Lysine Acetyltransferase 8: A Target for Natural Compounds in Cancer Therapy
Abstract
1. Introduction
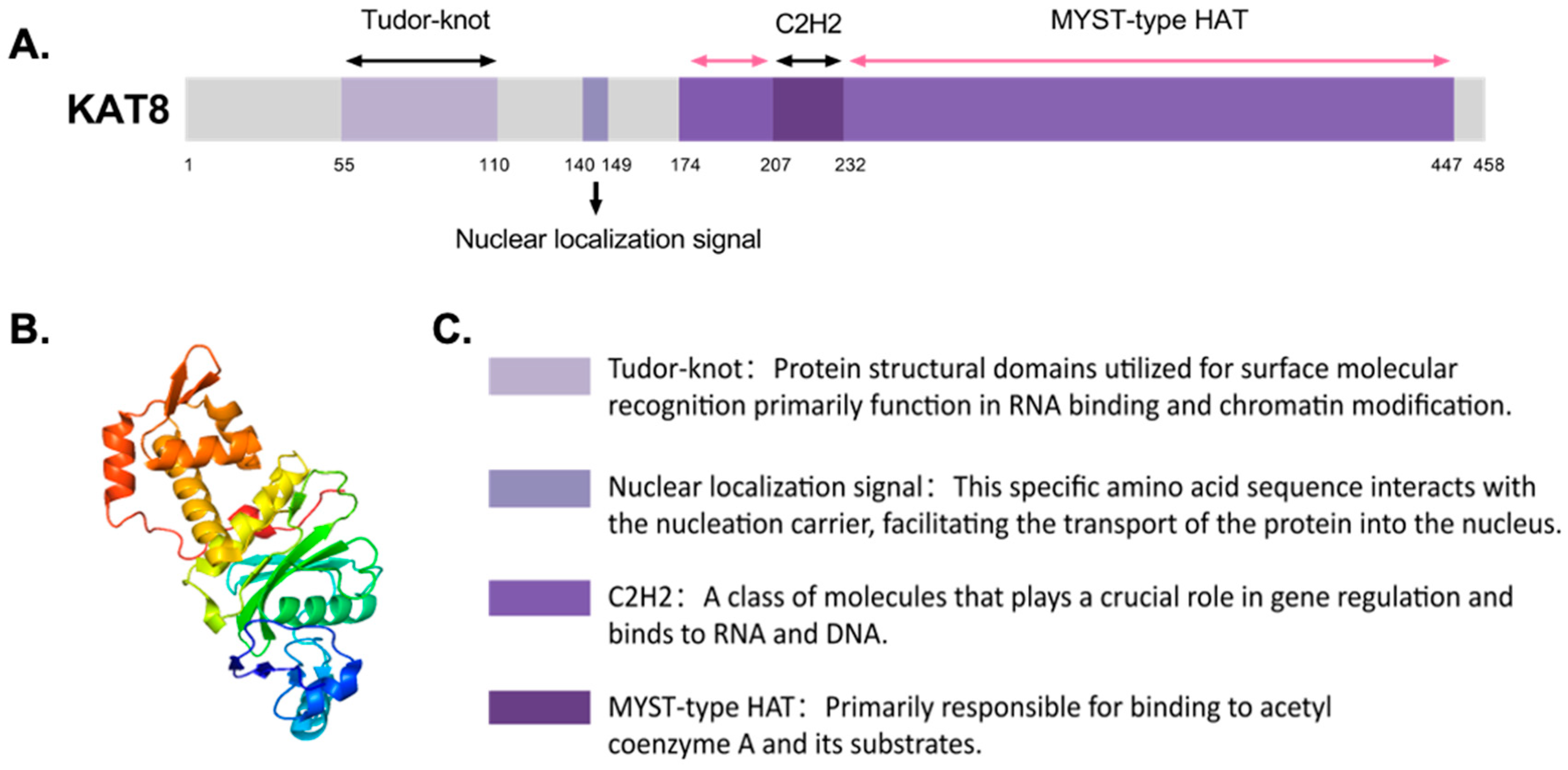
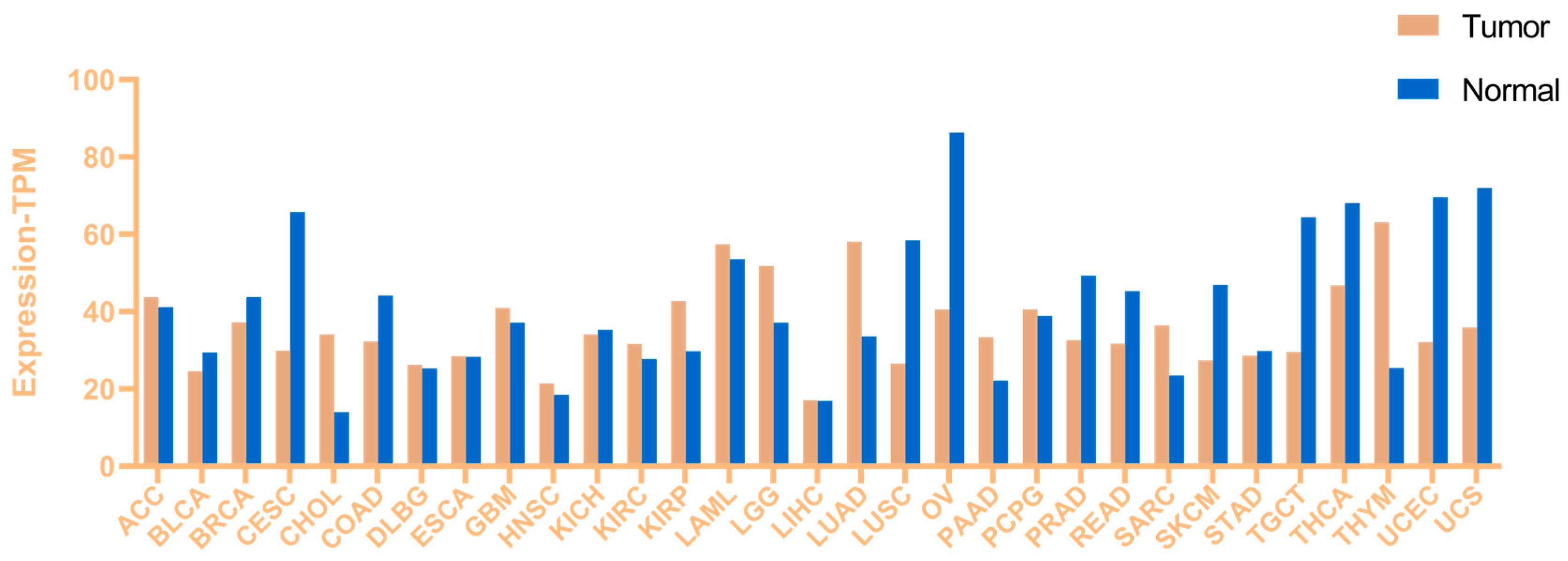
2. Aberrant Expression of Lysine Acetyltransferase 8 (KAT8) in Cancer
3. The Role of Lysine Acetyltransferase 8 (KAT8) in Cancer Progression
3.1. Lysine Acetyltransferase 8 (KAT8) and Apoptosis
3.2. Lysine Acetyltransferase 8 (KAT8) and Cell Proliferation
3.3. Lysine Acetyltransferase 8 (KAT8) and DeoxyriboNucleic Acid Damage and Repair
3.4. Lysine Acetyltransferase 8 (KAT8) and Autophagy
3.5. Lysine Acetyltransferase 8 (KAT8) and Invasion/Migration
4. Small-Molecule Drugs Targeting Lysine Acetyltransferase 8 (KAT8)
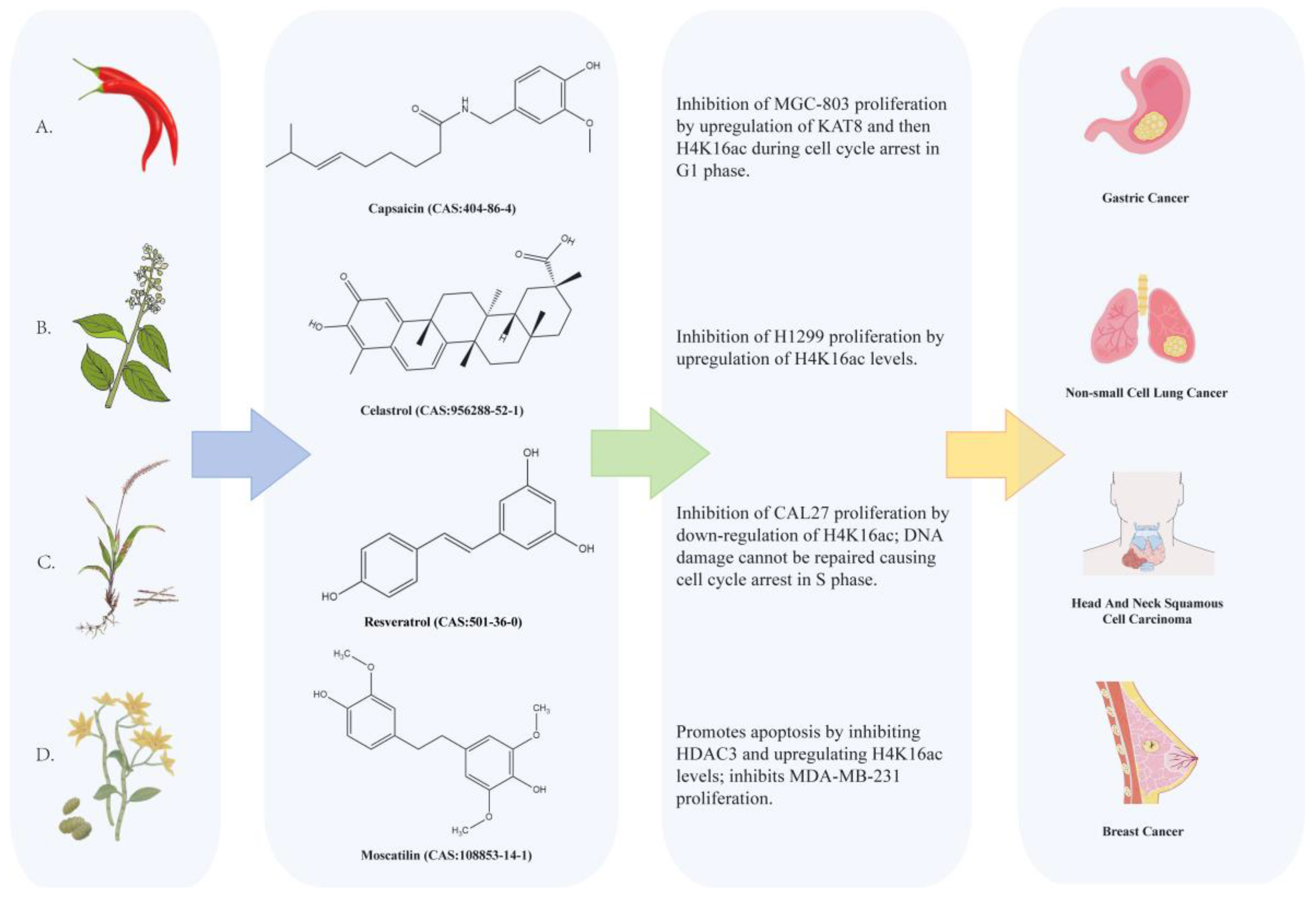
4.1. Natural Compounds Targeting Lysine Acetyltransferase 8 (KAT8)
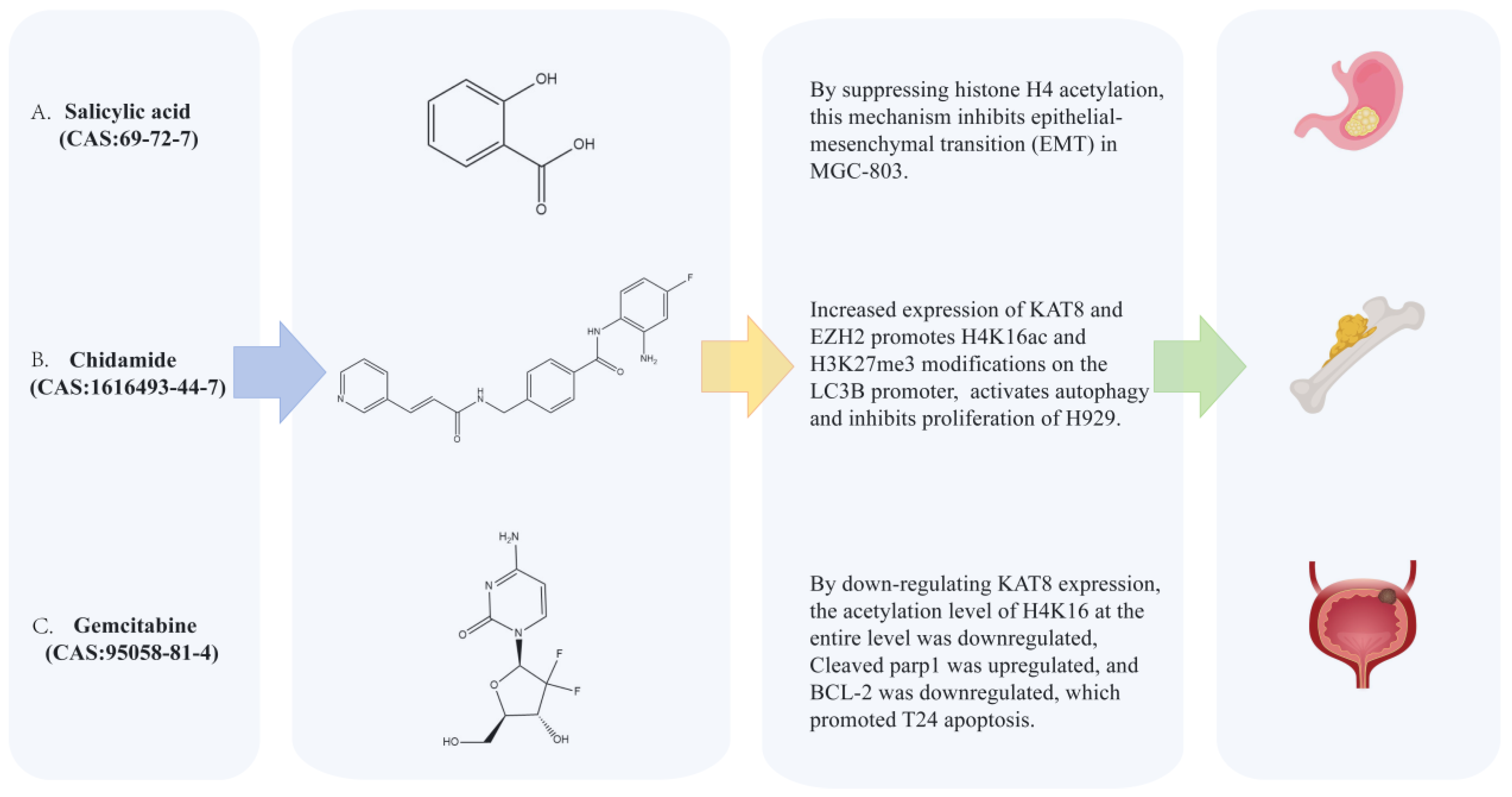
4.2. Synthetic Small-Molecule Compounds Targeting Lysine Acetyltransferase 8 (KAT8)
4.3. Metal-Based Agents Targeting Lysine Acetyltransferase 8 (KAT8)
4.4. Non-Coding RNA
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Kiri, S.; Ryba, T. Cancer, Metastasis, and the Epigenome. Mol. Cancer 2024, 23, 154. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Tang, X.; Wang, Y. Emerging Strategies to Investigate the Biology of Early Cancer. Nat. Rev. Cancer 2024, 24, 850–866. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Y.; Li, Y.; Li, S.; Liu, J.; Yang, X.; Xia, G.; Wang, G. Natural Products and Derivatives for Breast Cancer Treatment: From Drug Discovery to Molecular Mechanism. Phytomedicine 2024, 129, 155600. [Google Scholar] [CrossRef] [PubMed]
- Rea, S.; Xouri, G.; Akhtar, A. Males Absent on the First (MOF): From Flies to Humans. Oncogene 2007, 26, 5385–5394. [Google Scholar] [CrossRef]
- Xuan, H.; Xu, L.; Li, K.; Xuan, F.; Xu, T.; Wen, H.; Shi, X. Hotspot Cancer Mutation Impairs KAT8-Mediated Nucleosomal Histone Acetylation. J. Mol. Biol. 2024, 436, 168413. [Google Scholar] [CrossRef]
- An, H.-M.; Dai, Y.-F.; Zhu, J.; Liu, W.; Wang, X.-P. MYST Family Histone Acetyltransferases Regulate Reproductive Diapause Initiation. Int. J. Biol. Macromol. 2024, 256, 128269. [Google Scholar] [CrossRef]
- Chen, C.; Pawley, S.B.; Cote, J.M.; Carter, J.; Wang, M.; Xu, C.; Buesking, A.W. Identification of Triazolyl KAT6 Inhibitors via a Templated Fragment Approach. Bioorganic Med. Chem. Lett. 2024, 113, 129948. [Google Scholar] [CrossRef]
- Yang, Y.; Han, X.; Guan, J.; Li, X. Regulation and Function of Histone Acetyltransferase MOF. Front. Med. 2014, 8, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Seyfferth, J.; Lucci, J.; Gilsbach, R.; Preissl, S.; Böttinger, L.; Mårtensson, C.U.; Panhale, A.; Stehle, T.; Kretz, O.; et al. MOF Acetyl Transferase Regulates Transcription and Respiration in Mitochondria. Cell 2016, 167, 722–738.e23. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, D.-L.; Liu, Y.; Chen, S.; Sun, F.-L. Histone Acetyltransferase hMOF Promotes S Phase Entry and Tumorigenesis in Lung Cancer. Cell. Signal. 2013, 25, 1689–1698. [Google Scholar] [CrossRef]
- Yoo, L.; Mendoza, D.; Richard, A.J.; Stephens, J.M. KAT8 beyond Acetylation: A Survey of Its Epigenetic Regulation, Genetic Variability, and Implications for Human Health. Genes 2024, 15, 639. [Google Scholar] [CrossRef] [PubMed]
- Karoutas, A.; Szymanski, W.; Rausch, T.; Guhathakurta, S.; Rog-Zielinska, E.A.; Peyronnet, R.; Seyfferth, J.; Chen, H.-R.; De Leeuw, R.; Herquel, B.; et al. The NSL Complex Maintains Nuclear Architecture Stability via Lamin A/C Acetylation. Nat. Cell Biol. 2019, 21, 1248–1260. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Liu, W.; Sun, N.; Wu, Y.; Song, H.; Wang, C.; Wang, S.; Zou, R.; Lin, L.; Zeng, K.; et al. MOF Upregulates the Estrogen Receptor α Signaling Pathway by Its Acetylase Activity in Hepatocellular Carcinoma. Cancer Sci. 2021, 112, 1865–1877. [Google Scholar] [CrossRef]
- Nie, Q.; Huan, X.; Kang, J.; Yin, J.; Zhao, J.; Li, Y.; Zhang, Z. MG149 Inhibits MOF-Mediated P53 Acetylation to Attenuate X-Ray Radiation-Induced Apoptosis in H9c2 Cells. Radiat. Res. 2022, 198, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Zhou, L.; Li, T.; Lin, Y.; Zhang, R.; Zheng, X.; Zeng, C.; Zheng, L.; Zhong, L.; Huang, X.; et al. Targeting the KAT8/YEATS4 Axis Represses Tumor Growth and Increases Cisplatin Sensitivity in Bladder Cancer. Adv. Sci. 2024, 11, 2310146. [Google Scholar] [CrossRef]
- Xian, Q.; Song, Y.; Gui, C.; Zhou, Y. Mechanistic Insights into Genomic Structure and Functions of a Novel Oncogene YEATS4. Front. Cell Dev. Biol. 2023, 11, 1192139. [Google Scholar] [CrossRef]
- Hariharan, S.; Dharmaraj, S. Selenium and Selenoproteins: It’s Role in Regulation of Inflammation. Inflammopharmacology 2020, 28, 667–695. [Google Scholar] [CrossRef]
- Zhu, Z.; Nie, G.; Peng, X.; Zhan, X.; Ding, D. KAT8 Catalyzes the Acetylation of SEPP1 at Lysine 247/249 and Modulates the Activity of CD8+ T Cells via LRP8 to Promote Anti-Tumor Immunity in Pancreatic Cancer. Cell Biosci. 2025, 15, 24. [Google Scholar] [CrossRef]
