Quorum Signaling Molecules: Interactions Between Plants and Associated Pathogens
Abstract
1. Introduction
2. QS Definition and Core Mechanisms
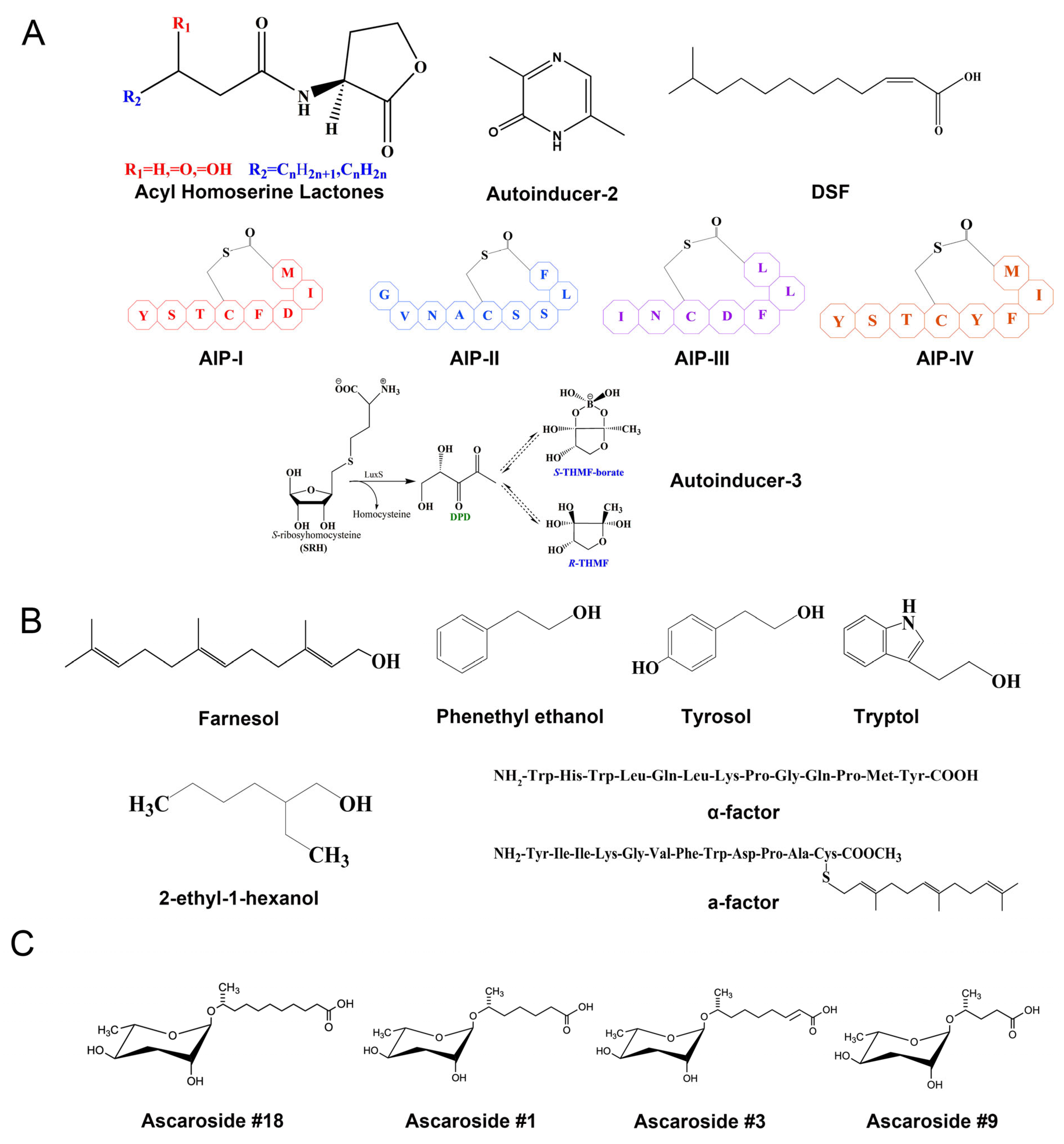
3. Major Classes of QSMs and Their Functions
3.1. Bacterial QSMs
3.2. Fungal QSMs
3.3. Parasitic Nematode QSMs
| QSMs | Producing Pathogens | Plant | Functions/Effect | Reference |
|---|---|---|---|---|
| C4-HSL(RhlI), 3-oxo-C12-HSL (LasI) | P. aeruginosa | Arabidopsis | growth promotion | [51] |
| 3-hydroxy-C4-HSL | Vibrio harveyi | Tobacco | plant resistance | [52] |
| C6-HSL | P. aeruginosa | A. thaliana, wheat | root growth, enhances cereal crop resistance to pathogens and abiotic stress | [29,53] |
| C10-HSL to C14-HSL | P. aeruginosa | Barley and Arabidopsis | resistance toward biotrophic and hemibiotrophic pathogens | [54,55] |
| C8-HSL, C7-HSL | Castellaniella defragrans, Cryobacterium sp. | Mortierella alpine A-178 | colonization | [56] |
| C6-and C8- HSL | S. liquefaciens, Pseudomonas putida | Tomato | ISR-like response | [57] |
| 3-OH-C10-HSL | Acidovorax radicis N35 | Barley | colonization of roots | |
| 3-oxo-C14-HSL | Sinorhizobium meliloti | M. truncatula | nodulation in roots | [58] |
| C12-HSL and C16-HSL | Agrobacterium vitis | M. truncatula, A. thaliana, Hordeum vulgare | AHL-priming | [59] |
| oxo-C12-HSL, oxo-C16-HSL | Ensifer meliloti P. aeruginosa | M. truncatula | auxin-responsive and flavonoid synthesis; mimicking QS secretion | [60] |
| oxo-C14-HSL | S. meliloti, Ensifer melilot | Arabidopsis barley, wheat, and tomato M. truncatula | AHL-priming for agriculture root nodulation in M. truncatula | [61,62] |
| 1-aminocyclopropane-1-carboxylate indole-3-acetic acid | Burkholderia phytofirmans | Phaseolus vulgaris | endophytically colonizes and promotes plant growth, forms symbiotic nodules and fix nitrogen | [63] |
| furanosyl borate diester (AI-2) | Pasteurella, Photorhabdus, Haemophilus, and Bacillus | Zoosporic plants | promoting plant infection | [64] |
| pyrazinone derivative (AI-3) | E. coli, Shigella sp. and Salmonella sp. | Animals | virulence | [65] |
| cyclic dipeptides | H. marmoreus | Arabidopsis | triggers plant immunity | [66,67] |
| CAI-1 | Vibrio cholerae | / | / | [68,69] |
| (R)-3-OH PAME, (R)-3-OH MAME | R. solanacearum | Tomato, tobacco, and potato | pathogenicity | [70,71] |
| indole-3-acetic acid | Azospirillum, Rhizobacteria | Citrus cinensis Arabidopsis | root formation | [72] |
| N-3-oxo-hexanoyl-homoserine | M. truncatula | Arabidopsis and wheat | enhances salt tolerance, primary root elongation | [73] |
| 3-oxo-C6-HSL | Pantoea stewartii | Mung beans, Arabidopsis | plant pathogen | [74] |
| 3-oxo-C14-HSL | S. meliloti | Mung beans barley, wheat, and tomato | nitrogen-fixing symbiont, plant immunity | [75] |
| 3-oxo-9-cis-C16-HSL | S. Meliloti, P. aeruginosa | Mung beans | nitrogen-fixing symbiont; induces auxin response and flavonoid synthesis | [76,77] |
| 3-hydroxy-7-cis-C14-HSL | Rhizobium leguminosarum | Mung beans | nitrogen-fixing symbiont | [78] |
| 9-cis-C16-HSL | Sinorhizobium melioti | Medicago | nitrogen-fixing symbiont | [79,80] |
| farnesol | Candida albicans, Trichoderma harzianum | Tomato | plant defense, regulates morphogenesis, biofilm development, sporulation, mating, drug efflux, and apoptosis, | [38,41,81] |
| Phenylethanol, tryptophol | Yeast | A. thaliana and tomato | drives filamentation | [38,39] |
| 2-ethyl-1-hexanol | F. oxysporum | A. thaliana and tomato | enhances plant growth | [40] |
| α-factor | Saccharomyces cerevisiae, Aspergillus fumigatus | Tomato | infection | [42] |
| ascr#1, ascr#3, ascr#9, ascr#10, ascr#18, oscr#9 | M. Incognita, M. javanica, M. hapla, H. glycines, Pratylenchus brachyurus | Arabidopsis, tomato, potato and barley | resistance to plant pathogens | [49,50] |
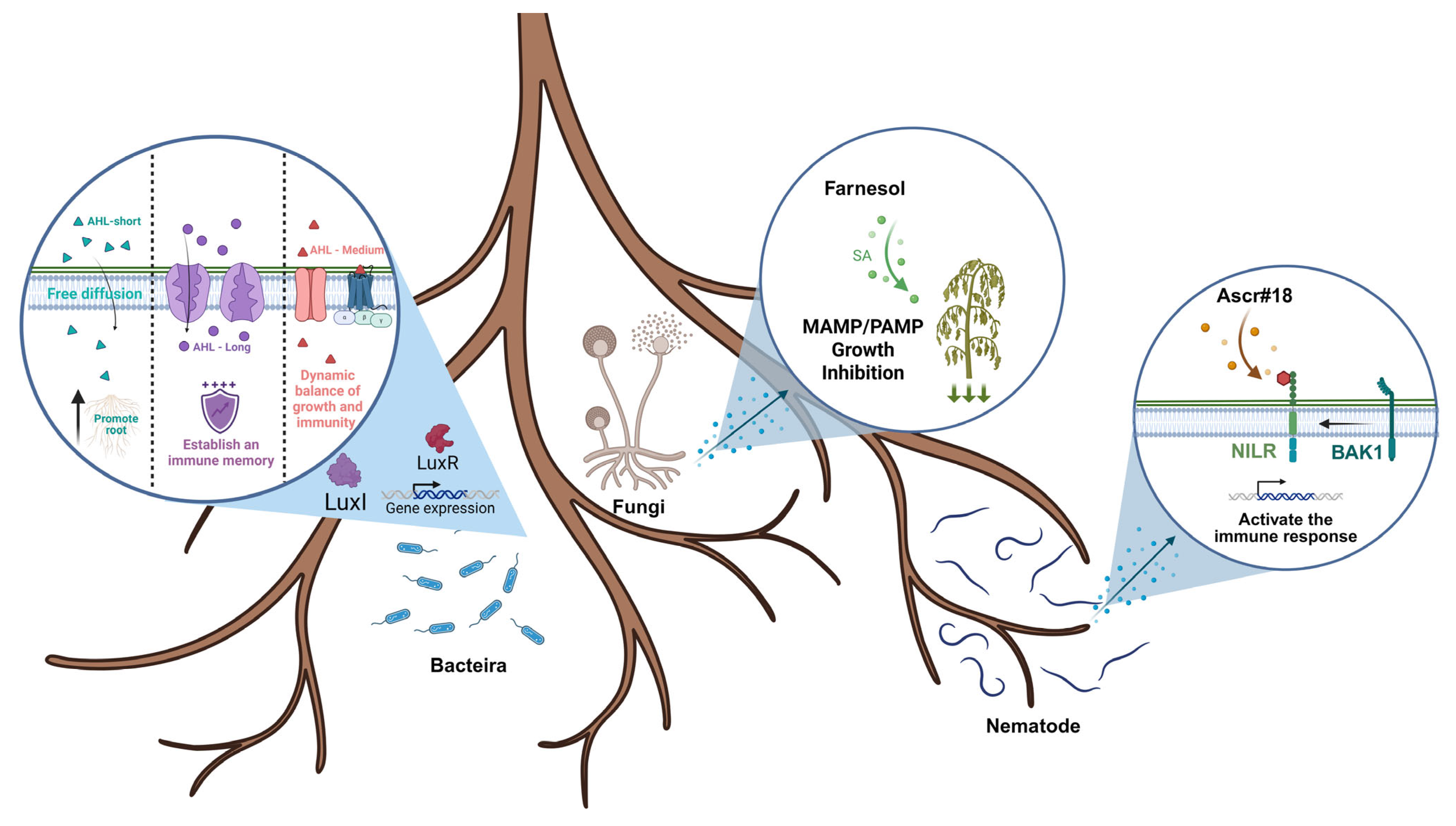
4. Perception of and Responses to QSMs
4.1. Molecular Mechanisms of QSM Perception in Plants
4.2. Plant Responses to AHLs
4.3. Plant Responses to Fungal QSMs
4.4. Plant Responses to Nematode QSMs
5. Strategies Used by Plants to Disrupt Pathogen QSMs
5.1. Metabolic Modification of Pathogen QSMs
5.2. QS Mimics Enabling Receptor Interference
5.3. Quorum Quenching
6. QS Plant Immunity and Sustainable Solutions
6.1. Metabolic Responses
6.2. Ecological Impacts
6.3. Agricultural Applications
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Term | Definition |
| DSF | Diffusible signaling factor |
| QS | Quorum sensing |
| QSM | Quorum sensing molecular |
| QSI | Quorum sensing inhibitor |
| PTI | Pattern-triggered immunity |
| MAPK Cascades | Mitogen-activated protein kinase signaling pathways |
| WRKY/MYB | Plant transcription factor families |
| AHL | Acyl-homoserine lactone |
| MAMP | Microbe-associated molecular pattern |
| PAMP | Pathogen-associated molecular patterns |
| JA | Jasmonic acid |
| SA | Salicylic acid |
References
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory mechanisms and promising applications of quorum sensing-inhibiting agents in control of bacterial biofilm formation. Front. Microbiol. 2020, 11, 589640. [Google Scholar] [CrossRef]
