TBEV NS1 Induces Tissue-Specific Microvascular Endothelial Cell Permeability by Activating the TNF-α Signaling Pathway
Abstract
1. Introduction
2. Results
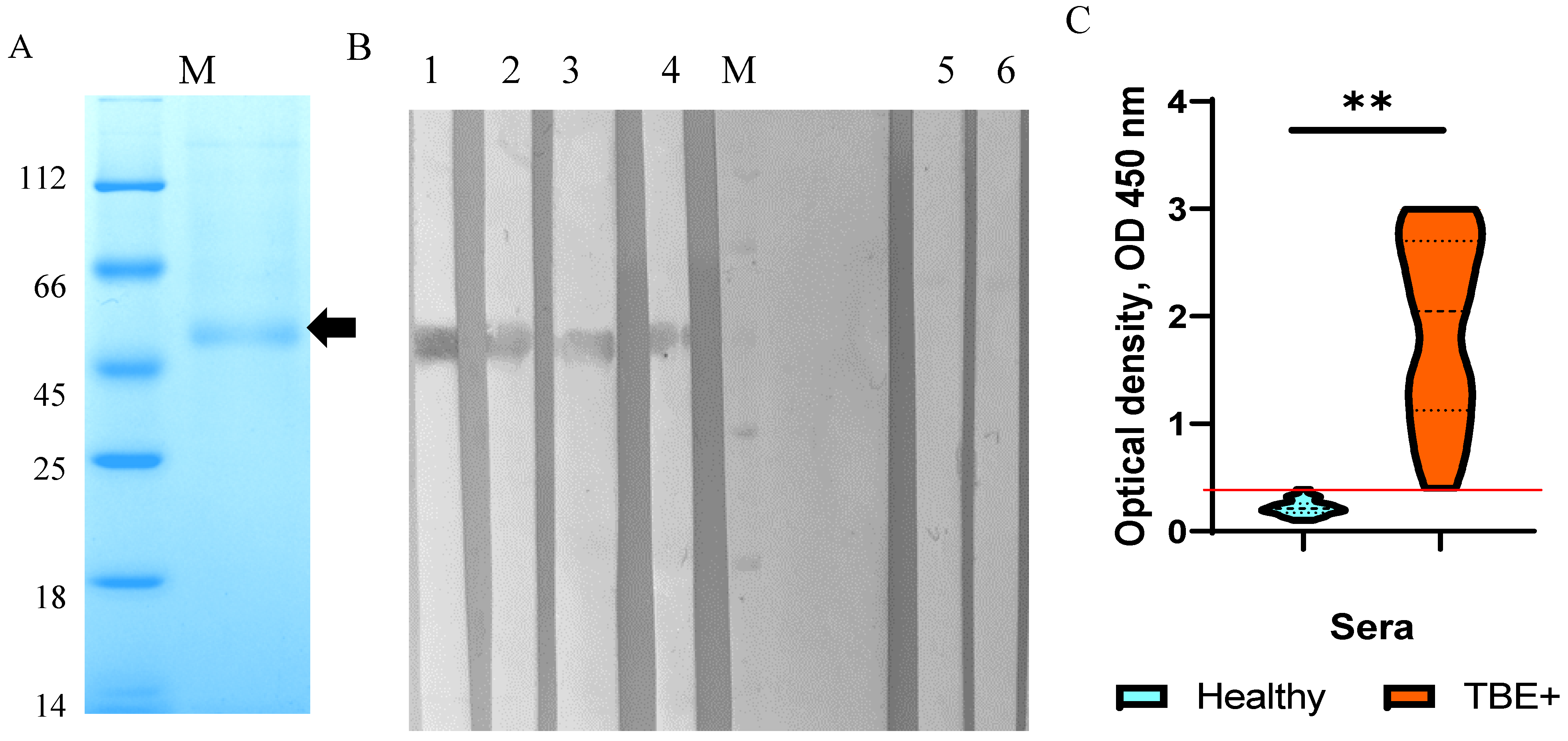
2.1. Production and Purification of the TBEV NS1 Protein
2.2. Characterization and Immunological Properties of the Recombinant TBEV NS1 Protein
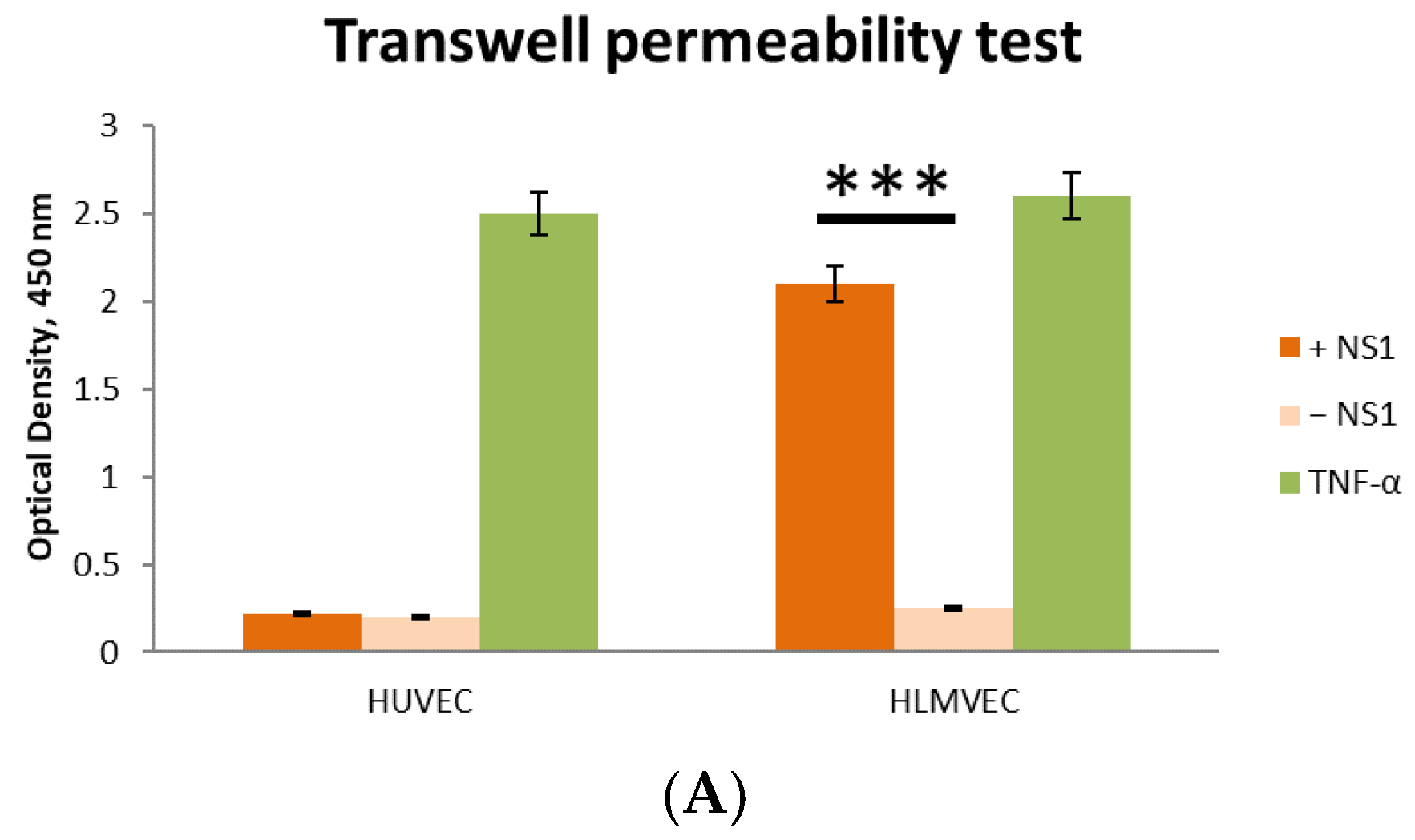
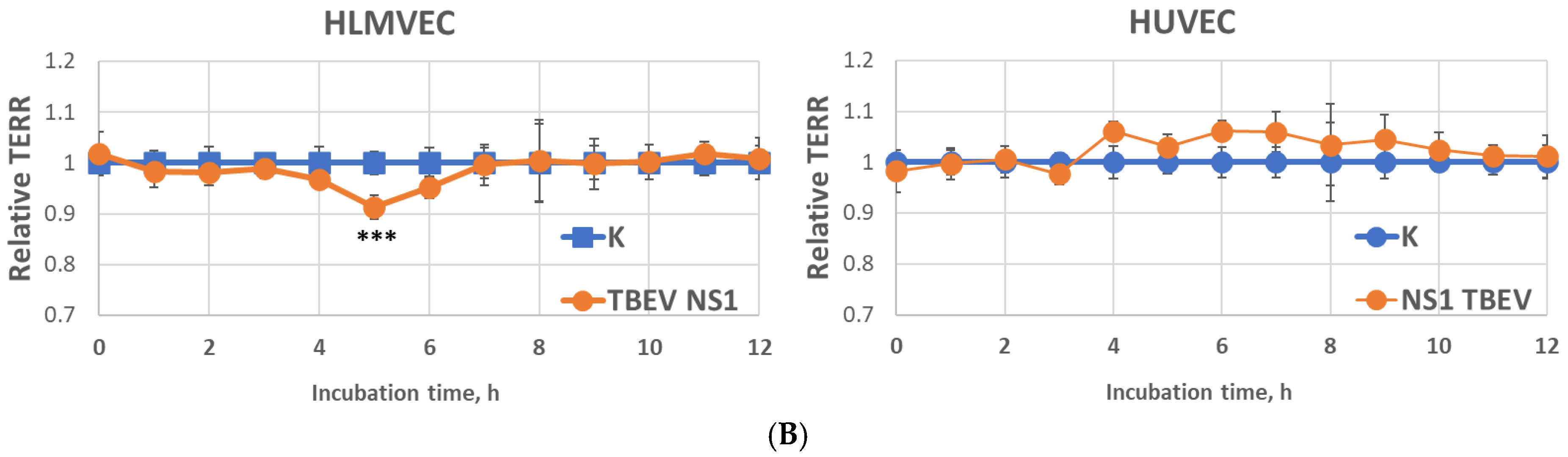
2.3. Effect of Recombinant TBEV NS1 Protein on Human Endothelial Permeability In Vitro
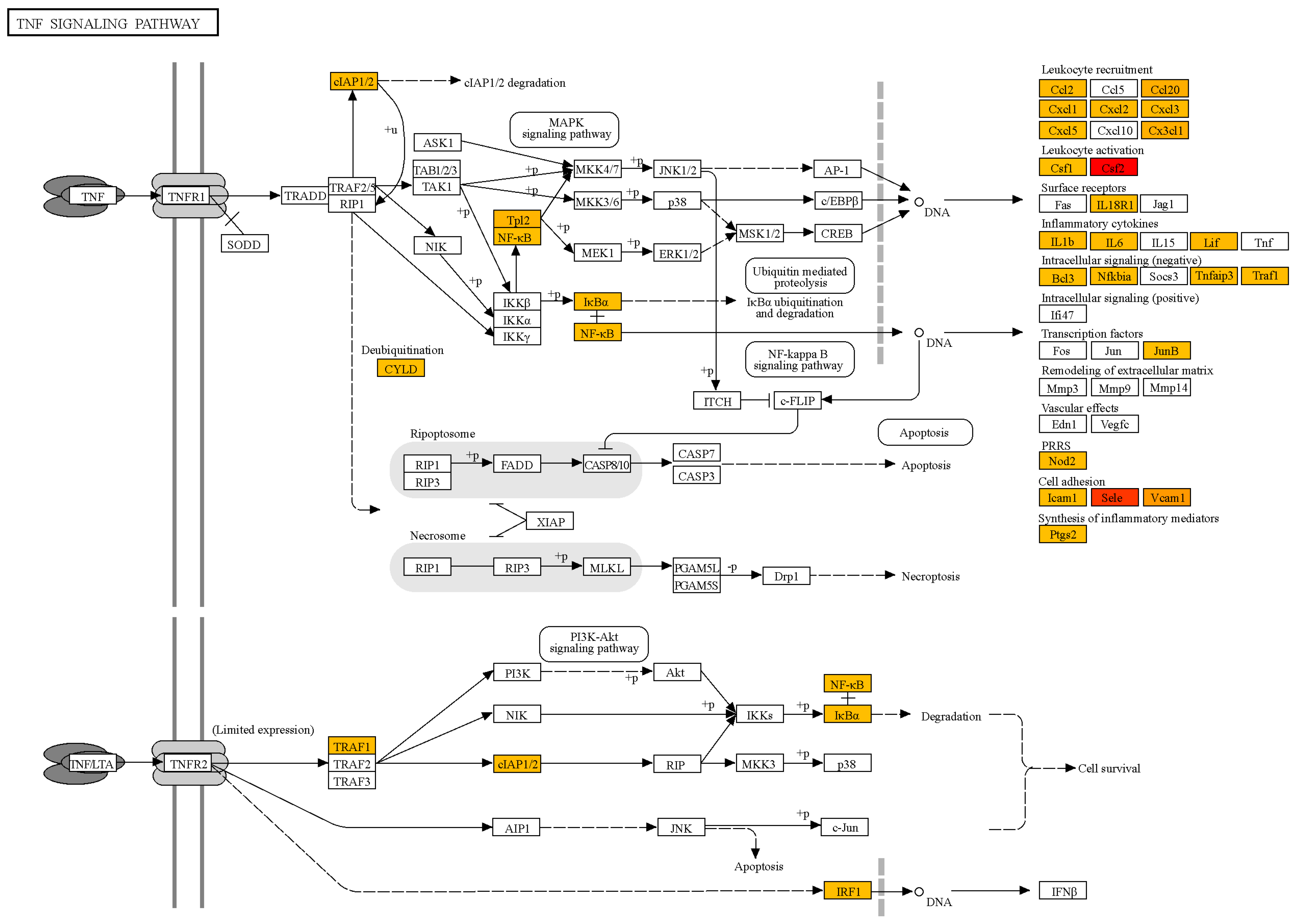
2.4. Transcription Profile of Human Lung Capillary Endothelial Cells Treated with the TBEV NS1 Recombinant Protein
3. Discussion
4. Materials and Methods
4.1. Sera, Plasmids, and Bacterial and Mammalian Cells
4.2. Construction of a Shuttle Plasmid Encoding the TBEV NS1 Protein
4.3. TBEV NS1 Protein Production and Purification
4.4. ELISA and Western Blot Analysis
4.5. Solute Flux Assay
4.6. Endothelial Permeability Test by Measuring Trans-Endothelial Electrical Resistance (TEER)
4.7. RNA-Seq
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TBEV | Tick-borne encephalitis virus |
| TBE | Tick-borne encephalitis |
| NS1 | Non-structural protein 1 of flaviviruses |
| EGL | Endothelial glycocalyx components |
| HUVECs | Human umbilical vein endothelial cells |
| HLMVECs | Human lung microvascular endothelial cells |
| sNS1 | secreted form of NS1 |
| GPI | Glycosyl-phosphatidylinositol |
References
- Ruzek, D.; Avšič Županc, T.; Borde, J.; Chrdle, A.; Eyer, L.; Karganova, G.; Kholodilov, I.; Knap, N.; Kozlovskaya, L.; Matveev, A.; et al. Tick-Borne Encephalitis in Europe and Russia: Review of Pathogenesis, Clinical Features, Therapy, and Vaccines. Antivir. Res. 2019, 164, 23–51. [Google Scholar] [CrossRef] [PubMed]