- Dong, Z.; Zou, J.; Li, J.; Pang, Y.; Liu, Y.; Deng, C.; Chen, F.; Cui, H. MYST1/KAT8 Contributes to Tumor Progression by Activating EGFR Signaling in Glioblastoma Cells. Cancer Med. 2019, 8, 7793–7808. [Google Scholar] [CrossRef]
- Liu, Y.; Du, J.; Liu, X.; Wang, L.; Han, Y.; Huang, C.; Liang, R.; Zheng, F.; Shi, G.; Li, B. MG149 Inhibits Histone Acetyltransferase KAT8-Mediated IL-33 Acetylation to Alleviate Allergic Airway Inflammation and Airway Hyperresponsiveness. SIGNAL Transduct. Target. Ther. 2021, 6, 321. [Google Scholar] [CrossRef]
- Pessoa Rodrigues, C.; Chatterjee, A.; Wiese, M.; Stehle, T.; Szymanski, W.; Shvedunova, M.; Akhtar, A. Histone H4 Lysine 16 Acetylation Controls Central Carbon Metabolism and Diet-Induced Obesity in Mice. Nat. Commun. 2021, 12, 6212. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.F.; Ballestar, E.; Villar-Garea, A.; Boix-Chornet, M.; Espada, J.; Schotta, G.; Bonaldi, T.; Haydon, C.; Ropero, S.; Petrie, K.; et al. Loss of Acetylation at Lys16 and Trimethylation at Lys20 of Histone H4 Is a Common Hallmark of Human Cancer. Nat. Genet. 2005, 37, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Song, J.S.; Kim, Y.S.; Kim, D.K.; Park, S.I.; Jang, S.J. Global Histone Modification Pattern Associated with Recurrence and Disease-free Survival in Non-small Cell Lung Cancer Patients. Pathol. Int. 2012, 62, 182–190. [Google Scholar] [CrossRef]
- Zhang, S.; Sui, L.; Kong, X.; Huang, R.; Li, Z. HDAC6 Decreases H4K16 and α-Tubulin Acetylation during Porcine Oocyte Maturation. Cell Cycle 2023, 22, 2057–2069. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.-J.; Loewenstein, P.M.; Green, M. The Adenoviral E1A N-Terminal Domain Represses MYC Transcription in Human Cancer Cells by Targeting Both P300 and TRRAP and Inhibiting MYC Promoter Acetylation of H3K18 and H4K16. Genes. Cancer 2016, 7, 98–109. [Google Scholar] [CrossRef]
- Su, J.; Wang, F.; Cai, Y.; Jin, J. The Functional Analysis of Histone Acetyltransferase MOF in Tumorigenesis. Int. J. Mol. Sci. 2016, 17, 99. [Google Scholar] [CrossRef]
- Pfister, S.; Rea, S.; Taipale, M.; Mendrzyk, F.; Straub, B.; Ittrich, C.; Thuerigen, O.; Sinn, H.P.; Akhtar, A.; Lichter, P. The Histone Acetyltransferase hMOF Is Frequently Downregulated in Primary Breast Carcinoma and Medulloblastoma and Constitutes a Biomarker for Clinical Outcome in Medulloblastoma. Int. J. Cancer 2008, 122, 1207–1213. [Google Scholar] [CrossRef]
- Cao, L.; Zhu, L.; Yang, J.; Su, J.; Ni, J.; Du, Y.; Liu, D.; Wang, Y.; Wang, F.; Jin, J.; et al. Correlation of Low Expression of hMOF with Clinicopathological Features of Colorectal Carcinoma, Gastric Cancer and Renal Cell Carcinoma. Int. J. Oncol. 2014, 44, 1207–1214. [Google Scholar] [CrossRef]
- Bui, H.-T.; Yamaoka, E.; Miyano, T. Involvement of Histone H3 (Ser10) Phosphorylation in Chromosome Condensation Without Cdc2 Kinase and Mitogen-Activated Protein Kinase Activation in Pig Oocytes1. Biol. Reprod. 2004, 70, 1843–1851. [Google Scholar] [CrossRef]
- Fullgrabe, J.; Lynch-Day, M.A.; Heldring, N.; Li, W.; Struijk, R.B.; Ma, Q.; Hermanson, O.; Rosenfeld, M.G.; Klionsky, D.J.; Joseph, B. The Histone H4 Lysine 16 Acetyltransferase hMOF Regulates the Outcome of Autophagy. Nature 2013, 500, 468–471. [Google Scholar] [CrossRef]
- Gupta, A.; Guerin-Peyrou, T.G.; Sharma, G.G.; Park, C.; Agarwal, M.; Ganju, R.K.; Pandita, S.; Choi, K.; Sukumar, S.; Pandita, R.K.; et al. The Mammalian Ortholog of Drosophila MOF That Acetylates Histone H4 Lysine 16 Is Essential for Embryogenesis and Oncogenesis. Mol. Cell. Biol. 2008, 28, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Hao, Z.; Liu, Q.; Ding, R.; Chen, L.; Jin, M.; Wang, S. Oncolytic Virus Infection Modulates Lysine Acetyltransferase in Gliomas: Comprehensive Analysis and Experimental Validation of KAT8 in Glioma. J. Cell. Mol. Medi 2025, 29, e70558. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, H.; Pan, H.; Yang, Y.; Huang, G.; Yang, Y.; Zhou, W.-P.; Pan, Z.-Y. The Histone Acetyltransferase hMOF Suppresses Hepatocellular Carcinoma Growth. Biochem. Biophys. Res. Commun. 2014, 452, 575–580. [Google Scholar] [CrossRef]
- Chen, D.; Kluz, T.; Fang, L.; Zhang, X.; Sun, H.; Jin, C.; Costa, M. Hexavalent Chromium (Cr(VI)) Down-Regulates Acetylation of Histone H4 at Lysine 16 through Induction of Stressor Protein Nupr1. PLoS ONE 2016, 11, e0157317. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, R.; Wu, D.; Lu, Z.; Sun, W.; Cai, Y.; Wang, C.; Jin, J. Epigenetic Change in Kidney Tumor: Downregulation of Histone Acetyltransferase MYST1 in Human Renal Cell Carcinoma. J. Exp. Clin. Cancer Res. 2013, 32, 8. [Google Scholar] [CrossRef]
- Sykes, S.M.; Mellert, H.S.; Holbert, M.A.; Li, K.; Marmorstein, R.; Lane, W.S.; McMahon, S.B. Acetylation of the P53 DNA-Binding Domain Regulates Apoptosis Induction. Mol. Cell 2006, 24, 841–851. [Google Scholar] [CrossRef]
