S.; Yadava, U.; Gu Kang, S. Evaluation of Caffeine as Inhibitor against Collagenase, Elastase and Tyrosinase Using in Silico and in Vitro Approach. J. Enzym. Inhib. Med. Chem. 2019, 34, 927–936. [Google Scholar] [CrossRef]
- Handayani, S.; Ni’maturrohmah, D.; Indrianingsih, A.W.; Nisa, K.; Windarsih, A.; Darsih, C.; Sefrienda, A.R.; Suryani, A.E.; Haryanti, S. Molecular Docking Study of Aloesin and Its Derivatives as Potential Antiaging Agents. In Proceedings of the 1st International Conference for Health Research—BRIN (ICHR 2022), Jakarta, Indonesia, 23–24 November 2022; pp. 288–299. [Google Scholar] [CrossRef]
- Dymek, M.; Olechowska, K.; Hąc-Wydro, K.; Sikora, E. Liposomes as Carriers of GHK-Cu Tripeptide for Cosmetic Application. Pharmaceutics 2023, 15, 2485. [Google Scholar] [CrossRef]
- Liu, J.; Zheng, A.; Peng, B.; Xu, Y.; Zhang, N. Size-Dependent Absorption through Stratum Corneum by Drug-Loaded Liposomes. Pharm. Res. 2021, 38, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, L.P. IPC—Isoelectric Point Calculator. Biol. Direct 2016, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Perrotta, R.; Prieto, M.; Alonso, S.; Chiaramoni, N. DSPC Liposomes Improve Transport of L-Cysteine and Reduce Metabolic Activity. Br. Biotechnol. J. 2016, 12, 1–11. [Google Scholar] [CrossRef]
- Kamiloglu, S.; Sari, G.; Ozdal, T.; Capanoglu, E. Guidelines for Cell Viability Assays. Food Front. 2020, 1, 332–349. [Google Scholar] [CrossRef]
- Khalef, L.; Lydia, R.; Filicia, K.; Moussa, B. Cell Viability and Cytotoxicity Assays: Biochemical Elements and Cellular Compartments. Cell Biochem. Funct. 2024, 42, e4007. [Google Scholar] [CrossRef]
- Ochoa-Sánchez, C.; Rodríguez-León, E.; Iñiguez-Palomares, R.; Rodríguez-Beas, C. Brief Comparison of the Efficacy of Cationic and Anionic Liposomes as Nonviral Delivery Systems. ACS Omega 2024, 9, 46664–46678. [Google Scholar] [CrossRef]
- Elias, P.M. Structure and Function of the Stratum Corneum Permeability Barrier. Drug Dev. Res. 1988, 13, 97–105. [Google Scholar] [CrossRef]
