Deciphering the Overlapping Immune Mechanism Between Depression and Breast Cancer
Abstract
1. Introduction
2. Results
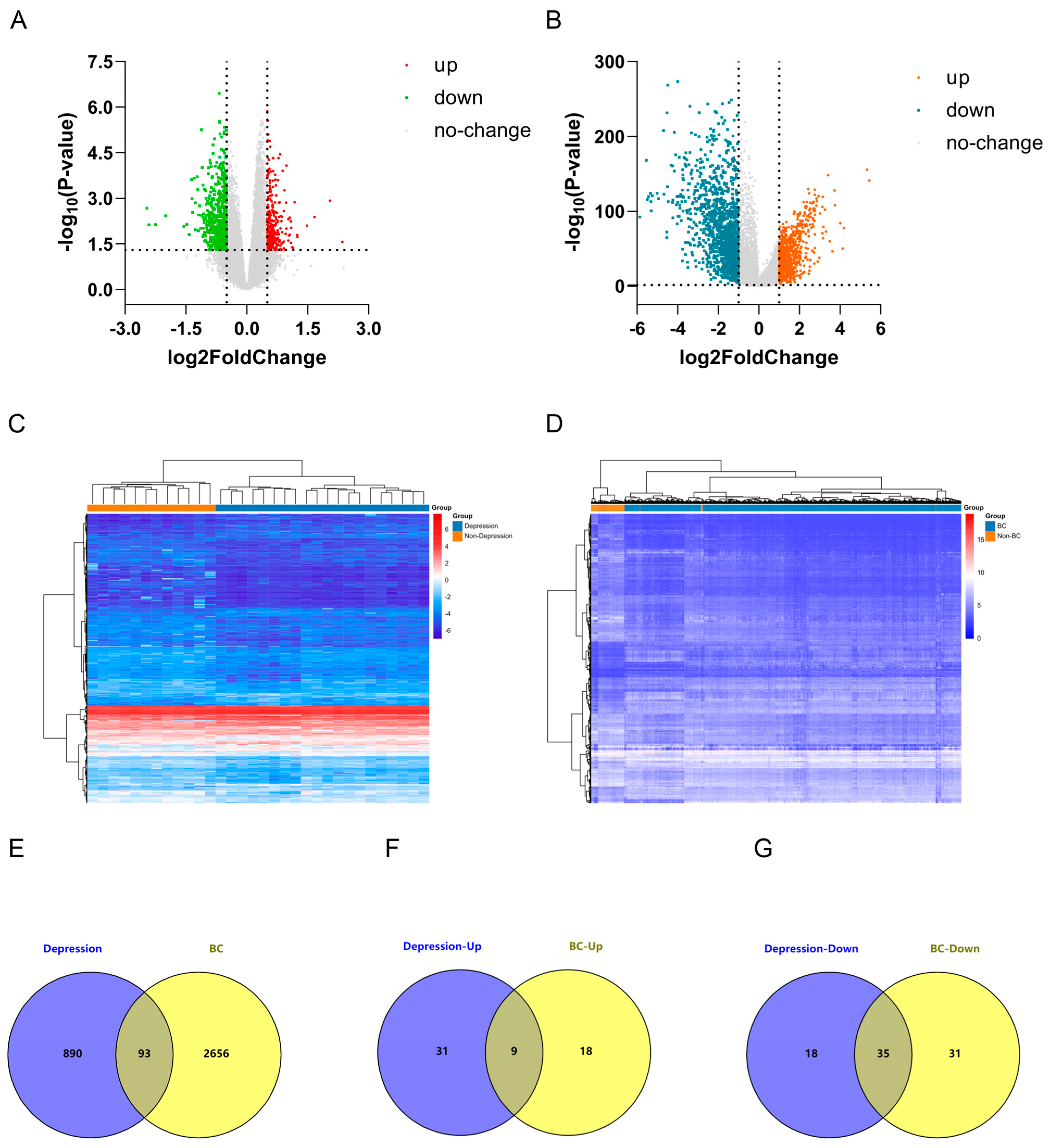
2.1. Identification of Differentially Expressed Genes (DEGs) in Depression and Breast Cancer (BC)
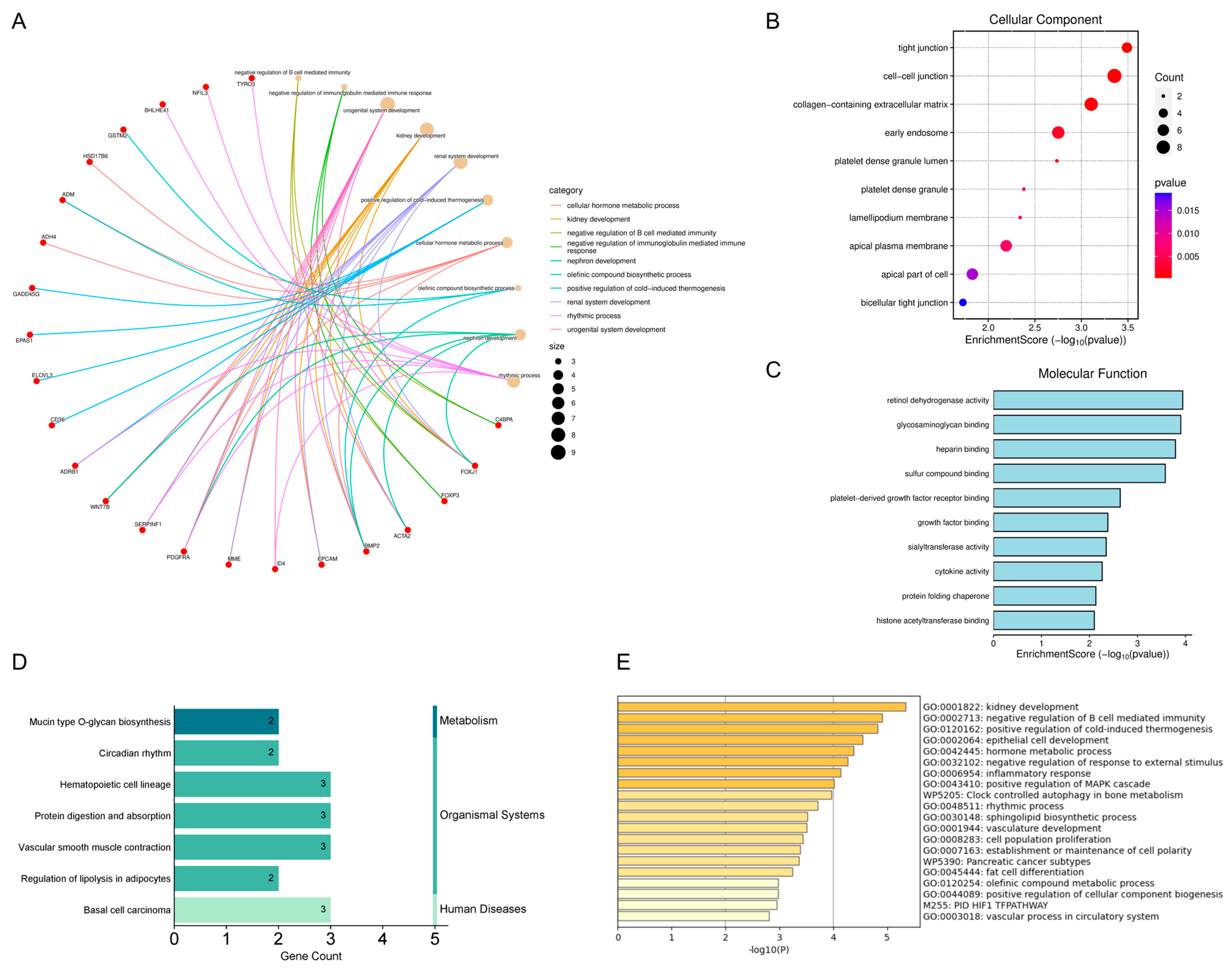
2.2. Results of Enrichment Analysis of Overlapping Genes
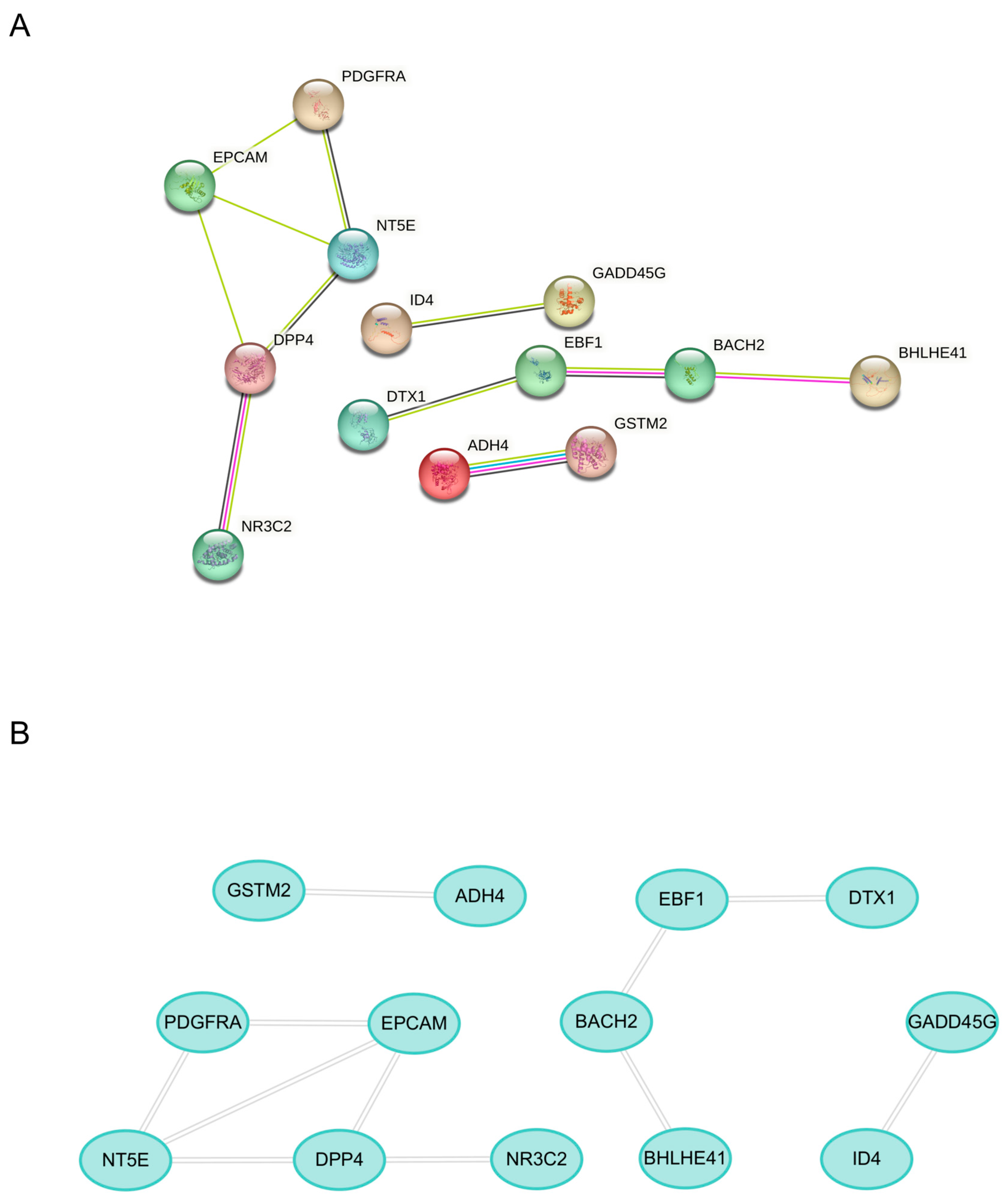
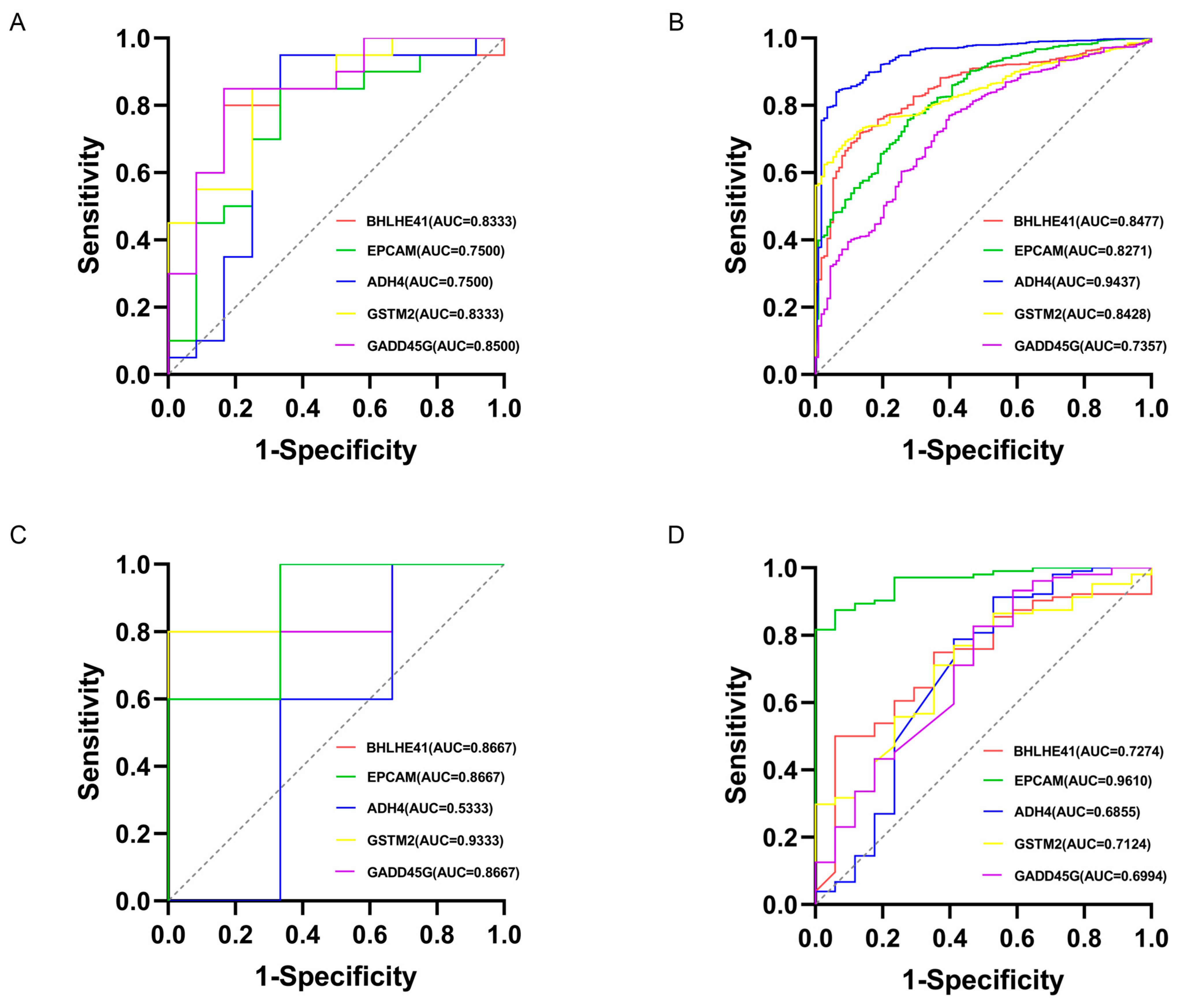
2.3. Machine Learning for Identification and Validation
2.4. Literature Validation
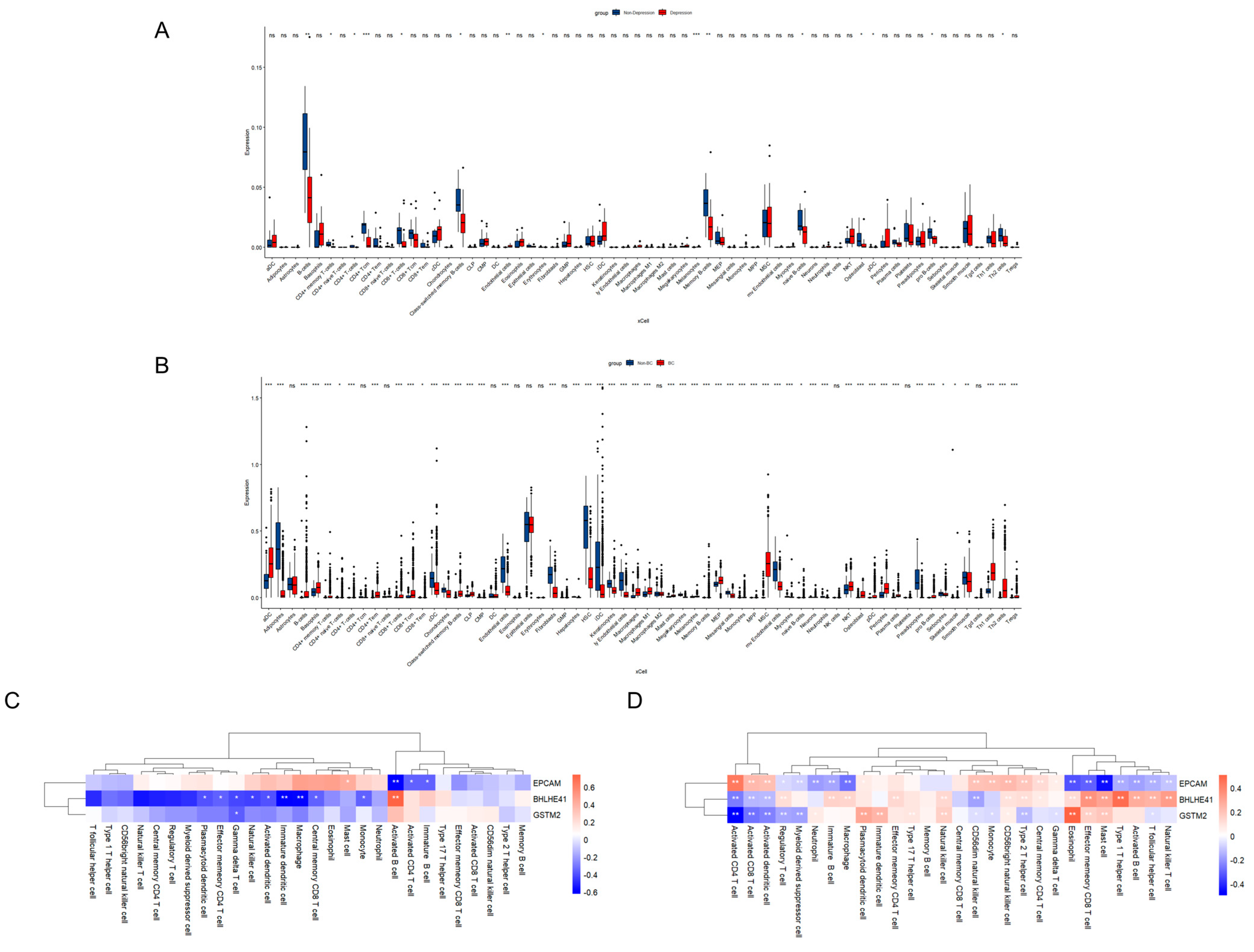
2.5. Immune Cell Infiltration and Correlation Analysis
3. Discussion
4. Materials and Methods
4.1. Enrichment Analysis of Overlapping Genes
4.2. Differential Expression Analysis
4.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
4.4. Construction of the Protein–Protein Interaction (PPI) Network
4.5. Mechanism-Associated Gene Selection and Validation
4.6. Immune Infiltration Analysis
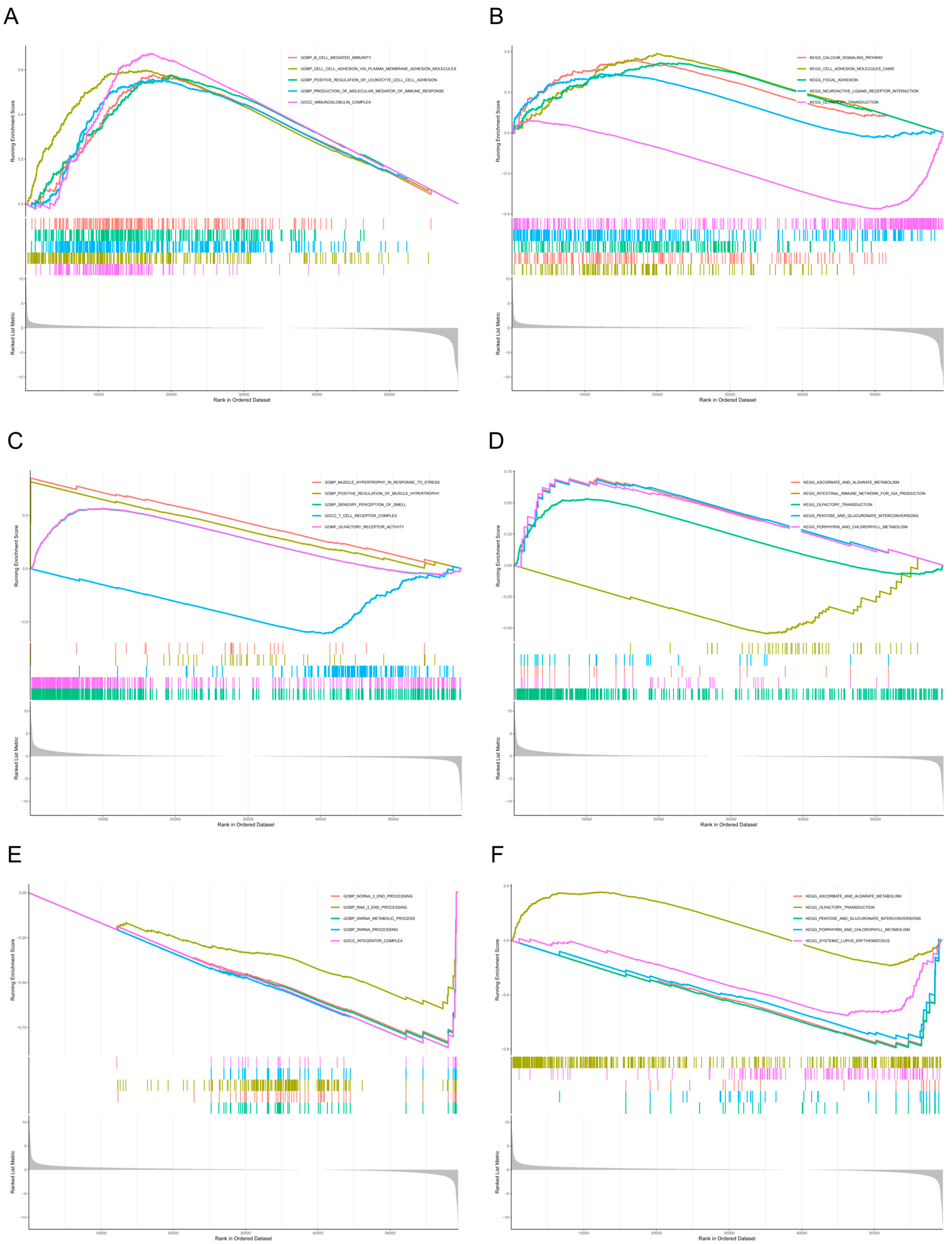
4.7. Single-Gene Gene Set Enrichment Analysis (GSEA) Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | area under the curve |
| BBB | blood–brain barrier |
| BC | Breast cancer |
| CNS | central nervous system |
| DEGs | differentially expressed genes |
| EMT | epithelial–mesenchymal transition |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| GSEA | Gene Set Enrichment Analysis |
| GWAS | genome-wide association studies |
| HPA | hypothalamic–pituitary–adrenal |
| IL-6 | interleukin-6 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LASSO | least absolute shrinkage and selection operator |
| MAPK | mitogen-activated protein kinase |
| PPI | protein-protein interaction |
| PRS | polygenic risk scores |
| ROC | receiver operating characteristic |
| TCGA | The Cancer Genome Atlas |
| TNF-α | tumor necrosis factor-α |
References
- Pitsillou, E.; Bresnehan, S.M.; Kagarakis, E.A.; Wijoyo, S.J.; Liang, J.; Hung, A.; Karagiannis, T.C. The cellular and molecular basis of major depressive disorder: Towards a unified model for understanding clinical depression. Mol. Biol. Rep. 2020, 47, 753–770. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, M.H.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 2020, 84, 106535. [Google Scholar] [CrossRef] [PubMed]