- Chen, J.; Liu, D.; Chen, B.; Yang, Y.; Zhu, H.; Li, D.; Liu, K.; Zhu, L.; Liu, H.; Li, M.; et al. The Histone Acetyltransferase Mof Regulates Runx2 and Osterix for Osteoblast Differentiation. Cell Tissue Res. 2023, 393, 265–279. [Google Scholar] [CrossRef]
- Kapoor-Vazirani, P.; Kagey, J.D.; Powell, D.R.; Vertino, P.M. Role of hMOF-Dependent Histone H4 Lysine 16 Acetylation in the Maintenance of TMS1/ASC Gene Activity. Cancer Res. 2008, 68, 6810–6821. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Li, L.; Lv, X.; Wu, X.-S.; Liu, D.-P.; Liang, C.-C. Modulations of hMOF Autoacetylation by SIRT1 Regulate hMOF Recruitment and Activities on the Chromatin. Cell Res. 2011, 21, 1182–1195. [Google Scholar] [CrossRef]
- Peng, L.; Ling, H.; Yuan, Z.; Fang, B.; Bloom, G.; Fukasawa, K.; Koomen, J.; Chen, J.; Lane, W.S.; Seto, E. SIRT1 Negatively Regulates the Activities, Functions, and Protein Levels of hMOF and TIP60. Mol. Cell. Biol. 2012, 32, 2823–2836. [Google Scholar] [CrossRef]
- Guo, R.; Liang, Y.; Zou, B.; Li, D.; Wu, Z.; Xie, F.; Zhang, X.; Li, X. The Histone Acetyltransferase MOF Regulates SIRT1 Expression to Suppress Renal Cell Carcinoma Progression. Front. Oncol. 2022, 12, 842967. [Google Scholar] [CrossRef] [PubMed]
- Jaganathan, A.; Chaurasia, P.; Xiao, G.-Q.; Philizaire, M.; Lv, X.; Yao, S.; Burnstein, K.L.; Liu, D.-P.; Levine, A.C.; Mujtaba, S. Coactivator MYST1 Regulates Nuclear Factor-κB and Androgen Receptor Functions during Proliferation of Prostate Cancer Cells. Mol. Endocrinol. 2014, 28, 872–885. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Lan, R.; Zhang, X.; Zhu, L.; Chen, F.; Xu, Z.; Liu, Y.; Ye, T.; Sun, H.; Lu, F.; et al. LSD1 Regulates Pluripotency of Embryonic Stem/Carcinoma Cells through Histone Deacetylase 1-Mediated Deacetylation of Histone H4 at Lysine 16. Mol. Cell. Biol. 2014, 34, 158–179. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, L.; Zou, Y.; Zhang, Y.; Zhang, M.; Xu, L.; Zheng, L.; He, W.; Yu, K.; Li, T.; et al. Disrupting the Phase Separation of KAT8-IRF1 Diminishes PD-L1 Expression and Promotes Antitumor Immunity. Nat. Cancer 2023, 4, 382–400. [Google Scholar] [CrossRef]
- Liang, J.; Cao, R.; Wang, X.; Zhang, Y.; Wang, P.; Gao, H.; Li, C.; Yang, F.; Zeng, R.; Wei, P.; et al. Mitochondrial PKM2 Regulates Oxidative Stress-Induced Apoptosis by Stabilizing Bcl2. Cell Res. 2017, 27, 329–351. [Google Scholar] [CrossRef]
- Wang, D.; Zhao, C.; Xu, F.; Zhang, A.; Jin, M.; Zhang, K.; Liu, L.; Hua, Q.; Zhao, J.; Liu, J.; et al. Cisplatin-Resistant NSCLC Cells Induced by Hypoxia Transmit Resistance to Sensitive Cells through Exosomal PKM2. Theranostics 2021, 11, 2860–2875. [Google Scholar] [CrossRef]
- Li, Z.; Lu, X.; Zhang, J.; Liu, T.; Xu, M.; Liu, S.; Liang, J. KAT8 Enhances the Resistance of Lung Cancer Cells to Cisplatin by Acetylation of PKM2. Anticancer. Drugs 2024, 35, 732–740. [Google Scholar] [CrossRef]
- Meunier, A.; Cornet, F.; Campos, M. Bacterial Cell Proliferation: From Molecules to Cells. FEMS Microbiol. Rev. 2021, 45, fuaa046. [Google Scholar] [CrossRef]
- Wang, F.; Zhao, J.; Liu, D.; Zhao, T.; Lu, Z.; Zhu, L.; Cao, L.; Yang, J.; Jin, J.; Cai, Y. Capsaicin Reactivates hMOF in Gastric Cancer Cells and Induces Cell Growth Inhibition. Cancer Biol. Ther. 2016, 17, 1117–1125. [Google Scholar] [CrossRef]
- Xu, A.; Yang, X.; Zhao, J.; Kong, S.; Tang, Q.; Li, X.; Qu, H.; Wang, G. KAT8 Facilitates the Proliferation of Cancer Cells through Enhancing E7 Function in HPV-Associated Cervical Cancer. bioRxiv 2025. [Google Scholar] [CrossRef]
- Qi, Y.; Tan, M.; Zheng, M.; Jin, S.; Wang, H.; Liu, J.; Wang, P.; Nie, X.; Gao, L.; Lin, B. Estrogen/Estrogen Receptor Promotes the Proliferation of Endometrial Carcinoma Cells by Enhancing hMOF Expression. Jpn. J. Clin. Oncol. 2020, 50, 241–253. [Google Scholar] [CrossRef]
- Kim, K.H.; Roberts, C.W.M. Targeting EZH2 in Cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, M.; Li, P.; Laster, K.V.; Zhao, D.; Zhi, Y.; Wei, H.; Nie, W.; Gao, Y.; Wu, Q.; et al. CHI-KAT8i5 Suppresses ESCC Tumor Growth by Inhibiting KAT8-Mediated c-Myc Stability. Cell Rep. 2025, 44, 115135. [Google Scholar] [CrossRef] [PubMed]
- Rojo De La Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ye, X.; Tang, N.; Shen, S.; Li, Z.; Niu, X.; Lu, S.; Xu, L. The Histone Acetylranseferase h MOF Acetylates Nrf 2 and Regulates Anti-drug Responses in Human Non-small Cell Lung Cancer. Br. J. Pharmacol. 2014, 171, 3196–3211. [Google Scholar] [CrossRef]
- De Coninck, S.; Berx, G.; Taghon, T.; Van Vlierberghe, P.; Goossens, S. ZEB2 in T-Cells and T-ALL. Adv. Biol. Regul. 2019, 74, 100639. [Google Scholar] [CrossRef]
- Zhao, K.; Zheng, M.; Su, Z.; Ghosh, S.; Zhang, C.; Zhong, W.; Ho, J.W.K.; Jin, G.; Zhou, Z. MOF-Mediated Acetylation of SIRT6 Disrupts SIRT6-FOXA2 Interaction and Represses SIRT6 Tumor-Suppressive Function by Upregulating ZEB2 in NSCLC. Cell Rep. 2023, 42, 112939. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yang, Y.; Chen, B.; Guo, X.; Gao, S.; Wang, M.; Duan, M.; Li, X. MOF Regulates TNK2 Transcription Expression to Promote Cell Proliferation in Thyroid Cancer. Front. Pharmacol. 2020, 11, 607605. [Google Scholar] [CrossRef]