- Tanet, L.; Tamburini, C.; Baumas, C.; Garel, M.; Simon, G.; Casalot, L. Bacterial bioluminescence: Light emission in Photobacterium phosphoreum is not under quorum-sensing control. Front. Microbiol. 2019, 10, 365. [Google Scholar] [CrossRef]
- Polke, M.; Jacobsen, I.D. Quorum sensing by farnesol revisited. Curr. Genet. 2017, 63, 791–797. [Google Scholar] [CrossRef] [PubMed]
- von Reuss, S.H. Exploring modular glycolipids involved in nematode chemical communication. Chimia 2018, 72, 297–303. [Google Scholar] [CrossRef]
- Duan, Y.; Han, M.; Grimm, M.; Ponath, J.; Reichelt, M.; Mithöfer, A.; Schikora, A. Combination of bacterial N-acyl homoserine lactones primes Arabidopsis defenses via jasmonate metabolism. Plant Physiol. 2023, 191, 2027–2044. [Google Scholar] [CrossRef]
- Schenk, S.T.; Hernández-Reyes, C.; Samans, B.; Stein, E.; Neumann, C.; Schikora, M.; Reichelt, M.; Mithöfer, A.; Becker, A.; Kogel, K.H. N-acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell 2014, 26, 2708–2723. [Google Scholar] [CrossRef]
- Nagi, M.; Chapple, I.L.; Sharma, P.; Kuehne, S.A.; Hirschfeld, J. Quorum sensing in oral biofilms: Influence on host cells. Microorganisms 2023, 11, 1688. [Google Scholar] [CrossRef] [PubMed]
- Holm, A.; Vikström, E. Quorum sensing communication between bacteria and human cells: Signals, targets, and functions. Front. Plant Sci. 2014, 5, 309. [Google Scholar] [CrossRef]
- Husain, F.M.; Ahmad, I.; Baig, M.H.; Khan, M.S.; Khan, M.S.; Hassan, I.; Al-Shabib, N.A. Broad-spectrum inhibition of AHL-regulated virulence factors and biofilms by sub-inhibitory concentrations of ceftazidime. RSC Adv. 2016, 6, 27952–27962. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- Bernabè, G.; Marzaro, G.; Di Pietra, G.; Otero, A.; Bellato, M.; Pauletto, A.; Scarpa, M.; Sut, S.; Chilin, A.; Dall’Acqua, S. A novel phenolic derivative inhibits AHL-dependent quorum sensing signaling in Pseudomonas aeruginosa. Front. Pharmacol. 2022, 13, 996871. [Google Scholar] [CrossRef] [PubMed]
- Brencic, A.; Winans, S.C. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 2005, 69, 155–194. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C.; Patel, S.K.; Kang, Y.C.; Lee, J.K. Quorum sensing inhibitors as antipathogens: Biotechnological applications. Biotechnol. Adv. 2019, 37, 68–90. [Google Scholar] [CrossRef]
- Zhang, Z.S.; Zhao, D.S.; Zhu, D.; Guan, M.; Xiong, L.T.; He, Z.; Li, Y.; Shi, Y.; Xu, Z.L.; Deng, X. Design, synthesis, and biological evaluation of asymmetrical disulfides based on garlic extract as Pseudomonas aeruginosa pqs quorum sensing inhibitors. J. Agric. Food Chem. 2025, 73, 5850–5859. [Google Scholar] [CrossRef]
- Escobar-Muciño, E.; Arenas-Hernández, M.M.; Luna-Guevara, M.L. Mechanisms of Inhibition of Quorum Sensing as an Alternative for the Control of E. coli and Salmonella. Microorganisms 2022, 10, 884. [Google Scholar] [CrossRef]
- Zeng, X.; Zou, Y.; Zheng, J.; Qiu, S.; Liu, L.; Wei, C. Quorum sensing-mediated microbial interactions: Mechanisms, applications, challenges and perspectives. Microbiol. Res. 2023, 273, 127414. [Google Scholar] [CrossRef]
- Wang, S.; Payne, G.F.; Bentley, W.E. Quorum sensing communication: Molecularly connecting cells, their neighbors, and even devices. Annu. Rev. Chem. Biomol. Eng. 2020, 11, 447–468. [Google Scholar] [CrossRef]
- Huang, S.; Liu, X.; Yang, W.; Ma, L.; Li, H.; Liu, R.; Qiu, J.; Li, Y. Insights into adaptive mechanisms of extreme acidophiles based on quorum sensing/quenching-related proteins. Msystems 2022, 7, e01491-21. [Google Scholar] [CrossRef]
- Li, K.; Ma, C.; Zhou, X.; Xiong, C.; Wang, B.; Wang, Y.; Liu, F. Regulatory effects of diverse DSF family quorum-sensing signals in plant-associated bacteria. Mol. Plant-Microbe Interact. 2024, 37, 6–14. [Google Scholar] [CrossRef]
- Belizário, J.E.; Sulca-Lopez, M.; Sircili, M.; Faintuch, J. Role of small volatile signaling molecules in the regulation of bacterial antibiotic resistance and quorum sensing systems. In Trends in Quorum Sensing and Quorum Quenching; CRC Press: Boca Raton, FL, USA, 2020; pp. 215–223. [Google Scholar]
- Yoshihara, A.; Shimatani, M.; Sakata, M.; Takemura, C.; Senuma, W.; Hikichi, Y.; Kai, K. Quorum sensing inhibition attenuates the virulence of the plant pathogen Ralstonia solanacearum species complex. ACS Chem. Biol. 2020, 15, 3050–3059. [Google Scholar] [CrossRef]
- Pérez-Velázquez, J.; Quiñones, B.; Hense, B.A.; Kuttler, C. A mathematical model to investigate quorum sensing regulation and its heterogeneity in Pseudomonas syringae on leaves. Ecol. Complex. 2015, 21, 128–141. [Google Scholar] [CrossRef]
- Ferluga, S.; Bigirimana, J.; Höfte, M.; Venturi, V. A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Mol. Plant Pathol. 2007, 8, 529–538. [Google Scholar] [CrossRef]
- Calatrava-Morales, N.; McIntosh, M.; Soto, M.J. Regulation mediated by N-acyl homoserine lactone quorum sensing signals in the rhizobium-legume symbiosis. Genes 2018, 9, 263. [Google Scholar] [CrossRef] [PubMed]