- Javan Biparva, A.; Raoofi, S.; Rafiei, S.; Masoumi, M.; Doustmehraban, M.; Bagheribayati, F.; Vaziri Shahrebabak, E.S.; Noorani Mejareh, Z.; Khani, S.; Abdollahi, B.; et al. Global depression in breast cancer patients: Systematic review and meta-analysis. PLoS ONE 2023, 18, e0287372. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.L.; Xu, C.; Yang, K.B.; Tang, S.Q.; Tang, L.L.; Chen, L.; Li, W.F.; Mao, Y.P.; Ma, J. Causal relationship between genetically predicted depression and cancer risk: A two-sample bi-directional mendelian randomization. BMC Cancer 2022, 22, 353. [Google Scholar] [CrossRef]
- Ren, Q.; Luo, F.; Ge, S.; Chen, P. Major depression disorder may causally associate with the increased breast cancer risk: Evidence from two-sample mendelian randomization analyses. Cancer Med. 2023, 12, 1984–1996. [Google Scholar] [CrossRef] [PubMed]
- Aboumrad, M.; Joshu, C.; Visvanathan, K. Impact of major depressive disorder on breast cancer outcomes: A national retrospective cohort study. J. Natl. Cancer Inst. 2025, 12, djae287. [Google Scholar] [CrossRef]
- Tian, R.; Ge, T.; Kweon, H.; Rocha, D.B.; Lam, M.; Liu, J.Z.; Singh, K.; Biogen Biobank Team; Levey, D.F.; Gelernter, J.; et al. Whole-exome sequencing in UK Biobank reveals rare genetic architecture for depression. Nat. Commun. 2024, 15, 1755. [Google Scholar] [CrossRef]
- Meng, X.; Navoly, G.; Giannakopoulou, O.; Levey, D.F.; Koller, D.; Pathak, G.A.; Koen, N.; Lin, K.; Adams, M.J.; Rentería, M.E.; et al. Multi-ancestry genome-wide association study of major depression aids locus discovery, fine mapping, gene prioritization and causal inference. Nat. Genet. 2024, 56, 222–233. [Google Scholar] [CrossRef]
- Wray, N.R.; Ripke, S.; Mattheisen, M.; Trzaskowski, M.; Byrne, E.M.; Abdellaoui, A.; Adams, M.J.; Agerbo, E.; Air, T.M.; Andlauer, T.M.F.; et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018, 50, 668–681. [Google Scholar] [CrossRef]
- Zhang, Z.; Fang, T.; Chen, L.; Ji, F.; Chen, J. Multi-omics Mendelian randomization integrating GWAS, eQTL, and mQTL data identified genes associated with breast cancer. Am. J. Cancer Res. 2024, 14, 1433–1445. [Google Scholar] [CrossRef]
- Gao, G.; McClellan, J.; Barbeira, A.N.; Fiorica, P.N.; Li, J.L.; Mu, Z.; Olopade, O.I.; Huo, D.; Im, H.K. A multi-tissue, splicing-based joint transcriptome-wide association study identifies susceptibility genes for breast cancer. Am. J. Hum. Genet. 2024, 111, 1100–1113. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, W.; Zhao, X.; Zhang, L.; Xu, M.; Hao, Y.; Xiao, J.; Zhang, B.; Li, J.; Kraft, P.; et al. Investigating the relationship between depression and breast cancer: Observational and genetic analyses. BMC Med. 2023, 21, 170. [Google Scholar] [CrossRef]
