Integrative Analysis of EPHX4 as a Novel Prognostic and Diagnostic Biomarker in Lung Adenocarcinoma
Abstract
1. Introduction
2. Results
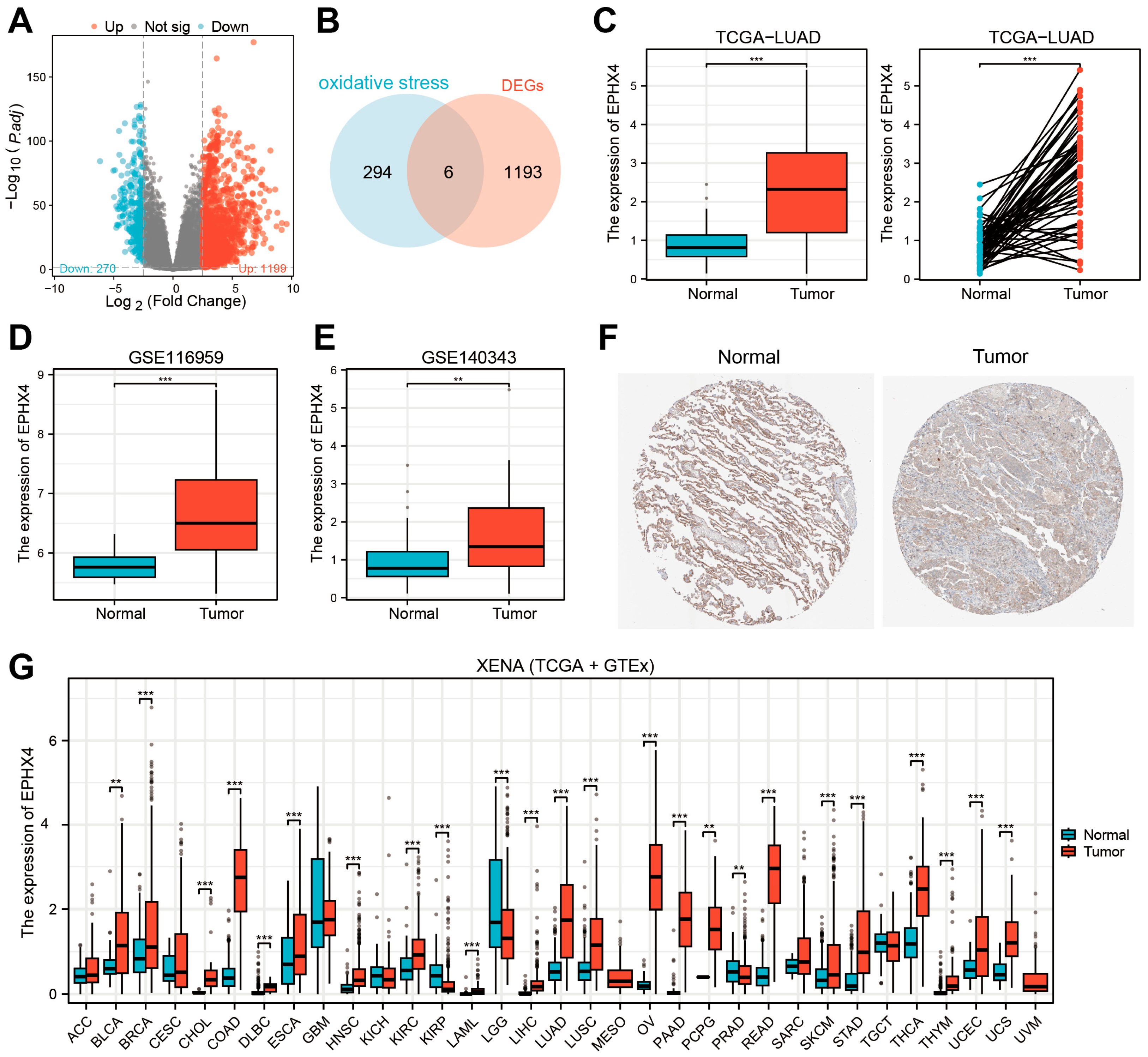
