Abstract
In plants, ferric-chelate reductase (FRO) plays a critical role in mediating extracellular iron (Fe) reduction, a process essential for cellular Fe homeostasis and abiotic stress tolerance. However, the biological functions and regulatory mechanisms of FRO proteins in fruit crops remain poorly characterized. Here, six VvFRO genes were identified in the table grape cultivar ‘Yanhong’. Transcriptional analysis revealed that root expression of these genes was mainly induced under Fe deficiency, Fe depletion, NaCl stress, and PEG-induced drought stress, respectively, but remained unchanged by low temperature (4 °C) or heat treatment (45 °C). Among them, VvFRO3 exhibited the highest constitutive expression, predominantly in leaves, and was significantly up-regulated under Fe deficiency, Fe depletion, or NaCl treatment. Functional complementation assays demonstrated that heterologous overexpression of VvFRO3 in the Arabidopsis thaliana fro2 knockout mutant rescued its growth retardation phenotype, particularly under Fe-deficient conditions. This study advances our understanding of Fe uptake, transport, and homeostasis mechanisms in perennial fruit crops.
1. Introduction
Iron (Fe) is an essential trace element in plant cells, functioning as a critical cofactor in iron–sulfur proteins and cytochromes. It participates in diverse metabolic pathways, including photosynthesis, respiration, phytohormone synthesis, amino acid and purine metabolism, and DNA repair [1,2,3,4]. Iron fertilization is closely associated with the growth, development, fruit quality formation, and yield of fruit trees. Iron deficiency in soil—particularly prevalent in calcareous soils—severely restricts plant growth and development, directly compromising crop productivity and quality [5,6,7,8].
In soil, the majority of iron exists as Fe3+, which is poorly bioavailable to plants [5,7,8]. Studies on the molecular mechanisms governing iron uptake, transport, and distribution have primarily focused on model plants, such as Arabidopsis thaliana and rice (Oryza sativa) [5,6,7,8,9,10,11,12], with limited attention given to perennial fruit crops. The optimal iron concentration for plant growth ranges from 10−9 to 10−4 mol/L. However, under typical soil pH conditions, dissolved Fe2+ and Fe3+ concentrations fall below 10−15 mol/L, insufficient to sustain robust plant development [6,8].
Romheld and Marschener [10] first delineated two distinct iron acquisition strategies in plants. Strategy I (employed by dicots and non-graminaceous monocots): Root plasma membrane-localized H+-ATPases secrete protons to acidify the rhizosphere, solubilizing Fe3+. Ferric-chelate reductase (FRO) subsequently reduces Fe3+ to Fe2+, which is absorbed via iron-regulated transporter (IRT) [6,8,9,10,11]. Strategy II (common in graminaceous monocots) relies on enzymatic synthesis and secretion of phytosiderophores (e.g., mugineic acids) that chelate Fe3+, enabling uptake via specialized iron transporter systems. Notably, rice utilizes both Strategy I and Strategy II for iron acquisition [6,8,9,10,11]. The MA-Fe³⁺ chelates are subsequently internalized through a phytosiderophore-dependent transport mechanism mediated by Yellow Stripe (YS) or Yellow Stripe-Like (YSL) transmembrane transporter systems [6].
Recent studies on the molecular mechanisms of iron uptake mediated by FROs has predominantly focused on annual model plants, such as Arabidopsis thaliana [10,12,13] and rice (Oryza sativa) [11]. The AtFRO2 gene (AT1G01580) was first cloned from Arabidopsis roots, and complementation of the Fe reductase deficient mutant fro2 with AtFRO2 alleviated iron deficiency-induced growth inhibition and significantly enhanced FRO activity at the root surface in transgenic lines [12,13]. In rice, OsFRO1 and OsFRO7 are highly expressed in flag leaves, with their transcript levels regulated by abiotic stressors, such as NaCl, polyethylene glycol (PEG), high temperature, and heavy metals. RNAi knockdown lines of OsFRO1 exhibited stunted growth, reduced iron and chlorophyll content, and diminished reactive oxygen species (ROS) scavenging capacity [11,14]. To date, FRO homologs have only been identified in Citrus junos cv. Ziyangxiangcheng [15], Malus xiaojinensis [16,17,18], and mango (Mangifera indica) [19], leaving the molecular basis of iron nutrition and utilization efficiency in perennial fruit trees poorly understood. In M. xiaojinensis, MxFRO4 confers iron and salt tolerance through up-regulating antioxidant capacity associated with the ROS scavenging [17] and MxFRO6 is implicated in iron and salt tolerance in A. thaliana [18].