- Bortolato, B.; Hyphantis, T.N.; Valpione, S.; Perini, G.; Maes, M.; Morris, G.; Kubera, M.; Köhler, C.A.; Fernandes, B.S.; Stubbs, B.; et al. Depression in cancer: The many biobehavioral pathways driving tumor progression. Cancer Treat. Rev. 2017, 52, 58–70. [Google Scholar] [CrossRef]
- Miller, A.H.; Raison, C.L. The role of inflammation in depression: From evolutionary imperative to modern treatment target. Nat. Rev. Immunol. 2016, 16, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Yirmiya, R. The inflammatory underpinning of depression: An historical perspective. Brain Behav. Immun. 2024, 122, 433–443. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Nagappa, A.N.; Patil, C.R. Role of oxidative stress in depression. Drug Discov. Today. 2020, 25, 1270–1276. [Google Scholar] [CrossRef]
- Gonzales, E.L.; Jeon, S.J.; Han, K.M.; Yang, S.J.; Kim, Y.; Remonde, C.G.; Ahn, T.J.; Ham, B.J.; Shin, C.Y. Correlation between immune-related genes and depression-like features in an animal model and in humans. Brain Behav. Immun. 2023, 113, 29–43. [Google Scholar] [CrossRef]
- Kokkosis, A.G.; Madeira, M.M.; Hage, Z.; Valais, K.; Koliatsis, D.; Resutov, E.; Tsirka, S.E. Chronic psychosocial stress triggers microglial-/macrophage-induced inflammatory responses leading to neuronal dysfunction and depressive-related behavior. Glia 2024, 72, 111–132. [Google Scholar] [CrossRef] [PubMed]
- Anuk, D.; Özkan, M.; Kizir, A.; Özkan, S. The characteristics and risk factors for common psychiatric disorders in patients with cancer seeking help for mental health. BMC Psychiatry 2019, 19, 269. [Google Scholar] [CrossRef] [PubMed]
- Maass, S.W.M.C.; Boerman, L.M.; Verhaak, P.F.M.; Du, J.; de Bock, G.H.; Berendsen, A.J. Long-term psychological distress in breast cancer survivors and their matched controls: A cross-sectional study. Maturitas 2019, 130, 6–12. [Google Scholar] [CrossRef]
- Villacampa, G.; Navarro, V.; Matikas, A.; Ribeiro, J.M.; Schettini, F.; Tolosa, P.; Martínez-Sáez, O.; Sánchez-Bayona, R.; Ferrero-Cafiero, J.M.; Salvador, F.; et al. Neoadjuvant Immune Checkpoint Inhibitors Plus Chemotherapy in Early Breast Cancer: A Systematic Review and Meta-Analysis. JAMA Oncol. 2024, 10, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Kennis, M.; Gerritsen, L.; van Dalen, M.; Williams, A.; Cuijpers, P.; Bockting, C. Prospective biomarkers of major depressive disorder: A systematic review and meta-analysis. Mol. Psychiatry 2020, 25, 321–338. [Google Scholar] [CrossRef]
- McEwen, B.S. The neurobiology of stress: From serendipity to clinical relevance. Brain Res. 2000, 886, 172–189. [Google Scholar] [CrossRef]