- Churchill, M.E.; Sibhatu, H.M.; Uhlson, C.L. Defining the structure and function of acyl-homoserine lactone autoinducers. In Quorum Sensing: Methods and Protocols; Humana Press: Totawa, NJ, USA, 2011; pp. 159–171. [Google Scholar]
- Churchill, M.E.; Chen, L. Structural basis of acyl-homoserine lactone-dependent signaling. Chem. Rev. 2011, 111, 68–85. [Google Scholar] [CrossRef]
- Thiel, V.; Kunze, B.; Verma, P.; Wagner-Döbler, I.; Schulz, S. New structural variants of homoserine lactones in bacteria. ChemBioChem 2009, 10, 1861–1868. [Google Scholar] [CrossRef] [PubMed]
- von Rad, U.; Klein, I.; Dobrev, P.I.; Kottova, J.; Zazimalova, E.; Fekete, A.; Hartmann, A.; Schmitt-Kopplin, P.; Durner, J. Response of Arabidopsis thaliana to N-hexanoyl-DL-homoserine-lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta 2008, 229, 73–85. [Google Scholar] [CrossRef]
- Vendeville, A.; Winzer, K.; Heurlier, K.; Tang, C.M.; Hardie, K.R. Making ‘sense’ of metabolism: Autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 2005, 3, 383–396. [Google Scholar] [CrossRef]
- Zhang, L.; Li, S.; Liu, X.; Wang, Z.; Jiang, M.; Wang, R.; Xie, L.; Liu, Q.; Xie, X.; Shang, D. Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat. Commun. 2020, 11, 5371. [Google Scholar] [CrossRef]
- Põllumaa, L.; Alamäe, T.; Mäe, A. Quorum sensing and expression of virulence in pectobacteria. Sensors 2012, 12, 3327–3349. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhang, H.; Shu, X.; Sun, X.; Feng, H.; Xu, Z.; Kovács, Á.T.; Zhang, R.; Liu, Y. Autoinducer-2 relieves soil stress-induced dormancy of Bacillus velezensis by modulating sporulation signaling. NPJ Biofilms Microbiomes 2024, 10, 117. [Google Scholar] [CrossRef]
- Jung, B.K.; Ibal, J.C.; Pham, H.Q.; Kim, M.C.; Park, G.S.; Hong, S.J.; Jo, H.W.; Park, C.E.; Choi, S.D.; Jung, Y. Quorum sensing system affects the plant growth promotion traits of Serratia fonticola GS2. Front. Microbiol. 2020, 11, 536865. [Google Scholar] [CrossRef] [PubMed]
- He, Y.W.; Deng, Y.; Miao, Y.; Chatterjee, S.; Tran, T.M.; Tian, J.; Lindow, S. DSF-family quorum sensing signal-mediated intraspecies, interspecies, and inter-kingdom communication. Trends Microbiol. 2023, 31, 36–50. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, F.; Song, C.; Zhai, T.; He, Z.; Ma, L.; Zhao, X.; Jia, Z.; Song, S. Diffusible signal factor primes plant immunity against Xanthomonas campestris pv. campestris (Xcc) via JA signaling in Arabidopsis and Brassica oleracea. Front. Cell. Infect. Microbiol. 2023, 13, 1203582. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, D.E.; Sintim, H.O. Quorum sensing autoinducer-3 finally yields to structural elucidation. ACS Cent. Sci. 2020, 6, 93–96. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, Y.G.; Khadke, S.K.; Lee, J. Antibiofilm and antifungal activities of medium-chain fatty acids against Candida albicans via mimicking of the quorum-sensing molecule farnesol. Microb. Biotechnol. 2021, 14, 1353–1366. [Google Scholar] [CrossRef]
- Ma, R.; Huang, Z.; Jiang, Y.; Zhao, A.; Yu, G.; Lin, C.; Zhu, L.; Zhang, X.; Li, X.; Wang, C. Regulation of ethanol production from anaerobic fermentation of food waste using aromatic alcohol-based quorum-sensing molecules. J. Environ. Manag. 2025, 376, 124382. [Google Scholar] [CrossRef] [PubMed]
- Veiga, F.F.; Marcomini, E.K.; Salvador, A.; Chiavelli, L.U.R.; Barros, I.L.E.; de Castro, L.V.; Lucca, D.L.; Ochikubo, L.M.K.; Baesso, M.L.; Pomini, A.M. Detection of 2-ethyl-1-hexanol and its modulating effect in biofilm of Fusarium oxysporum. Mol. Microbiol. 2024, 122, 630–642. [Google Scholar] [CrossRef]
- Hogan, D.A. Talking to themselves: Autoregulation and quorum sensing in fungi. Eukaryot. Cell 2006, 5, 613–619. [Google Scholar] [CrossRef]
- Wendland, J. Growth, Differentiation and Sexuality; Springer: Berlin/Heidelberg, Germany, 2016; Volume 1. [Google Scholar]
- Pungaliya, C.; Srinivasan, J.; Fox, B.W.; Malik, R.U.; Ludewig, A.H.; Sternberg, P.W.; Schroeder, F.C. A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2009, 106, 7708–7713. [Google Scholar] [CrossRef]
- Braendle, C. Pheromones: Evolving language of chemical communication in nematodes. Curr. Biol. 2012, 22, R294–R296. [Google Scholar] [CrossRef]
- Jeong, P.Y.; Jung, M.; Yim, Y.H.; Kim, H.; Park, M.; Hong, E.; Lee, W.; Kim, Y.H.; Kim, K.; Paik, Y.K. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature 2005, 433, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Wrobel, C.J.; Schroeder, F.C. Repurposing degradation pathways for modular metabolite biosynthesis in nematodes. Nat. Chem. Biol. 2023, 19, 676–686. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.R.; Ng, J.L.P.; Mathesius, U. Interaction of symbiotic rhizobia and parasitic root-knot nematodes in legume roots: From molecular regulation to field application. Mol. Plant-Microbe Interact. 2021, 34, 470–490. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, J.; Zheng, X.; Wang, X. Nematode pheromones: Structures and functions. Molecules 2023, 28, 2409. [Google Scholar] [CrossRef]