- Chiffi, G.; Grandgirard, D.; Leib, S.L.; Chrdle, A.; Růžek, D. Tick-Borne Encephalitis: A Comprehensive Review of the Epidemiology, Virology, and Clinical Picture. Rev. Med. Virol. 2023, 33, e2470. [Google Scholar] [CrossRef] [PubMed]
- Ličková, M.; Fumačová Havlíková, S.; Sláviková, M.; Klempa, B. Alimentary Infections by Tick-Borne Encephalitis Virus. Viruses 2022, 14, 56. [Google Scholar] [CrossRef] [PubMed]
- Michelitsch, A.; Wernike, K.; Klaus, C.; Dobler, G.; Beer, M. Exploring the Reservoir Hosts of Tick-Borne Encephalitis Virus. Viruses 2019, 11, 669. [Google Scholar] [CrossRef]
- Mittova, V.; Tsetskhladze, Z.R.; Motsonelidze, C.; Palumbo, R.; Vicidomini, C.; Roviello, G.N. Tick-Borne Encephalitis Virus (TBEV): Epidemiology, Diagnosis, Therapeutic Approaches and Some Molecular Aspects—An Updated Review. Microbiol. Res. 2024, 15, 2619–2649. [Google Scholar] [CrossRef]
- Formanová, P.; Černý, J.; Bolfíková, B.Č.; Valdés, J.J.; Kozlova, I.; Dzhioev, Y.; Růžek, D. Full Genome Sequences and Molecular Characterization of Tick-Borne Encephalitis Virus Strains Isolated from Human Patients. Ticks Tick-Borne Dis. 2015, 6, 38–46. [Google Scholar] [CrossRef]
- Shiryaev, S.A.; Strongin, A.Y. Structural and Functional Parameters of the Flaviviral Protease: A Promising Antiviral Drug Target. Future Virol. 2010, 5, 593–606. [Google Scholar] [CrossRef]
- Assenberg, R.; Mastrangelo, E.; Walter, T.S.; Verma, A.; Milani, M.; Owens, R.J.; Stuart, D.I.; Grimes, J.M.; Mancini, E.J. Crystal Structure of a Novel Conformational State of the Flavivirus NS3 Protein: Implications for Polyprotein Processing and Viral Replication. J. Virol. 2009, 83, 12895–12906. [Google Scholar] [CrossRef]
- Perera, D.R.; Ranadeva, N.D.; Sirisena, K.; Wijesinghe, K.J. Roles of NS1 Protein in Flavivirus Pathogenesis. ACS Infect. Dis. 2024, 10, 20–56. [Google Scholar] [CrossRef]