- Lutgendorf, S.K.; Cole, S.; Costanzo, E.; Bradley, S.; Coffin, J.; Jabbari, S.; Rainwater, K.; Ritchie, J.M.; Yang, M.; Sood, A.K. Stress-related mediators stimulate vascular endothelial growth factor secretion by two ovarian cancer cell lines. Clin. Cancer Res. 2003, 9, 4514–4521. [Google Scholar]
- Thaker, P.H.; Han, L.Y.; Kamat, A.A.; Arevalo, J.M.; Takahashi, R.; Lu, C.; Jennings, N.B.; Armaiz-Pena, G.; Bankson, J.A.; Ravoori, M.; et al. Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma. Nat. Med. 2006, 12, 939–944. [Google Scholar] [CrossRef]
- Ticher, A.; Haus, E.; Ron, I.G.; Sackett-Lundeen, L.; Ashkenazi, I.E. The pattern of hormonal circadian time structure (acrophase) as an assessor of breast-cancer risk. Int. J. Cancer 1996, 65, 591–593. [Google Scholar] [CrossRef]
- Su, F.; Ouyang, N.; Zhu, P.; Ouyang, N.; Jia, W.; Gong, C.; Ma, X.; Xu, H.; Song, E. Psychological stress induces chemoresistance in breast cancer by upregulating mdr1. Biochem. Biophys. Res. Commun. 2005, 329, 888–897. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Monje, F.J.; Cicvaric, A.; Acevedo Aguilar, J.P.; Elbau, I.; Horvath, O.; Diao, W.; Glat, M.; Pollak, D.D. Disrupted Ultradian Activity Rhythms and Differential Expression of Several Clock Genes in Interleukin-6-Deficient Mice. Front. Neurol. 2017, 8, 99. [Google Scholar] [CrossRef]
- Li, J.Z.; Bunney, B.G.; Meng, F.; Hagenauer, M.H.; Walsh, D.M.; Vawter, M.P.; Evans, S.J.; Choudary, P.V.; Cartagena, P.; Barchas, J.D.; et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc. Natl. Acad. Sci. USA 2013, 110, 9950–9955. [Google Scholar] [CrossRef] [PubMed]
- Montagner, M.; Enzo, E.; Forcato, M.; Zanconato, F.; Parenti, A.; Rampazzo, E.; Basso, G.; Leo, G.; Rosato, A.; Bicciato, S.; et al. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature 2012, 487, 380–384. [Google Scholar] [CrossRef]
- Piccolo, S.; Enzo, E.; Montagner, M. p63, Sharp1, and HIFs: Master regulators of metastasis in triple-negative breast cancer. Cancer Res. 2013, 73, 4978–4981. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sato, H.; Suzuki, T.; Yoshizawa, T.; Morohashi, S.; Seino, H.; Kawamoto, T.; Fujimoto, K.; Kato, Y.; Kijima, H. Involvement of c-Myc in the proliferation of MCF-7 human breast cancer cells induced by bHLH transcription factor DEC2. Int. J. Mol. Med. 2015, 35, 815–820. [Google Scholar] [CrossRef]
- Bernardi, R.; Gianni, L. Hallmarks of triple negative breast cancer emerging at last? Cancer Res. 2014, 24, 904–905. [Google Scholar] [CrossRef]