- Blum, A.; Wang, P.; Zenklusen, J.C. SnapShot: TCGA-Analyzed Tumors. Cell 2018, 173, 530. [Google Scholar] [CrossRef]
- Gorgoulis, V.G.; Vassiliou, L.-V.F.; Karakaidos, P.; Zacharatos, P.; Kotsinas, A.; Liloglou, T.; Venere, M.; DiTullio, R.A.; Kastrinakis, N.G.; Levy, B.; et al. Activation of the DNA Damage Checkpoint and Genomic Instability in Human Precancerous Lesions. Nature 2005, 434, 907–913. [Google Scholar] [CrossRef]
- Gaillard, H.; García-Muse, T.; Aguilera, A. Replication Stress and Cancer. Nat. Rev. Cancer 2015, 15, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Bacolla, A.; Chaudhary, S.; Hunt, C.R.; Pandita, S.; Chauhan, R.; Gupta, A.; Tainer, J.A.; Pandita, T.K. Histone Acetyltransferase MOF Orchestrates Outcomes at the Crossroad of Oncogenesis, DNA Damage Response, Proliferation, and Stem Cell Development. Mol. Cell. Biol. 2020, 40, e00232-20. [Google Scholar] [CrossRef] [PubMed]
- Valerio, D.G.; Xu, H.; Chen, C.-W.; Hoshii, T.; Eisold, M.E.; Delaney, C.; Cusan, M.; Deshpande, A.J.; Huang, C.-H.; Lujambio, A.; et al. Histone Acetyltransferase Activity of MOF Is Required for MLL-AF9 Leukemogenesis. Cancer Res. 2017, 77, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Huo, D.; Chen, H.; Cheng, Y.; Song, X.; Zhang, K.; Li, M.J.; Xuan, C. JMJD6 Modulates DNA Damage Response through Downregulating H4K16ac Independently of Its Enzymatic Activity. Cell Death Differ. 2020, 27, 1052–1066. [Google Scholar] [CrossRef]
- Li, X.; Corsa, C.A.S.; Pan, P.W.; Wu, L.; Ferguson, D.; Yu, X.; Min, J.; Dou, Y. MOF and H4 K16 Acetylation Play Important Roles in DNA Damage Repair by Modulating Recruitment of DNA Damage Repair Protein Mdc1. Mol. Cell. Biol. 2010, 30, 5335–5347. [Google Scholar] [CrossRef]
- Gupta, A.; Hunt, C.R.; Hegde, M.L.; Chakraborty, S.; Udayakumar, D.; Horikoshi, N.; Singh, M.; Ramnarain, D.B.; Hittelman, W.N.; Namjoshi, S.; et al. MOF Phosphorylation by ATM Regulates 53BP1-Mediated Double-Strand Break Repair Pathway Choice. Cell Rep. 2014, 8, 177–189. [Google Scholar] [CrossRef]
- Gupta, A.; Sharma, G.G.; Young, C.S.H.; Agarwal, M.; Smith, E.R.; Paull, T.T.; Lucchesi, J.C.; Khanna, K.K.; Ludwig, T.; Pandita, T.K. Involvement of Human MOF in ATM Function. Mol. Cell. Biol. 2005, 25, 5292–5305. [Google Scholar] [CrossRef]
- Hoege, C.; Pfander, B.; Moldovan, G.-L.; Pyrowolakis, G.; Jentsch, S. RAD6-Dependent DNA Repair Is Linked to Modification of PCNA by Ubiquitin and SUMO. Nature 2002, 419, 135–141. [Google Scholar] [CrossRef]
- Singh, D.K.; Pandita, R.K.; Singh, M.; Chakraborty, S.; Hambarde, S.; Ramnarain, D.; Charaka, V.; Ahmed, K.M.; Hunt, C.R.; Pandita, T.K. MOF Suppresses Replication Stress and Contributes to Resolution of Stalled Replication Forks. Mol. Cell. Biol. 2018, 38, e00484-17. [Google Scholar] [CrossRef]
- Punchihewa, C.; Inoue, A.; Hishiki, A.; Fujikawa, Y.; Connelly, M.; Evison, B.; Shao, Y.; Heath, R.; Kuraoka, I.; Rodrigues, P.; et al. Identification of Small Molecule Proliferating Cell Nuclear Antigen (PCNA) Inhibitor That Disrupts Interactions with PIP-Box Proteins and Inhibits DNA Replication. J. Biol. Chem. 2012, 287, 14289–14300. [Google Scholar] [CrossRef]
- Zhang, S.-Y.; Song, X.-Y.; Li, Y.; Ye, L.-L.; Zhou, Q.; Yang, W.-B. Tumor-Associated Macrophages: A Promising Target for a Cancer Immunotherapeutic Strategy. Pharmacol. Res. 2020, 161, 105111. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, S.; Ma, B. Autophagy and Autophagy-Related Proteins in Cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef]
- Turco, E.; Savova, A.; Gere, F.; Ferrari, L.; Romanov, J.; Schuschnig, M.; Martens, S. Reconstitution Defines the Roles of P62, NBR1 and TAX1BP1 in Ubiquitin Condensate Formation and Autophagy Initiation. Nat. Commun. 2021, 12, 5212. [Google Scholar] [CrossRef]
- Kocak, M.; Ezazi Erdi, S.; Jorba, G.; Maestro, I.; Farrés, J.; Kirkin, V.; Martinez, A.; Pless, O. Targeting Autophagy in Disease: Established and New Strategies. Autophagy 2022, 18, 473–495. [Google Scholar] [CrossRef] [PubMed]
- Meyer, N.; Henkel, L.; Linder, B.; Zielke, S.; Tascher, G.; Trautmann, S.; Geisslinger, G.; Münch, C.; Fulda, S.; Tegeder, I.; et al. Autophagy Activation, Lipotoxicity and Lysosomal Membrane Permeabilization Synergize to Promote Pimozide- and Loperamide-Induced Glioma Cell Death. Autophagy 2021, 17, 3424–3443. [Google Scholar] [CrossRef]
- Xu, Y.; Wan, W. Acetylation in the Regulation of Autophagy. Autophagy 2023, 19, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Zehender, A.; Li, Y.-N.; Lin, N.-Y.; Stefanica, A.; Nüchel, J.; Chen, C.-W.; Hsu, H.-H.; Zhu, H.; Ding, X.; Huang, J.; et al. TGFβ Promotes Fibrosis by MYST1-Dependent Epigenetic Regulation of Autophagy. Nat. Commun. 2021, 12, 4404. [Google Scholar] [CrossRef]