Table grape (Vitis vinifera), a globally significant fruit crop, has a well-annotated genome [20]. Iron, as the highest amount among trace elements in grape trees, regulates grape quality and yield [2,5,7]. Nevertheless, the biological functions of FROs in grape remain uncharacterized. In this study, we identified and cloned FRO family genes from the independently bred cultivar ‘Yanhong’. Tissue-specific expression patterns were analyzed via quantitative real-time PCR (qRT-PCR), and the functionality of VvFRO3 was validated through genetic complementation assays in the Arabidopsis fro2 mutant. This study provides a theoretical foundation for elucidating iron uptake and transport mechanisms in perennial woody fruit trees.
2. Results
2.1. Identification and Isolation of VvFRO Family Genes in Table Grape
Using the amino acid sequences of Arabidopsis AtFROs as references [6,12], six putative FRO family proteins were identified through mining the grape genome database. The coding sequences (CDS) of these FRO genes were downloaded, and their full-length CDS were amplified from the ‘Yanhong’ table grape using high-fidelity DNA polymerase. After sequencing validation, the genes were designated VvFRO1 to VvFRO6 and CDS sequences were submitted to Genbank via National Center for Biotechnology Information (NCBI) website to obtain relevant numbers (Figure 1 and Table 1). All encoded proteins were predicted to contain functional domains critical for iron reductase activity, including the FAD-binding site, NADPH-binding site, and oxidoreductase signature motif (Figure 1), confirming their classification as canonical iron reductase family members.

Figure 1.
Amino acid sequence alignment of VvFRO proteins.

Table 1.
Information for VvFRO family genes.
Notably, VvFRO genes are predominantly located on chromosomes 12 (VvFRO5), 15 (VvFRO3 and VvFRO4), 16 (VvFRO1 and VvFRO2), and 17 (VvFRO6) (Table 1), all of which contain at least 7 introns of varying lengths (Figure 2). In addition, the amino acid sequence identity of VvFRO proteins is 51.62% (Figure 1) and the nucleotide sequence identity is 48.65%. Homology analysis indicated that VvFRO members can be divided into two Groups, and VvFRO1–VvFRO3 clustered within Group I, whereas VvFRO4–VvFRO6 were assigned to Group II (Figure 2).
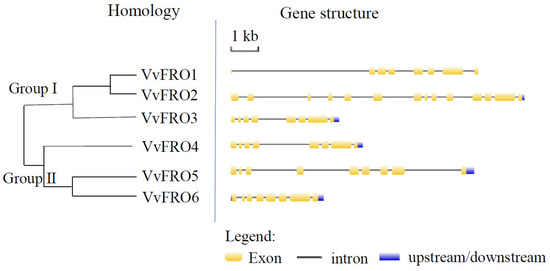
Figure 2.
Homology and gene structure analysis of VvFRO genes.
Comparative analysis of FRO proteins from 11 plant species across distinct families revealed high homology, with an amino acid sequence identity of 43.66%, and closely related homologs displayed sequence identities exceeding 67.35%. Phylogenetic tree analysis indicated that evolutionary relationships among FRO proteins from 10 plant species showed distinct clustering patterns. In particular, VvFROs exhibited closer phylogenetic proximity to homologs from C. junos CjFROs and M. xiaojinensis MxFROs (Figure 3). For instance, VvFRO1 and VvFRO4 clustered tightly with CjFRO1 and CjFRO5, respectively, while VvFRO6 grouped closely with CjFRO3 and CjFRO4. Similarly, VvFRO2 and VvFRO3 showed strong clustering with MxFRO6. In contrast, FRO homologs from legume species—soybean GmFRO2, barrel medic MtFRO1, and peanut AhFRO1, AhFRO2)—formed a distinct clade, reflecting their shared taxonomic lineage. Similarly, homologs from monocot species, rice OsFRO1 and maize ZmFRO2, exhibited the closest genetic distance (Figure 3).
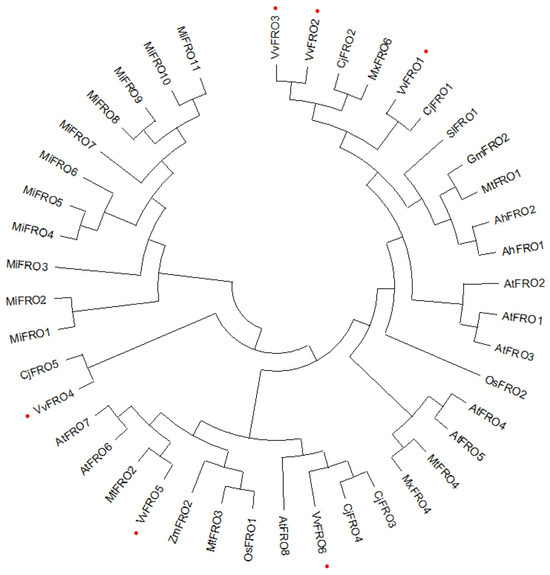
Figure 3.
Phylogenetic tree of plant FRO homologs. A phylogenetic tree was constructed using the Maximum Likelihood method in MEGA 13.0. Table grape VvFRO proteins are labelled with red dot.