- Huang, L.; Yuan, Y.; Lewis, C.; Kud, J.; Kuhl, J.C.; Caplan, A.; Dandurand, L.M.; Zasada, I.; Xiao, F. NILR1 perceives a nematode ascaroside triggering immune signaling and resistance. Curr. Biol. 2023, 33, 3992–3997.e3. [Google Scholar] [CrossRef]
- Manosalva, P.; Manohar, M.; Von Reuss, S.H.; Chen, S.; Koch, A.; Kaplan, F.; Choe, A.; Micikas, R.J.; Wang, X.; Kogel, K.H. Conserved nematode signalling molecules elicit plant defenses and pathogen resistance. Nat. Commun. 2015, 6, 7795. [Google Scholar] [CrossRef]
- Pesci, E.C.; Iglewski, B. Signalling in Pseudomonas aeruginosa. In Symposia-Society for General Microbiology; Cambridge University Press: Cambridge, UK, 1999; pp. 105–116. [Google Scholar]
- Milton, D.L.; Chalker, V.J.; Kirke, D.; Hardman, A.; Cámara, M.; Williams, P. The LuxM homologue VanM from Vibrio anguillarum directs the synthesis of N-(3-hydroxyhexanoyl) homoserine lactone and N-hexanoylhomoserine lactone. J. Bacteriol. 2001, 183, 3537–3547. [Google Scholar] [CrossRef]
- Moshynets, O.V.; Babenko, L.M.; Rogalsky, S.P.; Iungin, O.S.; Foster, J.; Kosakivska, I.V.; Potters, G.; Spiers, A.J. Priming winter wheat seeds with the bacterial quorum sensing signal N-hexanoyl-L-homoserine lactone (C6-HSL) shows potential to improve plant growth and seed yield. PLoS ONE 2019, 14, e0209460. [Google Scholar] [CrossRef]
- Lu, X.; Sun, H.; Li, X.; Li, C.; Wang, J.; Zhou, D. C14-HSL limits the mycelial morphology of pathogen Trichosporon cells but enhances their aggregation: Mechanisms and implications. Chin. Chem. Lett. 2024, 35, 108936. [Google Scholar] [CrossRef]
- Lu, X.; Wang, Y.; Feng, Z.; Fu, L.; Zhou, D. Bacterial signal C10-HSL stimulates spore germination of Galactomyces geotrichum by transboundary interaction. Chin. Chem. Lett. 2023, 34, 107617. [Google Scholar] [CrossRef]
- Kai, K.; Furuyabu, K.; Tani, A.; Hayashi, H. Production of the quorum-sensing molecules N-acylhomoserine lactones by endobacteria associated with Mortierella alpina A-178. ChemBioChem 2012, 13, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- Schuhegger, R.; Ihring, A.; Gantner, S.; Bahnweg, G.; Knappe, C.; Vogg, G.; Hutzler, P.; Schmid, M.; Van Breusegem, F.; Eberl, L. Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 2006, 29, 909–918. [Google Scholar] [CrossRef]
- Veliz-Vallejos, D.F.; Van Noorden, G.E.; Yuan, M.; Mathesius, U. A Sinorhizobium meliloti-specific N-acyl homoserine lactone quorum-sensing signal increases nodule numbers in Medicago truncatula independent of autoregulation. Front. Plant Sci. 2014, 5, 551. [Google Scholar] [CrossRef]
- Savka, M.A.; Le, P.T.; Burr, T.J. LasR receptor for detection of long-chain quorum-sensing signals: Identification of N-acyl-homoserine lactones encoded by the avsI locus of Agrobacterium vitis. Curr. Microbiol. 2011, 62, 101–110. [Google Scholar] [CrossRef]
- Mathesius, U.; Mulders, S.; Gao, M.; Teplitski, M.; Caetano-Anollés, G.; Rolfe, B.G.; Bauer, W.D. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci. USA 2003, 100, 1444–1449. [Google Scholar] [CrossRef]
- Schikora, A.; Schenk, S.T.; Hartmann, A. Beneficial effects of bacteria-plant communication based on quorum sensing molecules of the N-acyl homoserine lactone group. Plant Mol. Biol. 2016, 90, 605–612. [Google Scholar] [CrossRef]
- Shrestha, A.; Grimm, M.; Ojiro, I.; Krumwiede, J.; Schikora, A. Impact of quorum sensing molecules on plant growth and immune system. Front. Microbiol. 2020, 11, 1545. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.L.; Chang, Y.C.; Kurniawan, A.; Chang, P.C.; Liou, T.Y.; Wang, W.D.; Chuang, H.W. Employing Genomic Tools to Explore the molecular mechanisms behind the enhancement of plant growth and stress resilience facilitated by a Burkholderia Rhizobacterial strain. Int. J. Mol. Sci. 2024, 25, 6091. [Google Scholar] [CrossRef]
- Kong, P.; Tyler, B.M.; Richardson, P.A.; Lee, B.W.; Zhou, Z.S.; Hong, C. Zoospore interspecific signaling promotes plant infection by Phytophthora. BMC Microbiol. 2010, 10, 313. [Google Scholar] [CrossRef]
- Majdura, J.; Jankiewicz, U.; Gałązka, A.; Orzechowski, S. The role of quorum sensing molecules in bacterial–plant interactions. Metabolites 2023, 13, 114. [Google Scholar] [CrossRef] [PubMed]
- Prasad, C. Bioactive cyclic dipeptides. Peptides 1995, 16, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.J.; Liu, Y.C.; Weng, C.H.; Sun, S.W.; Li, F.; Li, H.; Zhu, H. Cyclic dipeptides mediating quorum sensing and their biological effects in Hypsizygus marmoreus. Biomolecules 2020, 10, 298. [Google Scholar] [CrossRef]
- Kelly, R.C.; Bolitho, M.E.; Higgins, D.A.; Lu, W.; Ng, W.L.; Jeffrey, P.D.; Rabinowitz, J.D.; Semmelhack, M.F.; Hughson, F.M.; Bassler, B.L. The Vibrio cholerae quorum-sensing autoinducer CAI-1: Analysis of the biosynthetic enzyme CqsA. Nat. Chem. Biol. 2009, 5, 891–895. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.J.; Ng, W.L.; Marano, P.; Brook, K.; Bassler, B.L.; Semmelhack, M.F. Role of the CAI-1 fatty acid tail in the Vibrio cholerae quorum sensing response. J. Med. Chem. 2012, 55, 9669–9681. [Google Scholar] [CrossRef]