- Wu, Q.; Luo, Y.; Lin, N.; Zheng, S.; Xie, X. Prognostic Value and Immune Signatures of Anoikis-related Genes in Breast Cancer. J. Immunother. 2024, 47, 328–341. [Google Scholar] [CrossRef]
- Kim, R.S.; Avivar-Valderas, A.; Estrada, Y.; Bragado, P.; Sosa, M.S.; Aguirre-Ghiso, J.A.; Segall, J.E. Dormancy signatures and metastasis in estrogen receptor positive and negative breast cancer. PLoS ONE 2012, 7, e35569. [Google Scholar] [CrossRef]
- Mal, A.; Bukhari, A.B.; Singh, R.K.; Kapoor, A.; Barai, A.; Deshpande, I.; Wadasadawala, T.; Ray, P.; Sen, S.; De, A. EpCAM-Mediated Cellular Plasticity Promotes Radiation Resistance and Metastasis in Breast Cancer. Front. Cell Dev. Biol. 2021, 8, 597673. [Google Scholar] [CrossRef]
- Kurian, A.W.; Ward, K.C.; Abrahamse, P.; Bondarenko, I.; Hamilton, A.S.; Deapen, D.; Morrow, M.; Berek, J.S.; Hofer, T.P.; Katz, S.J. Time Trends in Receipt of Germline Genetic Testing and Results for Women Diagnosed With Breast Cancer or Ovarian Cancer, 2012–2019. J. Clin. Oncol. 2021, 39, 1631–1640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xie, X.; Yeganeh, P.N.; Lee, D.J.; Valle-Garcia, D.; Meza-Sosa, K.F.; Junqueira, C.; Su, J.; Luo, H.R.; Hide, W.; et al. Immunotherapy for breast cancer using EpCAM aptamer tumor-targeted gene knockdown. Proc. Natl. Acad. Sci. USA 2021, 118, e2022830118. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.Z.; He, Y.F.; Wen, J.; Niu, Y.N.; Gao, Y.; Liu, L.H.; Zhang, X.P.; Wang, Y.; Zhang, X.L.; Zhang, H.F.; et al. Epithelial cell adhesion molecule promotes breast cancer resistance protein-mediated multidrug resistance in breast cancer by inducing partial epithelial-mesenchymal transition. Cell Biol. Int. 2021, 45, 1644–1653. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, Q.; Zhao, Q.; Yang, F.; Liu, T.; Huang, X.; Yan, Q.; Yang, X. Deglycosylated EpCAM regulates proliferation by enhancing autophagy of breast cancer cells via PI3K/Akt/mTOR pathway. Aging 2022, 14, 316–329. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, H.; Xu, T.; Tian, J.; Qin, X. Identification of Crucial Genes and Diagnostic Value Analysis in Major Depressive Disorder Using Bioinformatics Analysis. Comb. Chem. High Throughput Screen. 2022, 25, 13–20. [Google Scholar] [CrossRef]
- Lee, S.W.; Kim, S.; Chang, Y.; Cha, H.; Noeske, R.; Choi, C.; Lee, S.J. Quantification of Glutathione and Its Associated Spontaneous Neuronal Activity in Major Depressive Disorder and Obsessive-Compulsive Disorder. Biol. Psychiatry 2025, 97, 279–289. [Google Scholar] [CrossRef]