2.2. Expression Profiles of VvFRO Genes
qRT-PCR analysis revealed differential expression levels of VvFRO1–VvFRO6 in various tissues of ‘Yanhong’ tissue-cultured seedlings and adult trees (Figure 4). VvFRO3 exhibited the highest overall expression across tissues, and the highest levels were observed in young leaves of adult trees and seedling leaves, higher than that of the other tested tissues (Figure 4). VvFRO2 showed a similar expression profile to VvFRO3. Moreover, VvFRO5 displayed higher expression in fruits (either young or mature fruit) compared to other tissues. The remaining three genes (VvFRO1, VvFRO4, and VvFRO6) exhibited relatively low expression levels (all < 0.1 and values are closely comparable) across different table grape tissues (Figure 4).
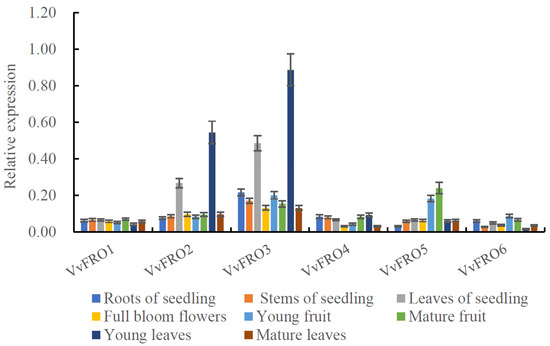
Figure 4.
Tissue specific expression analysis of VvFRO genes.
At the transcriptional level, the responses of VvFRO1–VvFRO6 in the roots of ‘Yanhong’ seedlings to eight different abiotic stresses exhibited distinct variations (Figure 5). The VvFRO family genes were most sensitive to Fe deficiency, Fe depletion, and NaCl stress. With the exception of VvFRO6, the expression levels of the other five VvFRO genes in roots were significantly induced under Fe deficiency, Fe depletion, and NaCl-treated conditions. Three genes (VvFRO1, VvFRO3, and VvFRO5) showed marked sensitivity to PEG treatment, with their root expression levels significantly up-regulated. Notably, only VvFRO3 responded to abscisic acid (ABA) treatment, displaying a significant induction in root expression, and it was also the sole gene suppressed under Fe toxicity, with its expression significantly down-regulated. In contrast, the VvFRO family genes were insensitive to temperature fluctuations, as no significant changes in expression were observed under either low-temperature (4 °C) or heat (45 °C) stress.
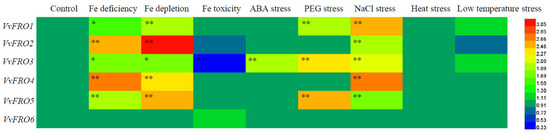
Figure 5.
Expression analysis of VvFRO genes under different abiotic stresses. One-month-old tissue cultured seedlings were subjected to Fe deficiency (50% Fe was removed from the MS solution), Fe depletion (100% Fe was removed from the MS solution), Fe toxicity (500 μmol∙L−1 FeCl3), ABA (200 µmol∙L−1 ABA), drought (10% PEG6000, w/v), NaCl (150 mmol∙L−1 NaCl), low temperature (4 °C), high temperature (4 °C) treatment for 48 h before q-RT-PCR analysis. Asterisks indicate statistical differences found between the control and abiotic stress treatment using Student’s t-test in SPSS 13.0 software (* p < 0.05, ** p < 0.01).
Specifically, three genes (VvFRO1, VvFRO3, and VvFRO5) exhibited transcriptional plasticity in roots, responding to at least three stress treatments. Among these, VvFRO3 displayed the highest overall expression levels in grape roots and responded to five stress conditions (excluding temperature extremes). Although VvFRO6 exhibited relatively low basal expression, its transcriptional stability was striking, showing no response to any of the seven stress treatments tested in this study.
2.3. VvFRO3 Rescued the Impaired Growth of Arabidopsis fro2 Mutant
In Arabidopsis, the growth of fro2 knockout mutant was severely impaired [9,14]. To determine whether VvFRO3 could restore the normal growth of fro2 mutant, VvFRO3 was introduced into the binary expression vector pHB (Figure 6A). At least seven putative (#1, #2, #4, #7, #9, #10, and #13) T1 generation fro2/35S::VvFRO3 lines were validated using reverse transcription PCR for the presence of a 2082 bp fragment of VvFRO3 (Figure 6B). Purified T3 generation of #1 and #13 fro2/35S::VvFRO3 lines were randomly selected for further physiological analysis. Given that #1 and #13 fro2/35S::VvFRO3 lines exhibited similar growth status, data of #1 fro2/35S::VvFRO3 lines are shown in this present work.