- Kai, K.; Ohnishi, H.; Shimatani, M.; Ishikawa, S.; Mori, Y.; Kiba, A.; Ohnishi, K.; Tabuchi, M.; Hikichi, Y. Methyl 3-hydroxymyristate, a diffusible signal mediating phc quorum sensing in Ralstonia solanacearum. ChemBioChem 2015, 16, 2309–2318. [Google Scholar] [CrossRef] [PubMed]
- Ujita, Y.; Sakata, M.; Yoshihara, A.; Hikichi, Y.; Kai, K. Signal production and response specificity in the phc quorum sensing systems of Ralstonia solanacearum species complex. ACS Chem. Biol. 2019, 14, 2243–2251. [Google Scholar]
- Spaepen, S.; Dobbelaere, S.; Croonenborghs, A.; Vanderleyden, J. Effects of Azospirillum brasilense indole-3-acetic acid production on inoculated wheat plants. Plant Soil 2008, 312, 15–23. [Google Scholar] [CrossRef]
- Zhao, Q.; Yang, X.Y.; Li, Y.; Liu, F.; Cao, X.Y.; Jia, Z.H.; Song, S.S. N-3-oxo-hexanoyl-homoserine lactone, a bacterial quorum sensing signal, enhances salt tolerance in Arabidopsis and wheat. Bot. Stud. 2020, 61, 8. [Google Scholar] [CrossRef]
- Carlier, A.L. Regulation of Surface Polysaccharides in Pantoea stewartii subsp. Stewartia. Ph.D. Thesis, University of Connecticut, Storrs, CT, USA, 2008. [Google Scholar]
- Gao, M.; Chen, H.; Eberhard, A.; Gronquist, M.R.; Robinson, J.B.; Rolfe, B.G.; Bauer, W.D. sinI- and expR-Dependent Quorum Sensing in Sinorhizobium meliloti. J. Bacteriol. 2005, 187, 7931–7944. [Google Scholar] [CrossRef]
- Marketon, M.M.; Gronquist, M.R.; Eberhard, A.; González, J.E. Characterization of the Sinorhizobium meliloti sinR/sinI Locus and the Production of Novel N-Acyl Homoserine Lactones. J. Bacteriol. 2002, 184, 5686–5695. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Hernández-Reyes, C.; Grimm, M.; Krumwiede, J.; Stein, E.; Schenk, S.T.; Schikora, A. AHL-priming protein 1 mediates N-3-oxo-tetradecanoyl-homoserine lactone priming in Arabidopsis. BMC Biol. 2022, 20, 268. [Google Scholar] [CrossRef] [PubMed]
- Schripsema, J.; De Rudder, K.; Van Vliet, T.; Lankhorst, P.P.; De Vroom, E.; Kijne, J.W.; Van Brussel, A. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-L-homoserine lactone molecules, known as autoinducers and as quorum sensing co-transcription factors. J. Bacteriol. 1996, 178, 366–371. [Google Scholar] [CrossRef]
- Wagner-Döbler, I.; Thiel, V.; Eberl, L.; Allgaier, M.; Bodor, A.; Meyer, S.; Ebner, S.; Hennig, A.; Pukall, R.; Schulz, S. Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine alphaproteobacteria. Chembiochem 2005, 6, 2195–2206. [Google Scholar] [CrossRef] [PubMed]
- Teplitski, M.; Eberhard, A.; Gronquist, M.R.; Gao, M.; Robinson, J.B.; Bauer, W.D. Chemical identification of N-acyl homoserine lactone quorum-sensing signals produced by Sinorhizobium meliloti strains in defined medium. Arch. Microbiol. 2003, 180, 494–497. [Google Scholar] [CrossRef]
- Kües, U.; Fischer, R. Growth, Differentiation and Sexuality; Springer: Berlin/Heidelberg, Germany, 2006; Volume 1. [Google Scholar]
- Liu, L.; Zeng, X.; Zheng, J.; Zou, Y.; Qiu, S.; Dai, Y. AHL-mediated quorum sensing to regulate bacterial substance and energy metabolism: A review. Microbiol. Res. 2022, 262, 127102. [Google Scholar] [CrossRef]
- Huang, Y.L.; Ki, J.S.; Case, R.J.; Qian, P.Y. Diversity and acyl-homoserine lactone production among subtidal biofilm-forming bacteria. Aquat. Microb. Ecol. 2008, 52, 185–193. [Google Scholar] [CrossRef]
- Jin, G.; Liu, F.; Ma, H.; Hao, S.; Zhao, Q.; Bian, Z.; Jia, Z.; Song, S. Two G-protein-coupled-receptor candidates, Cand2 and Cand7, are involved in Arabidopsis root growth mediated by the bacterial quorum-sensing signals N-acyl-homoserine lactones. Biochem. Biophys. Res. Commun. 2012, 417, 991–995. [Google Scholar] [CrossRef]
- Singh, K.; Chandra, R.; Purchase, D. Unraveling the secrets of rhizobacteria signaling in rhizosphere. Rhizosphere 2022, 21, 100484. [Google Scholar] [CrossRef]
- Sieper, T.; Forczek, S.; Matucha, M.; Krämer, P.; Hartmann, A.; Schröder, P. N-acyl-homoserine lactone uptake and systemic transport in barley rest upon active parts of the plant. New Phytol. 2014, 201, 545–555. [Google Scholar] [CrossRef]
- Vesty, E.F.; Whitbread, A.L.; Needs, S.; Tanko, W.; Jones, K.; Halliday, N.; Ghaderiardakani, F.; Liu, X.; Cámara, M.; Coates, J.C. Cross-kingdom signalling regulates spore germination in the moss Physcomitrella patens. Sci. Rep. 2020, 10, 2614. [Google Scholar] [CrossRef] [PubMed]
- Viswanath, G.; Sekar, J.; Prabavathy, V. Acyl homoserine lactone-producing rhizobacteria elicit systemic resistance in plants. In Microbial-Mediated Induced Systemic Resistance in Plants; Singapore: Springer Nature Singapore, 2016; pp. 135–146. [Google Scholar]
- Babenko, L.M.; Kosakivska, I.V.; Romanenko, K.O. Molecular mechanisms of N-acyl homoserine lactone signals perception by plants. Cell Biol. Int. 2022, 46, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Kariminik, A.; Baseri-Salehi, M.; Kheirkhah, B. Pseudomonas aeruginosa quorum sensing modulates immune responses: An updated review article. Immunol. Lett. 2017, 190, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Reyes, C.; Schenk, S.T.; Neumann, C.; Kogel, K.H.; Schikora, A. N-acyl-homoserine lactones-producing bacteria protect plants against plant and human pathogens. Microb. Biotechnol. 2014, 7, 580–588. [Google Scholar] [CrossRef]