- Fisher, R.; Lustig, Y.; Sklan, E.H.; Schwartz, E. The Role of NS1 Protein in the Diagnosis of Flavivirus Infections. Viruses 2023, 15, 572. [Google Scholar] [CrossRef]
- Somnuke, P.; Hauhart, R.E.; Atkinson, J.P.; Diamond, M.S.; Avirutnan, P. N-Linked Glycosylation of Dengue Virus NS1 Protein Modulates Secretion, Cell-Surface Expression, Hexamer Stability, and Interactions with Human Complement. Virology 2011, 413, 253–264. [Google Scholar] [CrossRef]
- Modhiran, N.; Watterson, D.; Muller, D.A.; Panetta, A.K.; Sester, D.P.; Liu, L.; Hume, D.A.; Stacey, K.J.; Young, P.R. Dengue Virus NS1 Protein Activates Cells via Toll-like Receptor 4 and Disrupts Endothelial Cell Monolayer Integrity. Sci. Transl. Med 2015, 7, 304ra142. [Google Scholar] [CrossRef] [PubMed]
- Avirutnan, P.; Fuchs, A.; Hauhart, R.E.; Somnuke, P.; Youn, S.; Diamond, M.S.; Atkinson, J.P. Antagonism of the Complement Component C4 by Flavivirus Nonstructural Protein NS1. J. Exp. Med. 2010, 207, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.N.; Silva, E.M.; Barbosa, A.S.; Mohana-Borges, R. The Complement System in Flavivirus Infections. Front. Microbiol. 2017, 8, 213. [Google Scholar] [CrossRef]
- Fan, J.; Liu, Y.; Yuan, Z. Critical Role of Dengue Virus NS1 Protein in Viral Replication. Virol. Sin. 2014, 29, 162–169. [Google Scholar] [CrossRef]
- Rastogi, M.; Sharma, N.; Singh, S.K. Flavivirus NS1: A Multifaceted Enigmatic Viral Protein. Virol. J. 2016, 13, 131. [Google Scholar] [CrossRef] [PubMed]
- Akey, D.L.; Brown, W.C.; Dutta, S.; Konwerski, J.; Jose, J.; Jurkiw, T.J.; DelProposto, J.; Ogata, C.M.; Skiniotis, G.; Kuhn, R.J.; et al. Flavivirus NS1 Crystal Structures Reveal a Surface for Membrane Association and Regions of Interaction with the Immune System. Science 2014, 343, 881–885. [Google Scholar] [CrossRef]