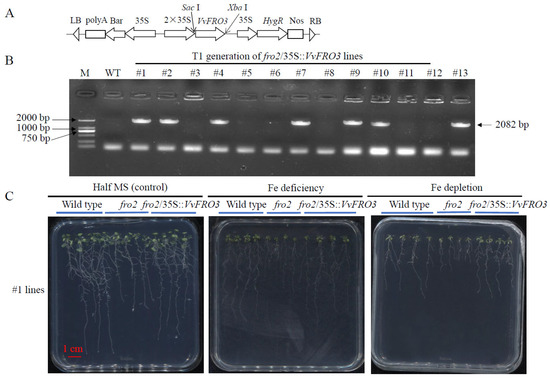
Figure 6.
Generation and phenotype analysis of VvFRO3 complementation Arabidopsis seedlings. (A) Construction of recombinant plasmid pBH-VvFRO3. (B) PCR verification of VvFRO3 in T1 generation fro2/35S::VvFRO3 lines. Note: M, standard DL2000 DNA ladder (Takara, Dalian, China). WT, wild type. (C) Phenotype analysis of T3 generation fro2/35S::VvFRO3 lines. Arabidopsis seedlings were germinated on half-strength MS solid medium and subjected to Fe deficiency (50% Fe was removed from the MS solution) or Fe depletion (100% Fe was removed from the MS solution) for 7 days before phenotype analysis. Data are presented as means ± SE (n = 20). Bar = 1 cm.
Compared to the wild type, the growth of fro2 lines was seriously hindered, accompanied by decreased fresh weight, dry weight, primary root length, lateral root numbers, and total leaf chlorophyll, under the control conditions, Fe deficiency, and Fe depletion, respectively (Figure 6C, Table 2). In contrast, #1 fro2/35S::VvFRO3 lines exhibited a better growth status than that of the fro2 mutant lines under all tested conditions. The fresh weight, dry weight, primary root length, lateral root numbers, and total leaf chlorophyll of #1 fro2/35S::VvFRO3 lines were significantly increased, compared to the fro2 mutant, similar to that of the wild type (Figure 6C, Table 2). These findings indicate that the complementation of VvFRO3 rescued the impaired growth of Arobidopsis fro2 mutants.

Table 2.
Physiological analysis of VvFRO3 complementation Arabidopsis seedlings.
In comparison to the wild type, the tissue Fe content, ACO activity, NiR activity, and SDH activity of fro2 mutant lines were significantly decreased under control conditions, Fe deficiency and Fe depletion, respectively (Table 2). Compared to the fro2 mutant, the tissue Fe concentration, ACO activity, NiR activity, and SDH activity of #1 fro2/35S::VvFRO3 lines were significantly induced (Table 2).
3. Discussion
Fe plays a critical role in fruit quality development and is closely associated with crop yield. However, the biological functions of genes involved in Fe uptake and translocation in fruit trees remain poorly characterized. As a Strategy I dicot species, grapevine absorbs Fe via rhizosphere acidification and FRO activity [5,6,8,9,10,11], yet the molecular mechanisms underlying Fe acquisition and transport remain largely unexplored. In this study, six FRO family genes were cloned and characterized from table grape (Figure 1). This number is slightly lower than homologs in Arabidopsis (8 FRO genes) [6,12] and mango (11 FRO genes) [19], suggesting lineage-specific divergence in FRO gene family size across plant taxa. Phylogenetic analysis further revealed distinct evolutionary relationships among FRO homologs from different plant families. Notably, VvFRO proteins clustered more closely with homologs from perennial woody species (C. junos and M. xiaojinensis) than with those from Poaceae, Fabaceae, Solanaceae, or Brassicaceae species (Figure 3), implying conserved biological roles of FRO enzymes in woody fruit crops. These findings suggest that, while FRO family members share close phylogenetic relationships, they may have undergone functional specialization during long-term evolution, highlighting adaptive evolution in plant Fe homeostasis mechanisms.
Previous studies have demonstrated high expression levels of FRO family genes in leaves, including OsFRO1 and OsFRO7 in rice [11], CjFRO1 in C. junos [15], MxFRO4 and MxFRO6 in M. xiaojinensis [16,17,18], and MiFRO4, MiFR8, and MiFR11 in mango [19]. Consistent with these findings, VvFRO2 and VvFRO3 exhibited elevated expression (2- to 4-fold higher than other tissues) in young leaves of mature grapevines and leaves of tissue-cultured seedlings (Figure 4), further supporting the critical role of FRO genes in leaf iron absorption and translocation. Notably, while no prior studies have reported FRO expression profiles in fruits, we observed relatively high expression of VvFRO5 in fruits, with levels significantly exceeding those in leaves, roots, and other tissues, suggesting its potential involvement in Fe homeostasis during pivotal developmental stages of fruit maturation.