- Cao, X.Y.; Zhao, Q.; Sun, Y.N.; Yu, M.X.; Liu, F.; Zhang, Z.; Jia, Z.H.; Song, S.S. Cellular messengers involved in the inhibition of the Arabidopsis primary root growth by bacterial quorum-sensing signal N-decanoyl-L-homoserine lactone. BMC Plant Biol. 2022, 22, 488. [Google Scholar] [CrossRef]
- Girard, L.; Lantoine, F.; Lami, R.; Vouvé, F.; Suzuki, M.T.; Baudart, J. Genetic diversity and phenotypic plasticity of AHL-mediated Quorum sensing in environmental strains of Vibrio mediterranei. ISME J. 2019, 13, 159–169. [Google Scholar] [CrossRef]
- Schenk, S.T.; Schikora, A. AHL-priming functions via oxylipin and salicylic acid. Front. Plant Sci. 2015, 5, 784. [Google Scholar] [CrossRef]
- Liu, F.; Bian, Z.; Jia, Z.; Zhao, Q.; Song, S. The GCR1 and GPA1 participate in promotion of Arabidopsis primary root elongation induced by N-acyl-homoserine lactones, the bacterial quorum-sensing signals. Mol. Plant-Microbe Interact. 2012, 25, 677–683. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, M.; Jia, Z.; Liu, F.; Ma, H.; Huang, Y.; Song, S. AtMYB44 positively regulates the enhanced elongation of primary roots induced by N-3-oxo-hexanoyl-homoserine lactone in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2016, 29, 774–785. [Google Scholar] [CrossRef]
- Li, Y.H.; Tian, X. Quorum sensing and bacterial social interactions in biofilms. Sensors 2012, 12, 2519–2538. [Google Scholar] [CrossRef]
- West, K.H.; Ma, S.V.; Pensinger, D.A.; Tucholski, T.; Tiambeng, T.N.; Eisenbraun, E.L.; Yehuda, A.; Hayouka, Z.; Ge, Y.; Sauer, J.D. Characterization of an autoinducing peptide signal reveals highly efficacious synthetic inhibitors and activators of quorum sensing and biofilm formation in Listeria monocytogenes. Biochemistry 2023, 62, 2878–2892. [Google Scholar] [CrossRef] [PubMed]
- Mohana Sheela, G.; Prathyusha, A.; Neelapu, N.R.R.; Bramhachari, P.V. Intra and inter-species communication in microbes: Living with complex and sociable neighbors. In Implication of Quorum Sensing System in Biofilm Formation and Virulence; Springer: Singapore, 2018; pp. 7–16. [Google Scholar]
- Velasco, P.; Lema, M.; Francisco, M.; Soengas, P.; Cartea, M.E. In vivo and in vitro effects of secondary metabolites against Xanthomonas campestris pv. campestris. Molecules 2013, 18, 11131–11143. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, J.; Zhang, Y.; Wang, N. Diffusible signal factor (DSF)-mediated quorum sensing modulates expression of diverse traits in Xanthomonas citri and responses of citrus plants to promote disease. BMC Genom. 2019, 20, 55. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Yu, J.; Pan, X.; Cao, H.; Song, T.; Liu, Y. Farnesol inhibits growth and development of Ustilaginoidea virens. New Plant Prot. 2024, 1, e17. [Google Scholar] [CrossRef]
- Cardoza, R.E.; McCormick, S.P.; Lindo, L.; Mayo-Prieto, S.; González-Cazón, D.; Martínez-Reyes, N.; Carro-Huerga, G.; Rodríguez-González, Á.; Proctor, R.H.; Casquero, P.A. Effect of farnesol in Trichoderma physiology and in fungal–plant interaction. J. Fungi 2022, 8, 1266. [Google Scholar] [CrossRef]
- Lindo, L.; Cardoza, R.E.; Lorenzana, A.; Casquero, P.A.; Gutiérrez, S. Identification of plant genes putatively involved in the perception of fungal ergosterol-squalene. J. Integr. Plant Biol. 2020, 62, 927–947. [Google Scholar] [CrossRef]
- La Camera, S.; Gouzerh, G.; Dhondt, S.; Hoffmann, L.; Fritig, B.; Legrand, M.; Heitz, T. Metabolic reprogramming in plant innate immunity: The contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 2004, 198, 267–284. [Google Scholar] [CrossRef]
- Shah, J. Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu. Rev. Phytopathol. 2005, 43, 229–260. [Google Scholar] [CrossRef]
- Christensen, S.A.; Kolomiets, M.V. The lipid language of plant–fungal interactions. Fungal Genet. Biol. 2011, 48, 4–14. [Google Scholar] [CrossRef]
- Manohar, M.; Tenjo-Castano, F.; Chen, S.; Zhang, Y.K.; Kumari, A.; Williamson, V.M.; Wang, X.; Klessig, D.F.; Schroeder, F.C. Plant metabolism of nematode pheromones mediates plant-nematode interactions. Nat. Commun. 2020, 11, 208. [Google Scholar] [CrossRef]
- Huang, L.; Yuan, Y.; Ramirez, C.; Zhao, Z.; Chen, L.; Griebel, T.; Kud, J.; Kuhl, J.C.; Caplan, A.; Dandurand, L.M. A receptor for dual ligands governs plant immunity and hormone response and is targeted by a nematode effector. Proc. Natl. Acad. Sci. USA 2024, 121, e2412016121. [Google Scholar] [CrossRef]
- Friesen, M.L. Social evolution and cheating in plant pathogens. Annu. Rev. Phytopathol. 2020, 58, 55–75. [Google Scholar] [CrossRef]
- Palmer, A.G.; Senechal, A.C.; Mukherjee, A.; Ané, J.M.; Blackwell, H.E. Plant responses to bacterial N-acyl L-homoserine lactones are dependent on enzymatic degradation to L-homoserine. ACS Chem. Biol. 2014, 9, 1834–1845. [Google Scholar] [CrossRef]