- Noisakran, S.; Dechtawewat, T.; Avirutnan, P.; Kinoshita, T.; Siripanyaphinyo, U.; Puttikhunt, C.; Kasinrerk, W.; Malasit, P.; Sittisombut, N. Association of Dengue Virus NS1 Protein with Lipid Rafts. J. Gen. Virol. 2008, 89, 2492–2500. [Google Scholar] [CrossRef]
- Falconar, A.K. The Dengue Virus Nonstructural-1 Protein (NS1) Generates Antibodies to Common Epitopes on Human Blood Clotting, Integrin/Adhesin Proteins and Binds to Human Endothelial Cells: Potential Implications in Haemorrhagic Fever Pathogenesis. Arch. Virol. 1997, 142, 897–916. [Google Scholar] [CrossRef]
- Chang, H.-H.; Shyu, H.-F.; Wang, Y.-M.; Sun, D.-S.; Shyu, R.-H.; Tang, S.-S.; Huang, Y.-S. Facilitation of Cell Adhesion by Immobilized Dengue Viral Nonstructural Protein 1 (NS1): Arginine-Glycine-Aspartic Acid Structural Mimicry within the Dengue Viral NS1 Antigen. J. Infect. Dis. 2002, 186, 743–751. [Google Scholar] [CrossRef]
- Puerta-Guardo, H.; Glasner, D.R.; Espinosa, D.A.; Biering, S.B.; Patana, M.; Ratnasiri, K.; Wang, C.; Beatty, P.R.; Harris, E. Flavivirus NS1 Triggers Tissue-Specific Vascular Endothelial Dysfunction Reflecting Disease Tropism. Cell Rep. 2019, 26, 1598–1613.e8. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Puerta-Guardo, H.; Biering, S.B.; Glasner, D.R.; Tran, E.B.; Patana, M.; Gomberg, T.A.; Malvar, C.; Lo, N.T.N.; Espinosa, D.A.; et al. Endocytosis of Flavivirus NS1 Is Required for NS1-Mediated Endothelial Hyperpermeability and Is Abolished by a Single N-Glycosylation Site Mutation. PLoS Pathog. 2019, 15, e1007938. [Google Scholar] [CrossRef] [PubMed]