- Romao, S.; Münz, C. LC3-Associated Phagocytosis. Autophagy 2014, 10, 526–528. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Berthier, A.; Seguin, S.; Sasco, A.J.; Bobin, J.Y.; De Laroche, G.; Datchary, J.; Saez, S.; Rodriguez-Lafrasse, C.; Tolle, F.; Fraichard, A.; et al. High Expression of Gabarapl1 Is Associated with a Better Outcome for Patients with Lymph Node-Positive Breast Cancer. Br. J. Cancer 2010, 102, 1024–1031. [Google Scholar] [CrossRef]
- Poillet-Perez, L.; Jacquet, M.; Hervouet, E.; Gauthier, T.; Fraichard, A.; Borg, C.; Pallandre, J.-R.; Gonzalez, B.J.; Ramdani, Y.; Boyer-Guittaut, M.; et al. GABARAPL1 Tumor Suppressive Function Is Independent of Its Conjugation to Autophagosomes in MCF-7 Breast Cancer Cells. Oncotarget 2017, 8, 55998–56020. [Google Scholar] [CrossRef] [PubMed]
- Boyer-Guittaut, M.; Poillet, L.; Liang, Q.; Bôle-Richard, E.; Ouyang, X.; Benavides, G.A.; Chakrama, F.-Z.; Fraichard, A.; Darley-Usmar, V.M.; Despouy, G.; et al. The Role of GABARAPL1/GEC1 in Autophagic Flux and Mitochondrial Quality Control in MDA-MB-436 Breast Cancer Cells. Autophagy 2014, 10, 986–1003. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Li, S.; Chen, X.; Yin, L.; Ma, P.; Ma, Y.; Su, B. GABARAPL1 Suppresses Metastasis by Counteracting PI3K/Akt Pathway in Prostate Cancer. Oncotarget 2017, 8, 4449–4459. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-W.; Zhao, Y.-M.; Jin, K.-Y.; Wang, J.-X.; Zhao, X.-F. KAT8 Is Upregulated and Recruited to the Promoter of Atg8 by FOXO to Induce H4 Acetylation for Autophagy under 20-Hydroxyecdysone Regulation. J. Biol. Chem. 2024, 300, 105704. [Google Scholar] [CrossRef]
- De Talhouët, C.; Esteras, N.; Soutar, M.P.M.; O’Callaghan, B.; Plun-Favreau, H. KAT8 Compound Inhibition Inhibits the Initial Steps of PINK1-Dependant Mitophagy. Sci. Rep. 2024, 14, 11721. [Google Scholar] [CrossRef]
- Yan, M.; Wang, J.; Wang, H.; Zhou, J.; Qi, H.; Naji, Y.; Zhao, L.; Tang, Y.; Dai, Y. Knockdown of NR3C1 Inhibits the Proliferation and Migration of Clear Cell Renal Cell Carcinoma through Activating Endoplasmic Reticulum Stress-Mitophagy. J. Transl. Med. 2023, 21, 701. [Google Scholar] [CrossRef]
- Van De Merbel, A.F.; Van Der Horst, G.; Buijs, J.T.; Van Der Pluijm, G. Protocols for Migration and Invasion Studies in Prostate Cancer. In Prostate Cancer; Culig, Z., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 1786, pp. 67–79. ISBN 978-1-4939-7843-4. [Google Scholar]
- Duff, D.; Long, A. Roles for RACK1 in Cancer Cell Migration and Invasion. Cell. Signal. 2017, 35, 250–255. [Google Scholar] [CrossRef]
- Li, D.; Cheng, Y.; Pan, J.; Guo, Z.; Wang, S.; Huang, Q.; Nie, P.; Shi, W.; Xu, X.; Wen, B.; et al. KAT8/SIRT7-mediated Fascin-K41 Acetylation/Deacetylation Regulates Tumor Metastasis in Esophageal Squamous Cell Carcinoma. J. Pathol. 2024, 263, 74–88. [Google Scholar] [CrossRef]
- Li, P.; Yang, L.; Park, S.Y.; Liu, F.; Li, A.H.; Zhu, Y.; Sui, H.; Gao, F.; Li, L.; Ye, L.; et al. Stabilization of MOF (KAT8) by USP10 Promotes Esophageal Squamous Cell Carcinoma Proliferation and Metastasis through Epigenetic Activation of ANXA2/Wnt Signaling. Oncogene 2024, 43, 899–917. [Google Scholar] [CrossRef]
- Qiu, B.; Li, S.; Li, M.; Wang, S.; Mu, G.; Chen, K.; Wang, M.; Zhu, W.; Wang, W.; Wang, J.; et al. KAT8 Acetylation-Controlled Lipolysis Affects the Invasive and Migratory Potential of Colorectal Cancer Cells. Cell Death Dis. 2023, 14, 164. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.-Y.; Wang, S.-Q.; Li, M.; Long, Y.-H.; Li, Y.-F.; Liu, Y.-K.; Li, Y.-H.; Wang, Y.-Q.; Mi, J.-S.; et al. WSTF Acetylation by MOF Promotes WSTF Activities and Oncogenic Functions. Oncogene 2020, 39, 5056–5067. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef]
- Chen, H.; Chen, X.; Pan, B.; Zheng, C.; Hong, L.; Han, W. KRT8 Serves as a Novel Biomarker for LUAD and Promotes Metastasis and EMT via NF-κB Signaling. Front. Oncol. 2022, 12, 875146. [Google Scholar] [CrossRef]
- Luo, H.; Shenoy, A.K.; Li, X.; Jin, Y.; Jin, L.; Cai, Q.; Tang, M.; Liu, Y.; Chen, H.; Reisman, D.; et al. MOF Acetylates the Histone Demethylase LSD1 to Suppress Epithelial-to-Mesenchymal Transition. Cell Rep. 2016, 15, 2665–2678. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, P.L.; Gray, N.S. Targeting Cancer with Small Molecule Kinase Inhibitors. Nat. Rev. Cancer 2009, 9, 28–39. [Google Scholar] [CrossRef]
- Roy, A. Plumbagin: A Potential Anti-Cancer Compound. Mini Rev. Med. Chem. 2021, 21, 731–737. [Google Scholar] [CrossRef]
- Choo, M.Z.Y.; Chua, J.A.T.; Lee, S.X.Y.; Ang, Y.; Wong, W.S.F.; Chai, C.L.L. Privileged Natural Product Compound Classes for Anti-Inflammatory Drug Development. Nat. Prod. Rep. 2025, 42, 856–875. [Google Scholar] [CrossRef]
- Kim, J.-D.; Kim, J.-M.; Pyo, J.-O.; Kim, S.-Y.; Kim, B.-S.; Yu, R.; Han, I.-S. Capsaicin Can Alter the Expression of Tumor Forming-Related Genes Which Might Be Followed by Induction of Apoptosis of a Korean Stomach Cancer Cell Line, SNU-1. Cancer Lett. 1997, 120, 235–241. [Google Scholar] [CrossRef]