- He, J.; Yu, Y.; He, Y.; He, J.; Ji, G.; Lu, H. Chemotherapy induces breast cancer stem cell enrichment through repression of glutathione S-transferase Mu. Genes Dis. 2023, 11, 528–531. [Google Scholar] [CrossRef]
- Rauscher, G.H.; Kresovich, J.K.; Poulin, M.; Yan, L.; Macias, V.; Mahmoud, A.M.; Al-Alem, U.; Kajdacsy-Balla, A.; Wiley, E.L.; Tonetti, D.; et al. Exploring DNA methylation changes in promoter, intragenic, and intergenic regions as early and late events in breast cancer formation. BMC Cancer 2015, 15, 816. [Google Scholar] [CrossRef]
- Gohari, N.; Abbasi, E.; Akrami, H. Comprehensive analysis of the prognostic value of glutathione S-transferases Mu family members in breast cancer. Cell Biol. Int. 2024, 48, 1313–1325. [Google Scholar] [CrossRef]
- Kresovich, J.K.; Gann, P.H.; Erdal, S.; Chen, H.Y.; Argos, M.; Rauscher, G.H. Candidate gene DNA methylation associations with breast cancer characteristics and tumor progression. Epigenomics 2018, 10, 367–378. [Google Scholar] [CrossRef]
- McGrath, J.; Panzica, L.; Ransom, R.; Withers, H.G.; Gelman, I.H. Identification of Genes Regulating Breast Cancer Dormancy in 3D Bone Endosteal Niche Cultures. Mol. Cancer Res. 2019, 17, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, X.; Qian, J. Silencing of circular RNA PTP4A2 ameliorates depressive-like behaviors by inhibiting microglia activation in mice. J. Neuroimmunol. 2024, 397, 578486. [Google Scholar] [CrossRef]
- Miller, A.H.; Haroon, E.; Raison, C.L.; Felger, J.C. Cytokine targets in the brain: Impact on neurotransmitters and neurocircuits. Depress. Anxiety 2013, 30, 297–306. [Google Scholar] [CrossRef]
- Falcicchia, C.; Tozzi, F.; Arancio, O.; Watterson, D.M.; Origlia, N. Involvement of p38 MAPK in Synaptic Function and Dysfunction. Int. J. Mol. Sci. 2020, 21, 5624. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhao, J.N.; Fang, Z.T.; Wang, X.; Zhang, B.G.; He, Y.; Liu, R.J.; Chen, J.; Liu, G.P. BGP-15 alleviates LPS-induced depression-like behavior by promoting mitophagy. Brain Behav. Immun. 2024, 119, 648–664. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zheng, Q.; Wang, C.; Zhao, N.; Liu, Y.; Wang, E. BHLHE41 suppresses MCF-7 cell invasion via MAPK/JNK pathway. J. Cell. Mol. Med. 2020, 24, 4001–4010. [Google Scholar] [CrossRef]
- Wen, S.; Hou, Y.; Fu, L.; Xi, L.; Yang, D.; Zhao, M.; Qin, Y.; Sun, K.; Teng, Y.; Liu, M. Cancer-associated fibroblast (CAF)-derived IL32 promotes breast cancer cell invasion and metastasis via integrin β3-p38 MAPK signalling. Cancer Lett. 2019, 442, 320–332. [Google Scholar] [CrossRef]