2.1. EPHX4 Emerged as the Key Oxidative Stress-Related DEG in LUAD
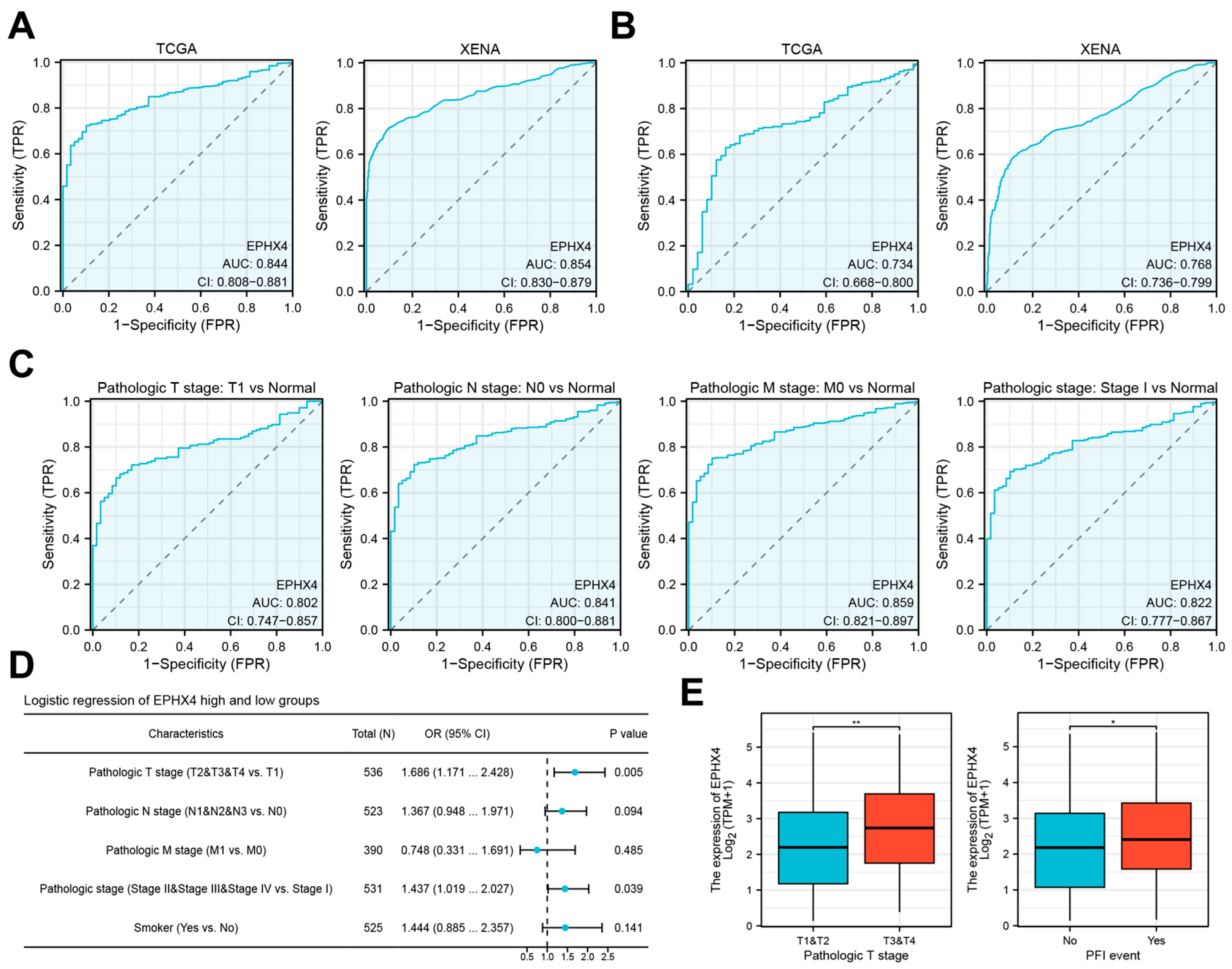
2.2. Elevated EPHX4 Expression