- Mandl, C.W. Steps of the Tick-Borne Encephalitis Virus Replication Cycle That Affect Neuropathogenesis. Virus. Res. 2005, 111, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Andrey, M.; Yana, K.; Olga, G.; Bogdana, K.; Sergey, T.; Lyudmila, E.; Nina, T. Tick-Borne Encephalitis Nonstructural Protein NS1 Expressed in E. Coli Retains Immunological Properties of the Native Protein. Protein Expr. Purif. 2022, 191, 106031. [Google Scholar] [CrossRef]
- Matveev, A.; Pyankov, O.; Khlusevich, Y.; Tyazhelkova, O.; Emelyanova, L.; Timofeeva, A.; Shipovalov, A.; Chechushkov, A.; Zaitseva, N.; Kudrov, G.; et al. Antibodies Capable of Enhancing SARS-CoV-2 Infection Can Circulate in Patients with Severe COVID-19. Int. J. Mol. Sci. 2023, 24, 10799. [Google Scholar] [CrossRef]
- Igolkina, Y.; Rar, V.; Krasnova, E.; Filimonova, E.; Tikunov, A.; Epikhina, T.; Tikunova, N. Occurrence and Clinical Manifestations of Tick-Borne Rickettsioses in Western Siberia: First Russian Cases of Rickettsia Aeschlimannii and Rickettsia Slovaca Infections. Ticks Tick-Borne Dis. 2022, 13, 101927. [Google Scholar] [CrossRef]
- Kravchuk, B.I.; Khlusevich, Y.A.; Chicherina, G.S.; Yakimenko, V.V.; Krasnova, E.I.; Tikunova, N.N.; Matveev, A.L. Cross-Reactive Antibodies to the NS1 Protein of Omsk Hemorrhagic Fever Virus Are Absent in the Sera of Patients with Tick-Borne Encephalitis. Viruses 2024, 16, 1032. [Google Scholar] [CrossRef]
- Labuda, M.; Austyn, J.M.; Zuffova, E.; Kozuch, O.; Fuchsberger, N.; Lysy, J.; Nuttall, P.A. Importance of Localized Skin Infection in Tick-Borne Encephalitis Virus Transmission. Virology 1996, 219, 357–366. [Google Scholar] [CrossRef]
- Prokopowicz, D.; Bobrowska, E.; Bobrowski, M.; Grzeszczuk, A. Prevalence of Antibodies against Tick-Borne Encephalitis among Residents of North-Eastern Poland. Scand. J. Infect. Dis. 1995, 27, 15–16. [Google Scholar] [CrossRef]