- Götz, C.; Fekete, A.; Gebefuegi, I.; Forczek, S.T.; Fuksová, K.; Li, X.; Englmann, M.; Gryndler, M.; Hartmann, A.; Matucha, M. Uptake, degradation and chiral discrimination of N-acyl-D/L-homoserine lactones by barley (Hordeum vulgare) and yam bean (Pachyrhizus erosus) plants. Anal. Bioanal. Chem. 2007, 389, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Roggatz, C.C.; Parsons, D.R. Potential climate change impacts on the abiotic degradation of acyl-homoserine lactones in the fluctuating conditions of marine biofilms. Front. Mar. Sci. 2022, 9, 882428. [Google Scholar] [CrossRef]
- Chen, A.Q.; Long, Z.Q.; Xiao, Y.; Feng, Y.M.; Zhou, Y.; Yang, S.; Liao, Y.M.; Zhou, X.; Liu, L.W.; Wu, Z.B. Application of natural product-based quorum sensing inhibitors in plant pathogen control: A review. Arab. J. Chem. 2024, 18, 106050. [Google Scholar] [CrossRef]
- See-Too, W.S.; Convey, P.; Pearce, D.A.; Chan, K.G. Characterization of a novel N-acylhomoserine lactonase, AidP, from Antarctic Planococcus sp. Microb. Cell Factories 2018, 17, 179. [Google Scholar] [CrossRef]
- Dong, Y.H.; Xu, J.L.; Li, X.Z.; Zhang, L.H. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 2000, 97, 3526–3531. [Google Scholar] [CrossRef]
- Sikdar, R.; Elias, M. Quorum quenching enzymes and their effects on virulence, biofilm, and microbiomes: A review of recent advances. Expert Rev. Anti Infect. Ther. 2020, 18, 1221–1233. [Google Scholar] [CrossRef]
- Vashistha, A.; Sharma, N.; Nanaji, Y.; Kumar, D.; Singh, G.; Barnwal, R.P.; Yadav, A.K. Quorum sensing inhibitors as Therapeutics: Bacterial biofilm inhibition. Bioorganic Chem. 2023, 136, 106551. [Google Scholar] [CrossRef]
- Naga, N.G.; Shaaban, M.I. Quorum Sensing and Quorum sensing inhibitors of natural origin. In Drug Discovery and Design Using Natural Products; Springer: Berlin/Heidelberg, Germany, 2023; pp. 395–416. [Google Scholar]
- Freckelton, M.L.; Høj, L.; Bowden, B.F. Quorum sensing interference and structural variation of quorum sensing mimics in Australian soft coral. Front. Mar. Sci. 2018, 5, 198. [Google Scholar] [CrossRef]
- Lima, E.M.F.; Winans, S.C.; Pinto, U.M. Quorum sensing interference by phenolic compounds – A matter of bacterial misunderstanding. Heliyon 2023, 9, e17657. [Google Scholar] [CrossRef]
- Qin, X.; Vila-Sanjurjo, C.; Singh, R.; Philipp, B.; Goycoolea, F.M. Screening of bacterial quorum sensing inhibitors in a vibrio fischeri LuxR-based synthetic fluorescent E. coli biosensor. Pharmaceuticals 2020, 13, 263. [Google Scholar] [CrossRef] [PubMed]
- Manefield, M.; Rasmussen, T.B.; Henzter, M.; Andersen, J.B.; Steinberg, P.; Kjelleberg, S.; Givskov, M. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 2002, 148, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Gurich, N.; González, J.E. Role of quorum sensing in Sinorhizobium meliloti-alfalfa symbiosis. J. Bacteriol. 2009, 191, 4372–4382. [Google Scholar] [CrossRef]
- Corral-Lugo, A.; Daddaoua, A.; Ortega, A.; Espinosa-Urgel, M.; Krell, T. Rosmarinic acid is a homoserine lactone mimic produced by plants that activates a bacterial quorum-sensing regulator. Sci. Signal. 2016, 9, ra1. [Google Scholar] [CrossRef]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rasamiravaka, T.; Stevigny, C.; Duez, P.; Rajaonson, S.; Diallo, B.; Mol, A.; Baucher, M.; El Jaziri, M. The flavanone naringenin reduces the production of quorum sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Microbiology 2011, 157, 2120–2132. [Google Scholar] [CrossRef]
- White, J.G.; Southgate, E.; Thomson, J.N.; Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. B 1986, 314, 1–340. [Google Scholar]
- Khan, M.S.A.; Zahin, M.; Hasan, S.; Husain, F.M.; Ahmad, I. Inhibition of quorum sensing regulated bacterial functions by plant essential oils with special reference to clove oil. J. Appl. Microbiol. 2009, 49, 354–360. [Google Scholar] [CrossRef]
- Keshavan, N.D.; Chowdhary, P.K.; Haines, D.C.; González, J.E. L-Canavanine made by Medicago sativa interferes with quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 2005, 187, 8427–8436. [Google Scholar] [CrossRef]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rajaonson, S.; Diallo, B.; Mol, A.; El Jaziri, M.; Baucher, M. Identification of catechin as one of the flavonoids from Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 2010, 76, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Garge, S.S.; Nerurkar, A.S. Attenuation of quorum sensing regulated virulence of Pectobacterium carotovorum subsp. carotovorum through an AHL lactonase produced by Lysinibacillus sp. Gs50. PLoS ONE 2016, 11, e0167344. [Google Scholar]
- Vesuna, A.; Nerurkar, A.S. Enzymatic quorum quenching for virulence attenuation of phytopathogenic bacteria. In Biotechnological Applications of Quorum Sensing Inhibitors; Springer: Singapore, 2018; pp. 447–473. [Google Scholar]
- Alramadhan, W.H.; Ejiofor, A.; Johnson, T. Degradation of N-Acyl Homoserine Lactone Quorum Sensing Signals by Bacillus thuringiensis AHL Lactonase. Adv. Microbiol. 2023, 13, 526–538. [Google Scholar] [CrossRef]
- Anandan, K.; Vittal, R.R. Quorum quenching strategies of endophytic Bacillus thuringiensis KMCL07 against soft rot pathogen Pectobacterium carotovorum subsp. carotovorum. Microb. Pathog. 2025, 200, 107356. [Google Scholar] [CrossRef]