Is Associated with Poor Prognosis in LUAD
2.3. EPHX4 Is a Potential Diagnostic Biomarker for LUAD
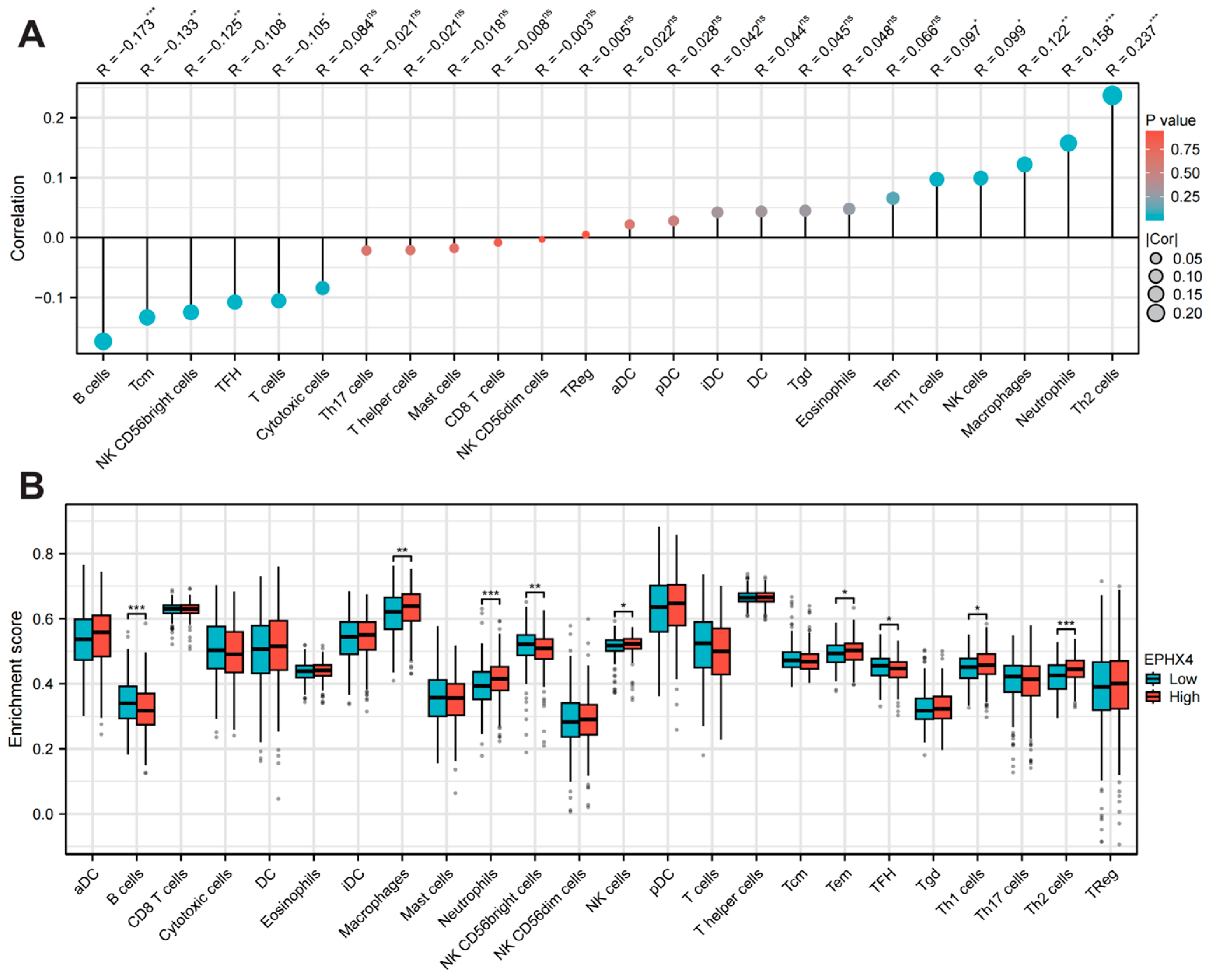