- Asanoma, K.; Liu, G.; Yamane, T.; Miyanari, Y.; Takao, T.; Yagi, H.; Ohgami, T.; Ichinoe, A.; Sonoda, K.; Wake, N.; et al. Regulation of the Mechanism of TWIST1 Transcription by BHLHE40 and BHLHE41 in Cancer Cells. Mol. Cell. Biol. 2015, 35, 4096–4109. [Google Scholar] [CrossRef]
- Li, N.Y.; Weber, C.E.; Wai, P.Y.; Cuevas, B.D.; Zhang, J.; Kuo, P.C.; Mi, Z. An MAPK-dependent pathway induces epithelial-mesenchymal transition via Twist activation in human breast cancer cell lines. Surgery 2013, 154, 404–410. [Google Scholar] [CrossRef]
- Brown, T.C.; Sankpal, N.V.; Gillanders, W.E. Functional Implications of the Dynamic Regulation of EpCAM during Epithelial-to-Mesenchymal Transition. Biomolecules 2021, 11, 956. [Google Scholar] [CrossRef]
- Chen, H.N.; Liang, K.H.; Lai, J.K.; Lan, C.H.; Liao, M.Y.; Hung, S.H.; Chuang, Y.T.; Chen, K.C.; Tsuei, W.W.; Wu, H.C. EpCAM Signaling Promotes Tumor Progression and Protein Stability of PD-L1 through the EGFR Pathway. Cancer Res. 2020, 80, 5035–5050. [Google Scholar] [CrossRef] [PubMed]
- Sankpal, N.V.; Mayfield, J.D.; Willman, M.W.; Fleming, T.P.; Gillanders, W.E. Activator protein 1 (AP-1) contributes to EpCAM-dependent breast cancer invasion. Breast Cancer Res. 2011, 13, R124. [Google Scholar] [CrossRef]
- Mahajan, G.J.; Vallender, E.J.; Garrett, M.R.; Challagundla, L.; Overholser, J.C.; Jurjus, G.; Dieter, L.; Syed, M.; Romero, D.G.; Benghuzzi, H.; et al. Altered neuro-inflammatory gene expression in hippocampus in major depressive disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 82, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.H.; Pyo, S.; Shin, Y.K.; Nam, B.G.; Kang, J.W.; Kim, K.P.; Lee, H.Y.; Cho, S.R. The Effect of Environmental Enrichment on Glutathione-Mediated Xenobiotic Metabolism and Antioxidation in Normal Adult Mice. Front. Neurol. 2018, 9, 425. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, D. Ginsenoside Rg5 induces G2/M phase arrest, apoptosis and autophagy via regulating ROS-mediated MAPK pathways against human gastric cancer. Biochem. Pharm. 2019, 168, 285–304. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Hu, Y.; Hu, F.; Li, H.; Chen, Y.; Zhang, J.; Yu, Y.; Jiang, S.; Weng, Q.; Tian, S.; et al. Hepatocyte glutathione S-transferase mu 2 prevents non-alcoholic steatohepatitis by suppressing ASK1 signaling. J. Hepatol. 2022, 76, 407–419. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kang, H.G.; Choi, Y.J.; Kim, H.M.; Jeong, H.J. Caudatin attenuates inflammatory reaction by suppressing JNK/AP-1/NF-κB/caspase-1 pathways in activated HMC-1 cells. Food Sci. Biotechnol. 2023, 32, 1101–1109. [Google Scholar] [CrossRef]