Previous studies have demonstrated that OsFRO1 and OsFRO7 in rice exhibit transcriptional responses to abiotic stressors, such as salt, drought, heat, and heavy metals [11], while MxFRO4 and MxFRO6 in M. xiaojinensis are modulated under NaCl stress [16,17,18]. Notably, OsFRO7 is transcriptionally induced by ABA treatment, suggesting its potential role in osmotic regulation in rice, whereas OsFRO1 lacks transcriptional responsiveness to ABA [11]. In contrast, our study revealed that only VvFRO3 in grapevine roots exhibited significant upregulation under ABA treatment, implying its involvement in osmotic adjustment and providing a foundation for further exploration of FRO enzyme functions and regulatory networks in grapevine. In addition, VvFRO1, VvFRO3, and VvFRO5 were transcriptionally induced in grapevine roots under PEG-mediated drought stress (Figure 5), suggesting potential roles in drought response pathways. Strikingly, despite grapevine’s limited adaptability to extreme climates, VvFRO genes exhibited no transcriptional changes under low-temperature (4 °C) or high-temperature (45 °C) treatment highlights. This expression stability may ensure the maintenance of essential iron metabolism and physiological processes, allowing grapevine to endure suboptimal thermal conditions. However, functional validation through targeted experiments is required to confirm these observations.
In the present study, the transcriptional regulation of VvFRO genes in table grape is responsive to Fe availability, with pronounced sensitivity to Fe deficiency and Fe depletion stresses. Specifically, five out of six VvFRO genes (excluding VvFRO6) exhibited significant up-regulation in roots under Fe-deficient conditions (Figure 5), suggesting that grape FRO family members are preferentially induced to maximize ferric-chelate reductase activity, thereby sustaining iron uptake, translocation, and Fe-dependent physiological processes in roots. The elevated expression of FRO genes is likely to serve as a critical signaling mechanism for grapevine adaptation to Fe-limited environments. Notably, VvFRO3 displayed the highest constitutive expression across tissues in ‘Yanhong’, with root expression levels 3- to 7-fold higher than other VvFRO genes. Its expression was markedly suppressed under Fe toxicity and induced under Fe-deficient stresses, implying that VvFRO3 encodes a highly active ferric-chelate reductase whose activity is directly modulated by external iron availability. These findings align with reported Fe-responsive regulation of FRO genes in Arabidopsis [12], C. junos [15], M. xiaojinensis [16,17,18], and mango [17]. In M. xiaojinensis, MxFRO4 enhances iron and salt stress tolerance by upregulating antioxidant systems that scavenge reactive oxygen species (ROS) [17]. Complementary studies demonstrate that heterologous expression of MxFRO6 in Arabidopsis similarly improves iron acquisition and salt tolerance in transgenic seedlings [18]. In this present study, heterologous expression of VvFRO3 in Arabidopsis fro2 mutant favorably rescued the retarded growth of fro2 mutants. In addition, tissue Fe content and activity of Fe-dependent enzymes (ACO, NiR, and SDH) were significantly enhanced in fro2/35S::VvFRO3 lines, which may partially account for the restored growth performance. Over-expression of VvFRO3 in fro2 mutant may positively strengthen the Fe uptake and transport capacity in fro2/35S::VvFRO3 lines, maintaining basic Fe-dependent metabolic processes, thereby preventing the transgenic seedlings from Fe-deficient stresses. Meanwhile, tissue Fe content and total leaf chlorophyll were indeed induced in fro2/35S::VvFRO3 lines.
To date, numerous transcription factors and Fe uptake and transport-related functional genes have been associated with iron uptake and transport mechanisms, and such regulatory mechanisms have been documented in Arabidopsis systems [4,6]. Our previous work utilized Arabidopsis fro2 mutants to functionally validate VvFRO3 through physiological phenotyping. Future investigations will employ these genetic resources to conduct integrated proteomic and transcriptomic profiling under Fe deprivation conditions to identify transcription factors coordinating with VvFRO3-mediated Fe homeostasis and characterize post-translational modifications influencing VvFRO3 function. Nonetheless, this study favors the proposition that ferric-chelate reductase FRO3 is implicated in modulating Fe transport and homeostasis and plant adaptation to undesired Fe-deficient stresses.
4. Materials and Methods
4.1. Plant Material and Growth Condition
The new variety of table grape ‘Yanhong’ cultivated independently was used throughout this study. One-month-old tissue-cultured seedlings and 5-year-old mature ‘Yanhong’ trees were grown in the National Grape Germplasm Repository (Yantai, China).