- Frias, B.; Merighi, A. Capsaicin, Nociception and Pain. Molecules 2016, 21, 797. [Google Scholar] [CrossRef]
- Hou, W.; Liu, B.; Xu, H. Celastrol: Progresses in Structure-Modifications, Structure-Activity Relationships, Pharmacology and Toxicology. Eur. J. Med. Chem. 2020, 189, 112081. [Google Scholar] [CrossRef] [PubMed]
- Contreras, S.M.; Ganuza, A.; Corvi, M.M.; Angel, S.O. Resveratrol Induces H3 and H4K16 Deacetylation and H2A.X Phosphorylation in Toxoplasma gondii. BMC Res. Notes 2021, 14, 19. [Google Scholar] [CrossRef]
- Lai, M.C.; Liu, W.Y.; Liou, S.-S.; Liu, I.-M. The Protective Effects of Moscatilin against Methylglyoxal-Induced Neurotoxicity via the Regulation of P38/JNK MAPK Pathways in PC12 Neuron-like Cells. Food Chem. Toxicol. 2020, 140, 111369. [Google Scholar] [CrossRef] [PubMed]
- Wallace, M.; Pappagallo, M. Qutenza®: A Capsaicin 8% Patch for the Management of Postherpetic Neuralgia. Expert. Rev. Neurother. 2011, 11, 15–27. [Google Scholar] [CrossRef]
- Wang, H.-M.; Chuang, S.-M.; Su, Y.-C.; Li, Y.-H.; Chueh, P.J. Down-Regulation of Tumor-Associated NADH Oxidase, tNOX (ENOX2), Enhances Capsaicin-Induced Inhibition of Gastric Cancer Cell Growth. Cell Biochem. Biophys. 2011, 61, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Thoennissen, N.H.; O’Kelly, J.; Lu, D.; Iwanski, G.B.; La, D.T.; Abbassi, S.; Leiter, A.; Karlan, B.; Mehta, R.; Koeffler, H.P. Capsaicin Causes Cell-Cycle Arrest and Apoptosis in ER-Positive and -Negative Breast Cancer Cells by Modulating the EGFR/HER-2 Pathway. Oncogene 2010, 29, 285–296. [Google Scholar] [CrossRef]
- Wang, C.; Dai, S.; Zhao, X.; Zhang, Y.; Gong, L.; Fu, K.; Ma, C.; Peng, C.; Li, Y. Celastrol as an Emerging Anticancer Agent: Current Status, Challenges and Therapeutic Strategies. Biomed. Pharmacother. 2023, 163, 114882. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, C.; Li, X.; Lu, J.; Wang, M. Recent Advances in Drug Delivery of Celastrol for Enhancing Efficiency and Reducing the Toxicity. Front. Pharmacol. 2024, 15, 1137289. [Google Scholar] [CrossRef]
- Kashyap, D.; Sharma, A.; Tuli, H.S.; Sak, K.; Mukherjee, T.; Bishayee, A. Molecular Targets of Celastrol in Cancer: Recent Trends and Advancements. Crit. Rev. Oncol. Hematol. 2018, 128, 70–81. [Google Scholar] [CrossRef]
- Chen, G.; Zhu, X.; Li, J.; Zhang, Y.; Wang, X.; Zhang, R.; Qin, X.; Chen, X.; Wang, J.; Liao, W.; et al. Celastrol Inhibits Lung Cancer Growth by Triggering Histone Acetylation and Acting Synergically with HDAC Inhibitors. Pharmacol. Res. 2022, 185, 106487. [Google Scholar] [CrossRef]
- Ren, B.; Kwah, M.X.-Y.; Liu, C.; Ma, Z.; Shanmugam, M.K.; Ding, L.; Xiang, X.; Ho, P.C.-L.; Wang, L.; Ong, P.S.; et al. Resveratrol for Cancer Therapy: Challenges and Future Perspectives. Cancer Lett. 2021, 515, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Luca, S.V.; Macovei, I.; Bujor, A.; Miron, A.; Skalicka-Woźniak, K.; Aprotosoaie, A.C.; Trifan, A. Bioactivity of Dietary Polyphenols: The Role of Metabolites. Crit. Rev. Food Sci. Nutr. 2020, 60, 626–659. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Bhardwaj, A.; Aggarwal, R.S.; Seeram, N.P.; Shishodia, S.; Takada, Y. Role of Resveratrol in Prevention and Therapy of Cancer: Preclinical and Clinical Studies. Anticancer. Res. 2004, 24, 2783–2840. [Google Scholar]
- Harikumar, K.B.; Kunnumakkara, A.B.; Sethi, G.; Diagaradjane, P.; Anand, P.; Pandey, M.K.; Gelovani, J.; Krishnan, S.; Guha, S.; Aggarwal, B.B. Resveratrol, a Multitargeted Agent, Can Enhance Antitumor Activity of Gemcitabine in Vitro and in Orthotopic Mouse Model of Human Pancreatic Cancer. Intl J. Cancer 2010, 127, 257–268. [Google Scholar] [CrossRef]
- Tomé-Carneiro, J.; Larrosa, M.; González-Sarrías, A.; Tomás-Barberán, F.; García-Conesa, M.; Espín, J. Resveratrol and Clinical Trials: The Crossroad from In Vitro Studies to Human Evidence. Curr. Pharm. Des. 2013, 19, 6064–6093. [Google Scholar] [CrossRef]
- Kundu, J.K.; Surh, Y.-J. Cancer Chemopreventive and Therapeutic Potential of Resveratrol: Mechanistic Perspectives. Cancer Lett. 2008, 269, 243–261. [Google Scholar] [CrossRef]
- Auti, A.; Alessio, N.; Ballini, A.; Dioguardi, M.; Cantore, S.; Scacco, S.; Vitiello, A.; Quagliuolo, L.; Rinaldi, B.; Santacroce, L.; et al. Protective Effect of Resveratrol against Hypoxia-Induced Neural Oxidative Stress. J. Pers. Med. 2022, 12, 1202. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Cao, Z.-X.; Peng, C.; Li, X.-H.; Xie, X.-F.; Zhang, T.-M.; Zhou, Q.-M.; Yang, L.; Guo, L. Phenolic Glucosides from Dendrobium aurantiacum Var. Denneanum and Their Bioactivities. Molecules 2013, 18, 6153–6160. [Google Scholar] [CrossRef]
- Silva-Reis, R.; Silva, V.L.M.; Cardoso, S.M.; Michalak, I.; Püsküllüoğlu, M.; Calina, D.; Sharifi-Rad, J. Moscatilin, a Potential Therapeutic Agent for Cancer Treatment: Insights into Molecular Mechanisms and Clinical Prospects. Med. Oncol. 2024, 41, 228. [Google Scholar] [CrossRef]