- Wagner, E.F.; Nebreda, A.R. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 2009, 9, 537–549. [Google Scholar] [CrossRef]
- Panja, S.; Ghate, N.B.; Mandal, N. A microalga, Euglena tuba induces apoptosis and suppresses metastasis in human lung and breast carcinoma cells through ROS-mediated regulation of MAPKs. Cancer Cell Int. 2016, 16, 51. [Google Scholar] [CrossRef]
- Xue, C.; Li, G.; Zheng, Q.; Gu, X.; Shi, Q.; Su, Y.; Chu, Q.; Yuan, X.; Bao, Z.; Lu, J.; et al. Tryptophan metabolism in health and disease. Cell Metab. 2023, 35, 1304–1326. [Google Scholar] [CrossRef]
- Hidalgo, J.V.; Bronsert, P.; Orlowska-Volk, M.; Díaz, L.B.; Stickeler, E.; Werner, M.; Schmitt-Graeff, A.; Kayser, G.; Malkovsky, M.; Fisch, P. Histological Analysis of γδ T Lymphocytes Infiltrating Human Triple-Negative Breast Carcinomas. Front. Immunol. 2014, 5, 632. [Google Scholar] [CrossRef] [PubMed]
- Chin Fatt, C.R.; Farrar, J.D.; Jha, M.K.; Minhajuddin, A.; Mayes, T.; Foster, J.A.; Trivedi, M.H. Immune characterization of suicidal behavior in female adolescents. Brain Behav. Immun. 2022, 25, 100499. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, D.L.; Church, D.M.; Lash, A.E.; Leipe, D.D.; Madden, T.L.; Pontius, J.U.; Schuler, G.D.; Schriml, L.M.; Tatusova, T.A.; Wagner, L.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2001, 29, 11–16. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Alhamzawi, R.; Ali, H.T.M. The Bayesian adaptive lasso regression. Math. Biosci. 2018, 303, 75–82. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Ming, Y.; Hou, Z.; Yu, Y.; Liu, J.; Wang, Z. Deciphering the Overlapping Immune Mechanism Between Depression and Breast Cancer. Int. J. Mol. Sci. 2025, 26, 5229. https://doi.org/10.3390/ijms26115229
Ma Y, Ming Y, Hou Z, Yu Y, Liu J, Wang Z. Deciphering the Overlapping Immune Mechanism Between Depression and Breast Cancer. International Journal of Molecular Sciences. 2025; 26(11):5229. https://doi.org/10.3390/ijms26115229
Chicago/Turabian StyleMa, Yiming, Yu Ming, Zhiyong Hou, Yanan Yu, Jun Liu, and Zhong Wang. 2025. "Deciphering the Overlapping Immune Mechanism Between Depression and Breast Cancer" International Journal of Molecular Sciences 26, no. 11: 5229. https://doi.org/10.3390/ijms26115229
APA StyleMa, Y., Ming, Y., Hou, Z., Yu, Y., Liu, J., & Wang, Z. (2025). Deciphering the Overlapping Immune Mechanism Between Depression and Breast Cancer. International Journal of Molecular Sciences, 26(11), 5229. https://doi.org/10.3390/ijms26115229