For the control treatments, 1-month-old tissue-cultured seedlings were grown in half-strength Murashige and Skoog (MS) liquid solution. For Fe deficiency treatments, 50% Fe was removed from the MS solution. For Fe depletion treatments, 100% Fe was deleted from the MS solution [4,19,21]. For Fe toxicity treatments, 500 μmol∙L−1 FeCl3 was added in half-strength MS solution [19,22]. For ABA treatments, 200 µmol∙L−1 ABA was supplied in half-strength MS medium [17,21]. For PEG-mediated drought stress treatments, 10% PEG6000 (w/v) was added in half-strength MS solution [19,22,23,24]. For salt stress treatments, 150 mmol∙L−1 NaCl was added in half-strength MS solution [19,22,23,24]. For low temperature treatments, seedlings were placed in the 4 °C incubator [4]. For high temperature treatments, seedlings were placed in the 45 °C incubator [19]. After being subjected to stress treatment for 48 h, root samples were collected and frozen in liquid nitrogen before further analyses.
The wild type Arabidopsis (Col-0), fro2 knockout mutants (purchased and verified from The Arabidopsis Information Resource, https://www.arabidopsis.org) [12,13], and fro2/35S::VvFRO2 lines were germinated in half-strength MS medium and exposed to the control, Fe deficiency, or Fe depletion treatment for 7 days before physiological analysis. Three independent biological repeats were conducted, each comprising 20 Arabidopsis seedlings.
4.2. Physiological Analysis
Fresh weight of stress-treated Arabidopsis seedlings was measured using an Analytical Balance (Thermo Electron, Waltham, MA, USA). Primary root length and lateral root number were manually quantified by direct visual inspection [4,19,21,22,23]. Enzyme activities of aconitase (ACO), nitrite reductase (NiR), and succinate dehydrogenase (SDH) were assayed using commercially available detection kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Total leaf chlorophyll content was quantified with a BioRad SmartSpec 3000 spectrophotometer (Wadsworth, IL, USA) following previously established protocol [21,22,23]. Tissue Fe content was determined by ICP–AES systems (IRIS Advantage, Thermo Electron, Waltham, MA, USA). Three independent biological replicates were executed, each comprising 20 seedlings.
4.3. Isolation and Cloning of VvFRO Family Genes from Table Grape
Putative grape FRO family genes (VvFROs) were identified by interrogating the Phytozome grape genome database (http://www.phytozome.net) using amino acid sequences of eight Arabidopsis AtFRO proteins as queries [6,12]. Functional domains of VvFRO proteins were confirmed through analysis on the Pfam online server (Accessed on 26 March 2023, http://pfam.xfam.org/search) [19,21,23]. Gene-specific primers (Table 1) were designed based on the coding sequences (CDS) of VvFRO genes. PCR amplification was performed with the PrimeSTARTM HS DNA Polymerase Kit (TaKaRa, Dalian, China) for high-fidelity amplification, and products were validated by sequencing at Sangon Biotech Co., Ltd. (Shanghai, China).
4.4. Phylogenetic Tree Analysis
Following the description of Gao et al. [23] and Zhang et al. [24], amino acid sequences of FRO homologous proteins were retrieved from 11 plant species: grape (VvFRO, Rosaceae), M. xiaojinensis (MxFRO, Rosaceae), C. junos (CjFRO, Rutaceae), mango (MiFRO, Anacardiaceae), Arabidopsis (AtFRO, Brassicaceae), tomato (SlFRO, Solanum lycopersicum, Solanaceae), maize (ZmFRO, Zea mays, Poaceae), rice (OsFRO, Poaceae), barrel medic (MtFRO, Medicago truncatula, Fabaceae), peanut (AhFRO, Arachis hypogaea, Fabaceae), and soybean (GmFRO, Glycine max, Fabaceae). Multiple sequence alignment was performed using ClustalX 2.0 to analyze amino acid sequence conservation. A phylogenetic tree of plant FRO homologs was constructed with the Maximum Likelihood method in MEGA 13.0 to elucidate the genetic evolutionary relationships among FRO proteins across these species [23,24].
4.5. Quantitative Real Time PCR (qRT-PCR)
Gene-specific primer pairs of VvFRO genes were designed using the NCBI/Primer-BLAST on-line server and are provided in Table 3. qRT-PCR analysis was conducted on the 7500 Real Time PCR System (Applied Biosystems, Foster City, CA, USA) with SYBR Premix Ex Taq (TaKaRa, Dalian, China). The Ubiquitin of wine grape was employed as the internal control [22,25,26]. The relative expression levels of VvFRO genes were calculated via normalization to Ubiquitin using data from three independent biological replicates.

Table 3.
Specific primer pairs used for qRT-PCR.