- Lazarus, H.P.S.; Easwaran, N. Molecular insights into PGPR Fluorescent Pseudomonads complex mediated intercellular and interkingdom signal transduction mechanisms in promoting plant’s immunity. Res. Microbiol. 2024, 175, 104218. [Google Scholar] [CrossRef]
- Liao, J.; Li, Z.; Xiong, D.; Shen, D.; Wang, L.; Lin, L.; Shao, X.; Liao, L.; Li, P.; Zhang, L.Q. Quorum quenching by a type IVA secretion system effector. ISME J. 2023, 17, 1564–1577. [Google Scholar] [CrossRef]
- Shrestha, A.; Schikora, A. AHL-priming for enhanced resistance as a tool in sustainable agriculture. FEMS Microbiol. Ecol. 2020, 96, fiaa226. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Han, M.; Schikora, A. The coordinated responses of host plants to diverse N-acyl homoserine lactones. Plant Signal. Behav. 2024, 19, 2356406. [Google Scholar] [CrossRef]
- Duan, Y.; Han, M.; Grimm, M.; Schikora, A. Network analysis uncovers the master role of WRKY transcription factors in Arabidopsis thaliana response to N-acyl homoserine lactones. CABI Agric. Biosci. Home 2024, 5, 6. [Google Scholar] [CrossRef]
- Deryabin, D.; Galadzhieva, A.; Kosyan, D.; Duskaev, G. Plant-derived inhibitors of AHL-mediated quorum sensing in bacteria: Modes of action. Int. J. Mol. Sci. 2019, 20, 5588. [Google Scholar] [CrossRef]
- Przybylska, A.; Obrępalska-Stęplowska, A. Plant defense responses in monocotyledonous and dicotyledonous host plants during root-knot nematode infection. Plant Soil 2020, 451, 239–260. [Google Scholar] [CrossRef]
- Kamboj, A.; Thielmann, J.; Delfan, S.; Kloppe, T.; Schulz, P.; Manohar, M.; Schroeder, F.C.; Klessig, D.F.; Kogel, K.H. The nematode signaling molecule ascr#18 induces prepenetration defenses in wheat against a leaf rust fungus. J. Plant Dis. Prot. 2024, 131, 2053–2062. [Google Scholar]
- Zhou, J.; Lyu, Y.; Richlen, M.L.; Anderson, D.M.; Cai, Z. Quorum sensing is a language of chemical signals and plays an ecological role in algal-bacterial interactions. Crit. Rev. Plant Sci. 2016, 35, 81–105. [Google Scholar] [CrossRef] [PubMed]
- Jube, S.; Borthakur, D. Expression of bacterial genes in transgenic tobacco: Methods, applications and future prospects. Electron. J. Biotechnol. 2007, 10, 452–467. [Google Scholar] [CrossRef]
- Ghitti, E.; Rolli, E.; Crotti, E.; Borin, S. Flavonoids are intra-and inter-kingdom modulator signals. Microorganisms 2022, 10, 2479. [Google Scholar] [CrossRef]
- del Carmen Orozco-Mosqueda, M.; Fadiji, A.E.; Babalola, O.O.; Santoyo, G. Bacterial elicitors of the plant immune system: An overview and the way forward. Plant Stress 2023, 7, 100138. [Google Scholar] [CrossRef]
- Ban, H.; Chai, X.; Lin, Y.; Zhou, Y.; Peng, D.; Zhou, Y.; Zou, Y.; Yu, Z.; Sun, M. Transgenic Amorphophallus konjac expressing synthesized acyl-homoserine lactonase (aiiA) gene exhibit enhanced resistance to soft rot disease. Plant Cell Rep. 2009, 28, 1847–1855. [Google Scholar] [CrossRef]
- Kumar, V.; Agrawal, S.; Bhat, S.A.; Américo-Pinheiro, J.H.P.; Shahi, S.K.; Kumar, S. Environmental impact, health hazards, and plant-microbes synergism in remediation of emerging contaminants. Clean. Chem. Eng. 2022, 2, 100030. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Guasch-Vidal, B.; González-Barroso, S.; López-Baena, F.J.; Cubo, T.; Ollero, F.J.; Gil-Serrano, A.M.; Rodríguez-Carvajal, M.Á.; Bellogín, R.A.; Espuny, M.R. Nodulation-gene-inducing flavonoids increase overall production of autoinducers and expression of N-acyl homoserine lactone synthesis genes in rhizobia. Res. Microbiol. 2011, 162, 715–723. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, W.J.; Bhatt, K.; Zhou, Z.; Huang, Y.; Zhang, L.H.; Chen, S.; Wang, J. Innovative microbial disease biocontrol strategies mediated by quorum quenching and their multifaceted applications: A review. Front. Plant Sci. 2023, 13, 1063393. [Google Scholar] [CrossRef]
- D’Aquila, P.; De Rose, E.; Sena, G.; Scorza, A.; Cretella, B.; Passarino, G.; Bellizzi, D. Quorum Quenching Approaches against Bacterial-Biofilm-Induced Antibiotic Resistance. Antibiotics 2024, 13, 619. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Liu, J.; Wang, X. Quorum Signaling Molecules: Interactions Between Plants and Associated Pathogens. Int. J. Mol. Sci. 2025, 26, 5235. https://doi.org/10.3390/ijms26115235
Zheng X, Liu J, Wang X. Quorum Signaling Molecules: Interactions Between Plants and Associated Pathogens. International Journal of Molecular Sciences. 2025; 26(11):5235. https://doi.org/10.3390/ijms26115235
Chicago/Turabian StyleZheng, Xi, Junjie Liu, and Xin Wang. 2025. "Quorum Signaling Molecules: Interactions Between Plants and Associated Pathogens" International Journal of Molecular Sciences 26, no. 11: 5235. https://doi.org/10.3390/ijms26115235
APA StyleZheng, X., Liu, J., & Wang, X. (2025). Quorum Signaling Molecules: Interactions Between Plants and Associated Pathogens. International Journal of Molecular Sciences, 26(11), 5235. https://doi.org/10.3390/ijms26115235