- Hawkins, B.T.; Davis, T.P. The Blood-Brain Barrier/Neurovascular Unit in Health and Disease. Pharmacol. Rev. 2005, 57, 173–185. [Google Scholar] [CrossRef]
- Oehler, E.; Watrin, L.; Larre, P.; Leparc-Goffart, I.; Lastere, S.; Valour, F.; Baudouin, L.; Mallet, H.; Musso, D.; Ghawche, F. Zika Virus Infection Complicated by Guillain-Barre Syndrome—Case Report, French Polynesia, December 2013. Euro Surveill. 2014, 19, 20720. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Nunez, N.V.; Gaudin, R. A Viral Journey to the Brain: Current Considerations and Future Developments. PLoS Pathog. 2020, 16, e1008434. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.T.; St John, A.L. Japanese Encephalitis Virus and Its Mechanisms of Neuroinvasion. PLoS Pathog. 2020, 16, e1008260. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, G. Immune Response and Blood-Brain Barrier Dysfunction during Viral Neuroinvasion. Innate Immun. 2021, 27, 109–117. [Google Scholar] [CrossRef]
- Watrin, L.; Ghawché, F.; Larre, P.; Neau, J.-P.; Mathis, S.; Fournier, E. Guillain-Barré Syndrome (42 Cases) Occurring During a Zika Virus Outbreak in French Polynesia. Medicine 2016, 95, e3257. [Google Scholar] [CrossRef]
- Morrison, T.E.; Diamond, M.S. Animal Models of Zika Virus Infection, Pathogenesis, and Immunity. J. Virol. 2017, 91, e00009-17. [Google Scholar] [CrossRef]
- Neal, J.W. Flaviviruses Are Neurotropic, but How Do They Invade the CNS? J. Infect. 2014, 69, 203–215. [Google Scholar] [CrossRef]
- Roe, K.; Kumar, M.; Lum, S.; Orillo, B.; Nerurkar, V.R.; Verma, S. West Nile Virus-Induced Disruption of the Blood–Brain Barrier in Mice Is Characterized by the Degradation of the Junctional Complex Proteins and Increase in Multiple Matrix Metalloproteinases. J. Gen. Virol. 