4.6. Overexpression of VvFRO3 in Arabidopsis fro2 Mutant
The CDS of VvFRO3 was cloned into the pBH vector [4,21,24] using the forward primer 5′-GACGAGCTCATGAGACCGCTTCTCTTGGTG-3′ (Sac I site underlined) and the reverse primer 5′ GAGTCTAGATCACCAGCTGAAGCTTATGGA-3′ (Xba I site underlined), yielding the recombinant plasmid pBH-VvFRO3. Both the empty pBH vector and pBH-VvFRO3 were transformed into Agrobacterium tumefa-ciens strain EHA 105 and subsequently introduced into Arabidopsis fro2 knockout homo-zygote mutants [9,13]. Transgenic T1 generation fro2/35S::VvFRO3 lines were screened on hygromycin-supplemented solid medium. Genomic DNA was extracted from T1 fro2/35S::VvFRO3 lines using the Universal Genomic DNA Extraction Kit (TaKaRa, Dalian, China), followed by PCR verification of a 2082 bp VvFRO3 fragment. Validated T3 seeds of fro2/35S::VvFRO3 lines were surface-sterilized and germinated on half-strength MS solid medium under controlled conditions for 7 days. Three biological replicates were conducted, each consisting of 20 seedlings.
4.7. Statistical Analysis
Statistical graphs were generated using OriginPro 12.0 software (OriginLab Corporation, Northampton, MA, USA). Significant differences were analyzed using Student’s t-test in SPSS 13.0 software (SPSS, Chicago, IL, USA) or ANOVA Fisher’s LSD test [4,19,21,23]. Please see details in the figure or table legends.
5. Conclusions
Six VvFRO genes were isolated from table grape ‘Yanhong’. VvFRO3 was the most abundantly expressed gene, which was mainly expressed in leaves and was up-regulated under Fe deficiency, Fe depletion, or NaCl treatment. Over-expressing of VvFRO3 rescued the retarded growth of Arabidopsis fro2 knockout mutant, especially under Fe deficiency and Fe depletion. VvFRO3 may be a crucial ferric-chelate reductase that is involved in Fe transport and homeostasis in table grape, especially under Fe-deficient conditions.
Author Contributions
Conceptualization, Z.S. and M.T.; methodology, Z.S., J.W. and M.T.; validation, C.W., Y.C. and M.S.; investigation, J.W., C.W., Y.C. and M.S.; data curation, Y.C. and M.S.; writing—original draft preparation, Z.S.; writing—review and editing, M.T.; project administration, M.T.; funding acquisition, Z.S. and M.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was jointly supported by the following grants: Key R&D Program of Shandong Province-Innovation Capability Enhancement Project for Technology based Small and Medium sized Enterprises (2024TSGC0718), Major Project of Science and Technology of Shandong Province (2022CXGC010605), China Agriculture Research System of MOF and MARA (CARS-29-17), the China Scholarship Council Fund (202208370080).
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Acknowledgments
The authors are grateful to Julia M. Davies, Department of Plant Sciences, University of Cambridge for critical reading and valuable suggestions.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lill, R. Function and biogenesis of iron-sulphur proteins. Nature 2009, 460, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Kermeur, N.; Pédrot, M.; Cabello-Hurtado, F. Iron availability and homeostasis in plants: A review of responses, adaptive mechanisms, and signaling. Methods Mol. Biol. 2023, 2642, 49–81. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Touraine, B.; Briat, J.F.; Gaymard, F.; Rouhier, N. The iron-sulfur cluster assembly machineries in plants: Current knowledge and open questions. Front. Plant Sci. 2013, 4, 259. [Google Scholar] [CrossRef]
- Song, Z.Z.; Lin, S.Z.; Fu, J.Y.; Chen, Y.H.; Zhang, H.X.; Li, J.Z.; Liang, M.X. Heterologous expression of ISU1 gene from Fragaria vesca enhances plant tolerance to Fe depletion in Arabidopsis. Plant Physiol. Biochem. 2022, 184, 65–74. [Google Scholar] [CrossRef]
- Tagliavini, M.; Rombolà, A.D. Iron deficiency and chlorosis in orchard and vineyard ecosystems. Eur. J. Agron. 2001, 15, 72–92. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nishizawa, N.K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef]
- Montejano-Ramírez, V.; Valencia-Cantero, E. Cross-talk between iron deficiency response and defense establishment in plants. Int. J. Mol. Sci. 2023, 24, 6263. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Sun, W.; Wang, T. The adaptive mechanism of plants to iron deficiency via iron uptake, transport, and homeostasis. Int. J. Mol. Sci. 2019, 20, 2424. [Google Scholar] [CrossRef]
- Fourcroy, P.; Tissot, N.; Gaymard, F.; Briat, J.F.; Dubos, C. Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe2+ transport system. Mol. Plant 2016, 9, 485. [Google Scholar] [CrossRef]