- El Maghraby, G.M.; Barry, B.W.; Williams, A.C. Liposomes and Skin: From Drug Delivery to Model Membranes. Eur. J. Pharm. Sci. 2008, 34, 203–222. [Google Scholar] [CrossRef]
- Loy, C.J.; Govindarajan, R. Biology of Skin Pigmentation and Cosmetic Skin Color Control. In Dermatologic, Cosmeceutic, and Cosmetic Development: Therapeutic and Novel Approaches; CRC Press: Boca Raton, FL, USA, 2007; pp. 61–96. ISBN 9780849375903. [Google Scholar]
- Hostynek, J.J.; Dreher, F.; Maibach, H.I. Human Skin Penetration of a Copper Tripeptide in Vitro as a Function of Skin Layer. Inflamm. Res. 2011, 60, 79–86. [Google Scholar] [CrossRef]
- Musakhanian, J.; Osborne, D.W.; Rodier, J.D. Skin Penetration and Permeation Properties of Transcutol in Complex Formulations. AAPS PharmSciTech 2024, 25, 201. [Google Scholar] [CrossRef] [PubMed]
- Csizmazia, E.; Eros, G.; Berkesi, O.; Berkó, S.; Szabó-Révész, P.; Csányi, E. Penetration Enhancer Effect of Sucrose Laurate and Transcutol on Ibuprofen. J. Drug Deliv. Sci. Technol. 2011, 21, 411–415. [Google Scholar] [CrossRef]
- Strusovskaya, O.G.; Poroiskii, S.V.; Strusovskaya, A.G. Chemical Enhancers or Transcutaneous Conductors: Transcutol. Pharm. Chem. J. 2019, 52, 879–884. [Google Scholar] [CrossRef]
- Souto, E.B.; Macedo, A.S.; Dias-Ferreira, J.; Cano, A.; Zielińska, A.; Matos, C.M. Elastic and Ultradeformable Liposomes for Transdermal Delivery of Active Pharmaceutical Ingredients (Apis). Int. J. Mol. Sci. 2021, 22, 9743. [Google Scholar] [CrossRef]
- Abdel-Messih, H.A.; Ishak, R.A.H.; Geneidi, A.S.; Mansour, S. Tailoring Novel Soft Nano-Vesicles ‘Flexosomes’ for Enhanced Transdermal Drug Delivery: Optimization, Characterization and Comprehensive Ex Vivo—In Vivo Evaluation. Int. J. Pharm. 2019, 560, 101–115. [Google Scholar] [CrossRef]
- Aljohani, A.A.; Alanazi, M.A.; Munahhi, L.A.; Hamroon, J.D.; Mortagi, Y.; Qushawy, M.; Soliman, G.M. Binary Ethosomes for the Enhanced Topical Delivery and Antifungal Efficacy of Ketoconazole. OpenNano 2023, 11, 100145. [Google Scholar] [CrossRef]