2012, 93, 1193–1203. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Yu, L.; Cao, S.; Wang, K.; Yuan, J.; Wang, C.; Wang, K.; Cui, M.; Fu, Z.F. Viral Infection of the Central Nervous System and Neuroinflammation Precede Blood-Brain Barrier Disruption during Japanese Encephalitis Virus Infection. J. Virol. 2015, 89, 5602–5614. [Google Scholar] [CrossRef]
- Růžek, D.; Salát, J.; Singh, S.K.; Kopecký, J. Breakdown of the Blood-Brain Barrier during Tick-Borne Encephalitis in Mice Is Not Dependent on CD8+ T-Cells. PLoS ONE 2011, 6, e20472. [Google Scholar] [CrossRef]
- Weber, E.; Finsterbusch, K.; Lindquist, R.; Nair, S.; Lienenklaus, S.; Gekara, N.O.; Janik, D.; Weiss, S.; Kalinke, U.; Överby, A.K.; et al. Type I Interferon Protects Mice from Fatal Neurotropic Infection with Langat Virus by Systemic and Local Antiviral Responses. J. Virol. 2014, 88, 12202–12212. [Google Scholar] [CrossRef] [PubMed]
- Kurhade, C.; Zegenhagen, L.; Weber, E.; Nair, S.; Michaelsen-Preusse, K.; Spanier, J.; Gekara, N.O.; Kröger, A.; Överby, A.K. Type I Interferon Response in Olfactory Bulb, the Site of Tick-Borne Flavivirus Accumulation, Is Primarily Regulated by IPS-1. J. Neuroinflamm. 2016, 13, 22. [Google Scholar] [CrossRef] [PubMed]
- Palus, M.; Vancova, M.; Sirmarova, J.; Elsterova, J.; Perner, J.; Ruzek, D. Tick-Borne Encephalitis Virus Infects Human Brain Microvascular Endothelial Cells without Compromising Blood-Brain Barrier Integrity. Virology 2017, 507, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Camarão, A.A.R.; Gern, O.L.; Stegmann, F.; Mulenge, F.; Costa, B.; Saremi, B.; Jung, K.; Lepenies, B.; Kalinke, U.; Steffen, I. Secreted NS1 Proteins of Tick-Borne Encephalitis Virus and West Nile Virus Block Dendritic Cell Activation and Effector Functions. Microbiol. Spectr. 2023, 11, e02192-23. [Google Scholar] [CrossRef]