- Romheld, V.; Marschener, H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol. 1986, 80, 175–180. [Google Scholar] [CrossRef]
- Muhammad, I.; Jin, X.Q.; Shalmani, A.; Ali, M.; Yi, S.; Gan, P.F.; Li, W.Q.; Liu, W.T.; Chen, K.M. Comparative in silico analysis of ferric reduction oxidase (FRO) genes expression patterns in response to abiotic stresses, metal and hormone applications. Molecules 2018, 23, 1163. [Google Scholar] [CrossRef] [PubMed]
- Wu, H. Molecular and biochemical characterization of the Fe (III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 1505–1514. [Google Scholar] [CrossRef] [PubMed]
- Connolly, E.L.; Campbell, N.H.; Grotz, N.; Prichard, C.L.; Guerinot, M.L. Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol. 2003, 133, 1102–1110. [Google Scholar] [CrossRef]
- Li, L.; Ye, L.X.; Kong, Q.H.; Shou, H.X. A vacuolar membrane ferric-chelate reductase, OsFRO1, alleviates Fe toxicity in rice (Oryza sativa L.). Front. Plant Sci. 2019, 10, 700. [Google Scholar] [CrossRef]
- Yao, X.L.; Chen, S.C.; He, Y.R. Cloning and analysis of ferric reduction oxidase family genes in Ziyangxiangcheng. In Proceedings of the 2014 Academic Annual Conference of the Chinese Society for Horticultural Science, Wuhan, China, 17–21 March 2014; Summary collection of papers. The Chinese Society for Horticultural Science, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences: Beijing, China, 2014. (In Chinese). [Google Scholar]
- Zhou, Z.Y. Isolation and functional analysis of ferric reductase gene MxFRO4 and MxFRO6 in Malus xiaojinensis. Master’s Thesis, Northeast Agricultural University, Harbin, China, 2021. (In Chinese). [Google Scholar]
- Ren, C.; Luo, G.; Li, X.; Yao, A.; Liu, W.; Zhang, L.; Wang, Y.; Li, W.; Han, D. MxFRO4 confers iron and salt tolerance through up-regulating antioxidant capacity associated with the ROS scavenging. J Plant Physiol. 2023, 285, 154001. [Google Scholar] [CrossRef]
- Li, Y.; Zhong, J.; Huang, P.; Shao, B.; Li, W.; Liu, W.; Wang, Y.; Xie, L.; Han, M.; Han, D. Overexpression of MxFRO6, a FRO gene from Malus xiaojinensis, increases iron and salt tolerance in Arabidopsis thaliana. In Vitro Cell. Dev. Biol.-Plant 2022, 58, 189–199. [Google Scholar] [CrossRef]
- Han, G.D.; Sun, F.P.; Ren, Z.Y.; Cao, J.W.; Gao, A.P.; Huang, J.F.; Song, Z.Z. Identification and expression pattern analysis of ferric reduction oxidase encoding genes in mango. Chin. J. Trop. Crops 2024, 45, 225–233. (In Chinese) [Google Scholar]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrabde, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef]
- Song, Z.Z.; Wang, X.; Li, M.Y.; Ning, Y.Z.; Shi, S.P.; Yang, G.R.; Zhang, H.X.; Tang, M.L.; Peng, B. Isolation, heterologous expression, and functional determination of an iron regulated transporter (IRT) gene involved in Fe2+ transport and tolerance to Fe2+ deficiency in Vitis vinifera. Plant Growth Regul. 2024, 156, 65. [Google Scholar] [CrossRef]
- Song, Z.Z.; Yang, Y.; Xu, J.L.; Ma, R.J.; YU, M.L. Physiological and transcriptional responses in the iron- sulphur cluster assembly pathway under abiotic stress in peach (Prunus persica L.) seedlings. Plant Cell Tiss. Org. 2014, 117, 419–430. [Google Scholar] [CrossRef]
- Gao, Y.C.; Yu, C.Y.; Zhang, K.; Zhang, H.X.; Zhang, S.Y.; Song, Z.Z. Identification and characterization of the strawberry KT/HAK/KUP transporter gene family in response to K+ deficiency. Acta Physiol. Plant. 2021, 43, 1. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, X.M.; Lin, S.Z.; Wang, J.P.; Tang, M.L.; Huang, J.F.; Gao, T.P.; Zhang, H.X.; Song, Z.Z. Heterologous expression of the MiHAK14 homologue from Mangifera indica enhances plant tolerance to K+ deficiency and salinity stress in Arabidopsis. Plant Growth Regul. 2022, 98, 39–49. [Google Scholar] [CrossRef]
- Peng, B.; Ran, J.G.; Li, Y.Y.; Tang, M.L.; Xiao, H.L.; Shi, S.P.; Ning, Y.Z.; Dark, A.; Guan, X.Q.; Song, Z.Z. Site-directed mutagenesis of VvCYP76F14 (cytochrome P450) unveils its potential for selection in wine grape varieties linked to the development of wine bouquet. J. Agric. Food Chem. 2024, 72, 3683–3694. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.Z.; Tang, M.L.; Xiao, H.L.; Xu, H.H.; Shi, M.; Dark, A.; Xie, Z.Q.; Peng, B. Unraveling the trisubstrate-triproduct reaction mechanisms of wine grape VvCYP76F14 to improve wine bouquet. Food Chem. 2025, 474, 143077. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).