- Chen, T.-H.; Pan, S.-L.; Guh, J.-H.; Liao, C.-H.; Huang, D.-Y.; Chen, C.-C.; Teng, C.-M. Moscatilin Induces Apoptosis in Human Colorectal Cancer Cells: A Crucial Role of c-Jun NH2-Terminal Protein Kinase Activation Caused by Tubulin Depolymerization and DNA Damage. Clin. Cancer Res. 2008, 14, 4250–4258. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, Y.; Xu, X.-F.; Jin, D.-Y. Moscatilin Induces Apoptosis of Pancreatic Cancer Cells via Reactive Oxygen Species and the JNK/SAPK Pathway. Mol. Med. Rep. 2017, 15, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Zeng, L.; Chen, W. Moscatilin Suppresses the Breast Cancer Both In Vitro and In Vivo by Inhibiting HDAC3. Dose-Response 2021, 19, 15593258211001251. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.R.; Black, J.A. Aspirin: Latest Evidence and Developments. Heart 2024, 110, 1069–1073. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberger, L.M.; Phan, T.; Fang, D.; Edler, S.; Philip, J.; Li-Geng, T.; Dial, E.J. Bioavailability of Aspirin in Rats Comparing the Drug’s Uptake into Gastrointestinal Tissue and Vascular and Lymphatic Systems: Implications on Aspirin’s Chemopreventive Action. J. Physiol. Pharmacol. 2016, 67, 635–642. [Google Scholar]
- Fernandez, H.R.; Lindén, S.K. The Aspirin Metabolite Salicylate Inhibits Lysine Acetyltransferases and MUC1 Induced Epithelial to Mesenchymal Transition. Sci. Rep. 2017, 7, 5626. [Google Scholar] [CrossRef]
- Chan, T.; Tse, E.; Kwong, Y.-L. Chidamide in the Treatment of Peripheral T-Cell Lymphoma. OncoTargets Ther. 2017, 10, 347–352. [Google Scholar] [CrossRef]
- Xu, L.; Feng, J.; Tang, H.; Dong, Y.; Shu, M.; Chen, X. Chidamide Epigenetically Represses Autophagy and Exerts Cooperative Antimyeloma Activity with Bortezomib. Cell Death Dis. 2020, 11, 297. [Google Scholar] [CrossRef]
- Miao, H.; Chen, X.; Luan, Y. Small Molecular Gemcitabine Prodrugs for Cancer Therapy. Curr. Med. Chem. 2020, 27, 5562–5582. [Google Scholar] [CrossRef]
- Thompson, B.R.; Shi, J.; Zhu, H.-J.; Smith, D.E. Pharmacokinetics of Gemcitabine and Its Amino Acid Ester Prodrug Following Intravenous and Oral Administrations in Mice. Biochem. Pharmacol. 2020, 180, 114127. [Google Scholar] [CrossRef]
- Wang, J.; Yang, J.; Narang, A.; He, J.; Wolfgang, C.; Li, K.; Zheng, L. Consensus, Debate, and Prospective on Pancreatic Cancer Treatments. J. Hematol. Oncol. 2024, 17, 92. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, Y.; Wei, T.; Zhao, X.; Li, F.; Li, Y.; Wang, F.; Cai, Y.; Jin, J. KAT8/MOF-Mediated Anti-Cancer Mechanism of Gemcitabine in Human Bladder Cancer Cells. Biomol. Ther. 2021, 29, 184–194. [Google Scholar] [CrossRef]
- Howe, C.G.; Gamble, M.V. Influence of Arsenic on Global Levels of Histone Posttranslational Modifications: A Review of the Literature and Challenges in the Field. Curr. Environ. Health Rep. 2016, 3, 225–237. [Google Scholar] [CrossRef]
- Chen, J.; Jin, Z.; Zhang, S.; Zhang, X.; Li, P.; Yang, H.; Ma, Y. Arsenic Trioxide Elicits Prophylactic and Therapeutic Immune Responses against Solid Tumors by Inducing Necroptosis and Ferroptosis. Cell Mol. Immunol. 2022, 20, 51–64. [Google Scholar] [CrossRef]
- Ma, P.; Schultz, R.M. Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse. PLoS Genet. 2013, 9, e1003377. [Google Scholar] [CrossRef]
- Guo, X.; Wang, W.; Hu, J.; Feng, K.; Pan, Y.; Zhang, L.; Feng, Y. Lentivirus-Mediated RNAi Knockdown of NUPR1 Inhibits Human Nonsmall Cell Lung Cancer Growth In Vitro and In Vivo. Anat. Rec. 2012, 295, 2114–2121. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA Therapeutics—Challenges and Potential Solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Gallant-Behm, C.L.; Piper, J.; Lynch, J.M.; Seto, A.G.; Hong, S.J.; Mustoe, T.A.; Maari, C.; Pestano, L.A.; Dalby, C.M.; Jackson, A.L.; et al. A MicroRNA-29 Mimic (Remlarsen) Represses Extracellular Matrix Expression and Fibroplasia in the Skin. J. Investig. Dermatol. 2019, 139, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Murata, A.; Otabe, T.; Zhang, J.; Nakatani, K. BzDANP, a Small-Molecule Modulator of Pre-miR-29a Maturation by Dicer. ACS Chem. Biol. 2016, 11, 2790–2796. [Google Scholar] [CrossRef]
- Jiang, N.-J.; Yin, Y.-N.; Lin, J.; Li, W.-Y.; Long, D.-R.; Mei, L. MicroRNA-21 in Gynecological Cancers: From Molecular Pathogenesis to Clinical Significance. Pathol.-Res. Pract. 2023, 248, 154630. [Google Scholar] [CrossRef]
- Chang, J.; Davis-Dusenbery, B.N.; Kashima, R.; Jiang, X.; Marathe, N.; Sessa, R.; Louie, J.; Gu, W.; Lagna, G.; Hata, A. Acetylation of P53 Stimulates miRNA Processing and Determines Cell Survival Following Genotoxic Stress: Acetylation of P53 Stimulates miRNA Processing and Determines Cell Survival. EMBO J. 2013, 32, 3192–3205. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA Signature Associated with Prognosis and Progression in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent Deletions and Down-Regulation of Micro- RNA Genes miR15 and miR16 at 13q14 in Chronic Lymphocytic Leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chen, H.; Jia, Y.; Lu, H.; Tan, Q.; Zhou, X. miR-149-5p Inhibition Reduces Alzheimer’s Disease β-amyloid Generation in 293/APPsw Cells by Upregulating H4K16ac via KAT8. Exp. Ther. Med. 2020, 20, 88. [Google Scholar] [CrossRef]