| № | Monoclonal Antibodies | KD Native TBEV NS1 | KD Recombinant TBEV NS1 |
|---|---|---|---|
| 1 | NS1-1.3 | 8.51 ± 0.01 nM | 8.21 ± 0.02 nM |
| 2 | NS1-2.299 | 0.20 ± 0.01 nM | 0.22 ± 0.01 nM |
| 3 | NS1-2.290 | 0.35 ± 0.01 nM | 0.36 ± 0.01 nM |
| 4 | NS1-2.44 | 0.68 ± 0.14 nM | 0.67 ± 0.11 nM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khlusevich, Y.; Kravchuk, B.; Kechin, A.; Stepanova, A.; Emelyanova, L.; Khachatryan, S.; Tikunova, N.; Matveev, A. TBEV NS1 Induces Tissue-Specific Microvascular Endothelial Cell Permeability by Activating the TNF-α Signaling Pathway. Int. J. Mol. Sci. 2025, 26, 5311. https://doi.org/10.3390/ijms26115311
Khlusevich Y, Kravchuk B, Kechin A, Stepanova A, Emelyanova L, Khachatryan S, Tikunova N, Matveev A. TBEV NS1 Induces Tissue-Specific Microvascular Endothelial Cell Permeability by Activating the TNF-α Signaling Pathway. International Journal of Molecular Sciences. 2025; 26(11):5311. https://doi.org/10.3390/ijms26115311
Chicago/Turabian StyleKhlusevich, Yana, Bogdana Kravchuk, Andrey Kechin, Alena Stepanova, Lyudmila Emelyanova, Sargis Khachatryan, Nina Tikunova, and Andrey Matveev. 2025. "TBEV NS1 Induces Tissue-Specific Microvascular Endothelial Cell Permeability by Activating the TNF-α Signaling Pathway" International Journal of Molecular Sciences 26, no. 11: 5311. https://doi.org/10.3390/ijms26115311
APA StyleKhlusevich, Y., Kravchuk, B., Kechin, A., Stepanova, A., Emelyanova, L., Khachatryan, S., Tikunova, N., & Matveev, A. (2025). TBEV NS1 Induces Tissue-Specific Microvascular Endothelial Cell Permeability by Activating the TNF-α Signaling Pathway. International Journal of Molecular Sciences, 26(11), 5311. https://doi.org/10.3390/ijms26115311