- Uchida, T.; Kadhum, W.R.; Kanai, S.; Todo, H.; Oshizaka, T.; Sugibayashi, K. Prediction of Skin Permeation by Chemical Compounds Using the Artificial Membrane, Strat-M. Eur. J. Pharm. Sci. 2015, 67, 113–118. [Google Scholar] [CrossRef]
- Thalluri, K.; Kou, B.; Yang, X.; Zaykov, A.N.; Mayer, J.P.; Gelfanov, V.M.; Liu, F.; DiMarchi, R.D. Synthesis of Relaxin-2 and Insulin-like Peptide 5 Enabled by Novel Tethering and Traceless Chemical Excision. J. Pept. Sci. 2017, 23, 455–465. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A Hierarchical Approach to All-Atom Protein Loop Prediction. Proteins Struct. Funct. Genet. 2004, 55, 351–367. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Friesner, R.A.; Xiang, Z.; Honig, B. On the Role of the Crystal Environment in Determining Protein Side-Chain Conformations. J. Mol. Biol. 2002, 320, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A Software Program for PKa Prediction and Protonation State Generation for Drug-like Molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Johnston, R.C.; Yao, K.; Kaplan, Z.; Chelliah, M.; Leswing, K.; Seekins, S.; Watts, S.; Calkins, D.; Chief Elk, J.; Jerome, S.V.; et al. Epik: PKa and Protonation State Prediction through Machine Learning. J. Chem. Theory Comput. 2023, 19, 2380–2388. [Google Scholar] [CrossRef]
- Yang, Y.; Yao, K.; Repasky, M.P.; Leswing, K.; Abel, R.; Shoichet, B.K.; Jerome, S.V. Efficient Exploration of Chemical Space with Docking and Deep Learning. J. Chem. Theory Comput. 2021, 17, 7106–7119. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Kandárová, H.; Hayden, P.; Klausner, M.; Kubilus, J.; Sheasgreen, J. An in Vitro Skin Irritation Test (SIT) Using the EpiDerm Reconstructed Human Epidermal (RHE) Model. J. Vis. Exp. 2009, 29, 1366. [Google Scholar] [CrossRef]
- Malinowska, M.; Miroslaw, B.; Sikora, E.; Ogonowski, J.; Wojtkiewicz, A.M.; Szaleniec, M.; Pasikowska-Piwko, M.; Eris, I. New Lupeol Esters as Active Substances in the Treatment of Skin Damage. PLoS ONE 2019, 14, e0214216. [Google Scholar] [CrossRef]
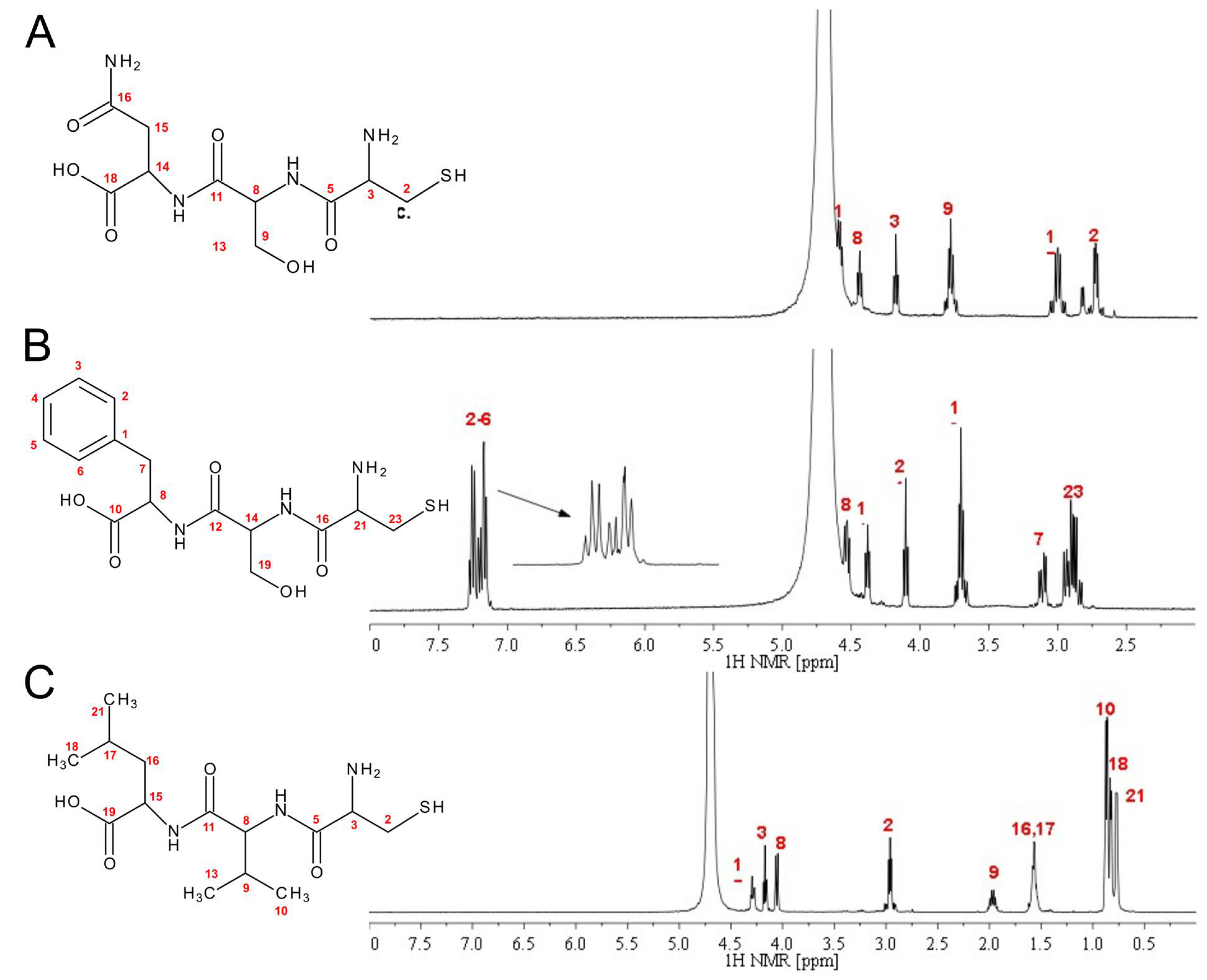