- Shao, I.; Peng, P.; Wu, H.; Chen, J.; Lai, J.C.; Chang, J.; Wu, H.; Wu, K.; Pang, S.; Hsu, K. RP11-367G18.1 V2 Enhances Clear Cell Renal Cell Carcinoma Progression via Induction of Epithelial–Mesenchymal Transition. Cancer Med. 2023, 12, 9788–9801. [Google Scholar] [CrossRef] [PubMed]
- Verheul, T.C.J.; Van Hijfte, L.; Perenthaler, E.; Barakat, T.S. The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1. Front. Cell Dev. Biol. 2020, 8, 592164. [Google Scholar] [CrossRef]
- Peng, P.-H.; Chen, J.-L.; Wu, H.-H.; Yang, W.-H.; Lin, L.-J.; Lai, J.C.-Y.; Chang, J.-S.; Syu, J.-L.; Wu, H.-T.; Hsu, F.-T.; et al. Interplay between lncRNA RP11-367G18.1 Variant 2 and YY1 Plays a Vital Role in Hypoxia-Mediated Gene Expression and Tumorigenesis. Cancer Cell Int. 2023, 23, 266. [Google Scholar] [CrossRef]
- Liu, G.; Huang, K.; Jie, Z.; Wu, Y.; Chen, J.; Chen, Z.; Fang, X.; Shen, S. CircFAT1 Sponges miR-375 to Promote the Expression of Yes-Associated Protein 1 in Osteosarcoma Cells. Mol. Cancer 2018, 17, 170. [Google Scholar] [CrossRef]
- Chen, J.; Liu, G.; Wu, Y.; Ma, J.; Wu, H.; Xie, Z.; Chen, S.; Yang, Y.; Wang, S.; Shen, P.; et al. CircMYO10 Promotes Osteosarcoma Progression by Regulating miR-370-3p/RUVBL1 Axis to Enhance the Transcriptional Activity of β-Catenin/LEF1 Complex via Effects on Chromatin Remodeling. Mol. Cancer 2019, 18, 150. [Google Scholar] [CrossRef]
| Mechanism | Target | Model | Reference |
|---|---|---|---|
| Apoptosis | KAT8 promotes SIRT1 expression to downregulate STAT3 expression to promote apoptosis. | HCC (HepG2) | [41] |
| KAT8 overexpression H4K16 acetylation level upregulation CA9 expression reduction promotes apoptosis. | RCC (786-0) | [36] | |
| KAT8 interacts with PKM2 and acetylates the PKM2 K433 site thereby upregulating Bcl-2 to restrain apoptosis. | PRAD (BHP10-3, TT2609) | [47] | |
| Cell Proliferation | KAT8 promotes upregulation of H4K16 acetylation levels, cell cycle arrest in G1 phase, and inhibition of cell proliferation. | STAD (MGC-803) | [52] |
| Knockdown of KAT8 inhibits tumorigenicity of UM1 cells by downregulating EZH2 expression. | TSCC (UM1) | [43] | |
| Knockdown of KAT8 downregulates acetylation of NRF2 588 site and inhibits NQO1 and HO-1 expression to suppress cell proliferation. | NSCLC (H1975) | [58] | |
| KAT8 mediates SIRT6 acetylation to inhibit SIRT6 and FOXA2 interactions, which in turn activates ZEB2 transcription, thereby promoting cell proliferation. | NSCLC (A549, H1299) | [59] | |
| KAT8 catalyzes the Skp2 promoter region H4K16ac, upregulates p27, promotes cell passage through the S phase, and inhibits cell proliferation. | NSCLC (A549) | [13] | |
| DNA Damage and Repair | KAT8 interacts with PCNA and ubiquitinates it to promote its recruitment to replication stress DNA damage sites for DNA damage repair. | Osteosarcoma (U2OS) | [71] |
| Autophagy | Downregulation of H4K16 acetylation levels by KAT8 promotes increased LC3-LC3II conversion and activates cellular autophagy. | NSCLC (U1810) | [79] |
| KAT8 downregulates H4K16 acetylation levels, PINK1 expression, inhibits p62 recruitment, and inhibits the mitochondrial-lysosomal autophagy pathway. | Renal cell carcinoma (ACNH) | [87] | |
| Invasion and Migration EMT | KAT8 catalyzes the acetylation of the bundle protein Fascin-K41 site and promotes cell invasiveness. | ESCC (KYSE150) | [91] |
| KAT8 acetylates the WSTF-K426 site and promotes WSTF-S158 phosphorylation, cell migration, and invasion ability. | Breast cancer (MDAMB-237) | [95] | |
| KAT8 inhibits LSD1 recruitment at the promoter region and restores the methylation of H3K4 and KRT8, and inhibits EMT and invasive capability. | NSCLC (A549) | [98] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zhao, L.; Lan, X.; Zhu, M.; Tan, Y.; Luo, H.; Wu, D. Lysine Acetyltransferase 8: A Target for Natural Compounds in Cancer Therapy. Int. J. Mol. Sci. 2025, 26, 5257. https://doi.org/10.3390/ijms26115257
Wang L, Zhao L, Lan X, Zhu M, Tan Y, Luo H, Wu D. Lysine Acetyltransferase 8: A Target for Natural Compounds in Cancer Therapy. International Journal of Molecular Sciences. 2025; 26(11):5257. https://doi.org/10.3390/ijms26115257
Chicago/Turabian StyleWang, Lei, Liting Zhao, Xintian Lan, Ming Zhu, Yiying Tan, Haoming Luo, and Donglu Wu. 2025. "Lysine Acetyltransferase 8: A Target for Natural Compounds in Cancer Therapy" International Journal of Molecular Sciences 26, no. 11: 5257. https://doi.org/10.3390/ijms26115257
APA StyleWang, L., Zhao, L., Lan, X., Zhu, M., Tan, Y., Luo, H., & Wu, D. (2025). Lysine Acetyltransferase 8: A Target for Natural Compounds in Cancer Therapy. International Journal of Molecular Sciences, 26(11), 5257. https://doi.org/10.3390/ijms26115257