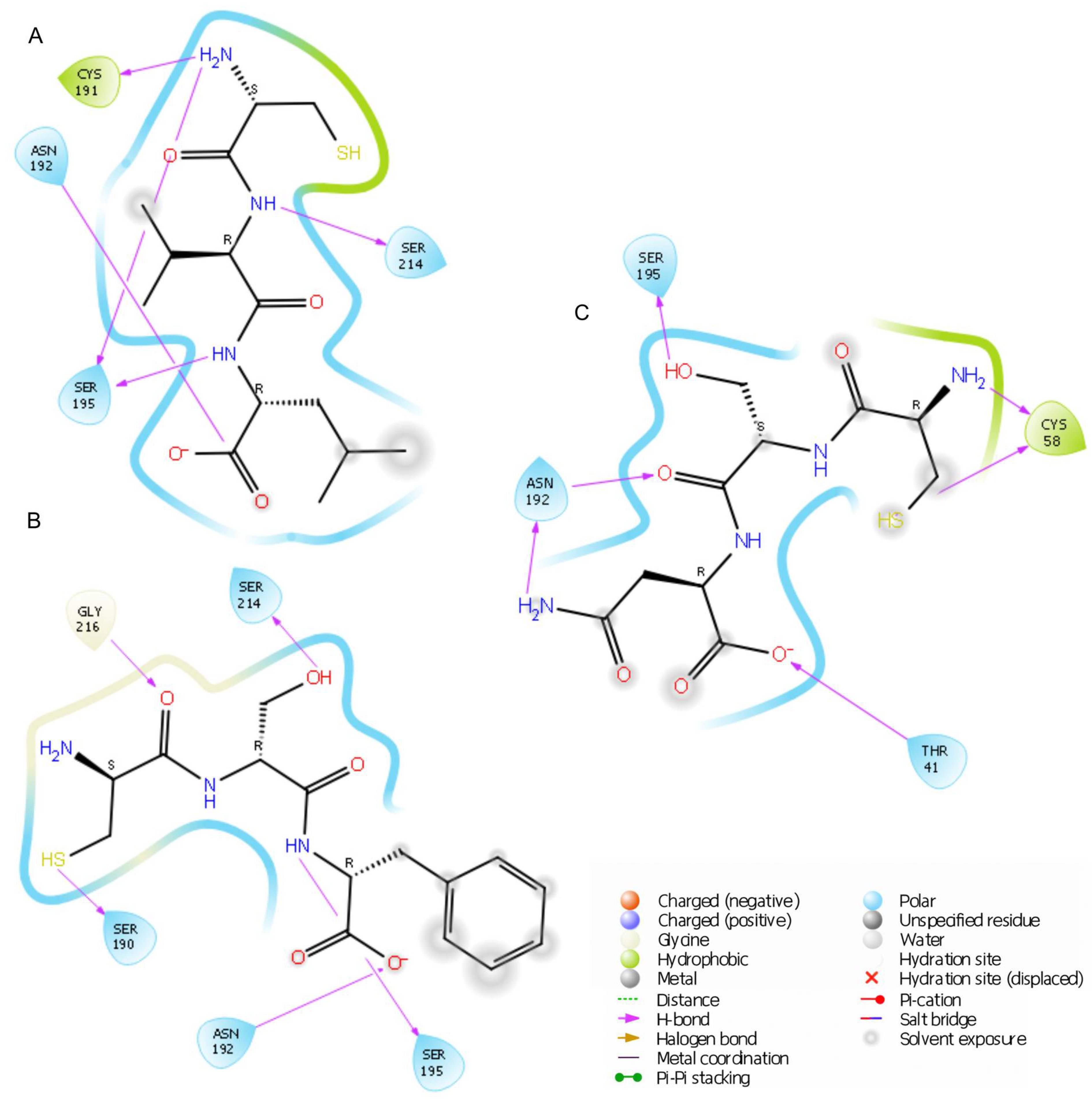

| Peptide | Inhibition Activity [%] ± SD | Docking Score [kcal/mol] |
|---|---|---|
| CSF | 27.6 ± 1.2 | −6.149 |
| CVL | 35.3 ± 8.6 | −4.755 |
| CSN | 28.9 ± 7.5 | −5.304 |
| Liposomes | Zeta Potential ± SD [mV] | EE ± SD [%] | LC ± SD [(mg of Peptide)/(mg of Total Lipids)] |
|---|---|---|---|
| CL | 42.7 ± 2.5 | - | - |
| CSF-CL | 34.2 ± 1.5 | 33.98 ± 6.90 | 0.68 ± 0.14 |
| CVL-CL | 36.0 ± 1.9 | 25.98 ± 9.58 | 0.52 ± 0.19 |
| CSN-CL | 44.7 ± 2.6 | 28.99 ± 5.67 | 0.58 ± 0.11 |
| AL | −38.6 ± 1.3 | - | - |
| CSF-AL | −38.7 ± 2.2 | 29.65 ± 6.91 | 0.59 ± 0.14 |
| CVL-AL | −37.4 ± 1.3 | 21.01 ± 2.49 | 0.42 ± 0.05 |
| CSN-AL | −35.1 ± 6.4 | 26.71 ± 4.08 | 0.53 ± 0.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dymek, M.; García-Celma, M.J.; Escribano-Ferrer, E.; Warszycki, D.; Kaźmierski, S.; Skoczylas, Ł.; Tabaszewska, M.; Sikora, E. Tripeptide-Loaded Liposomes as Multifunctional Components in Topical Formulations. Int. J. Mol. Sci. 2025, 26, 5321. https://doi.org/10.3390/ijms26115321
Dymek M, García-Celma MJ, Escribano-Ferrer E, Warszycki D, Kaźmierski S, Skoczylas Ł, Tabaszewska M, Sikora E. Tripeptide-Loaded Liposomes as Multifunctional Components in Topical Formulations. International Journal of Molecular Sciences. 2025; 26(11):5321. https://doi.org/10.3390/ijms26115321
Chicago/Turabian StyleDymek, Michał, Maria José García-Celma, Elvira Escribano-Ferrer, Dawid Warszycki, Sławomir Kaźmierski, Łukasz Skoczylas, Małgorzata Tabaszewska, and Elżbieta Sikora. 2025. "Tripeptide-Loaded Liposomes as Multifunctional Components in Topical Formulations" International Journal of Molecular Sciences 26, no. 11: 5321. https://doi.org/10.3390/ijms26115321
APA StyleDymek, M., García-Celma, M. J., Escribano-Ferrer, E., Warszycki, D., Kaźmierski, S., Skoczylas, Ł., Tabaszewska, M., & Sikora, E. (2025). Tripeptide-Loaded Liposomes as Multifunctional Components in Topical Formulations. International Journal of Molecular Sciences, 26(11), 5321. https://doi.org/10.3390/ijms26115321