2.4. EPHX4 Is Significantly Linked to Immune Cell Infiltration
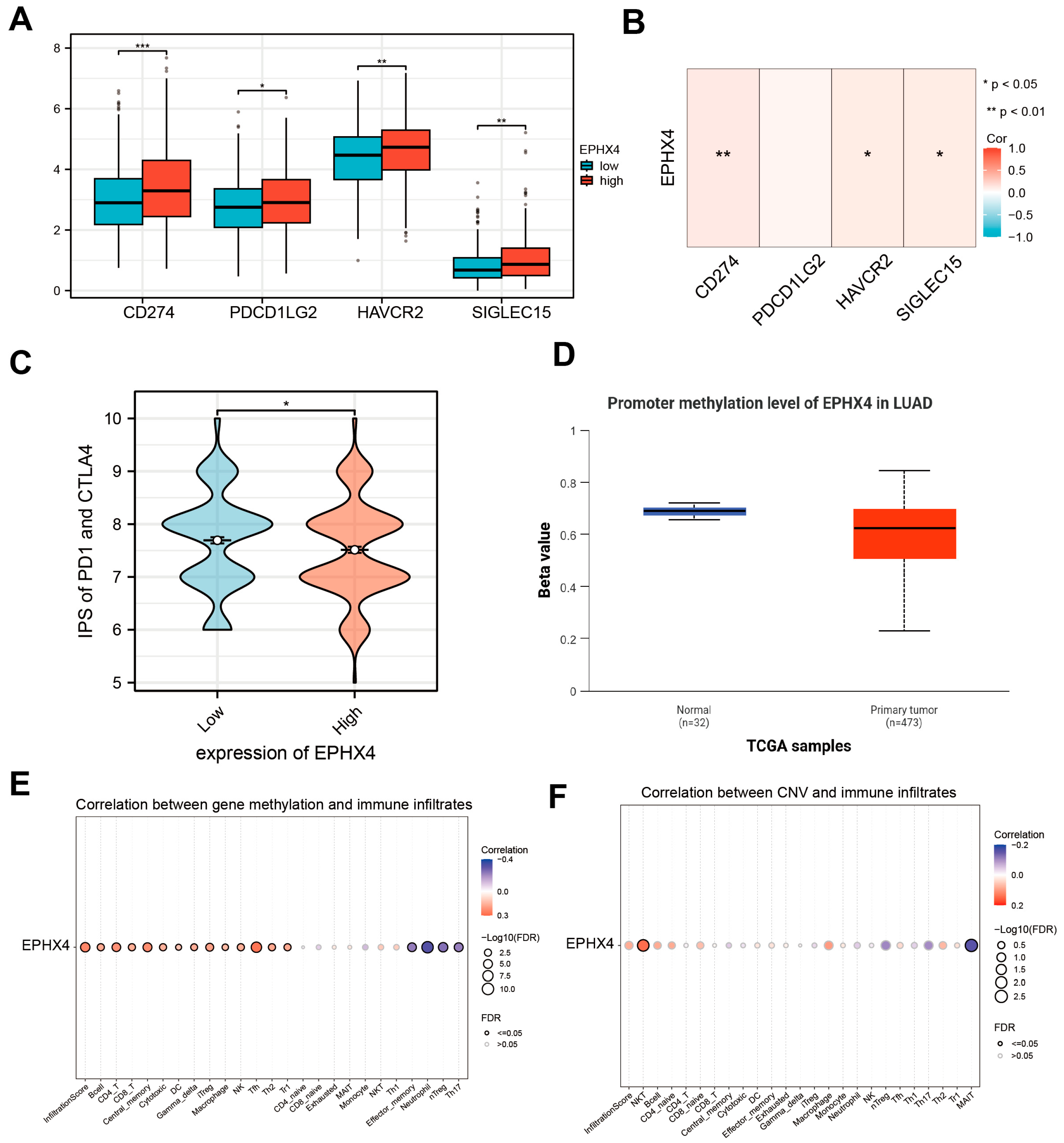
2.5. EPHX4 Is Associated with Immune Checkpoints and Immunotherapy Response
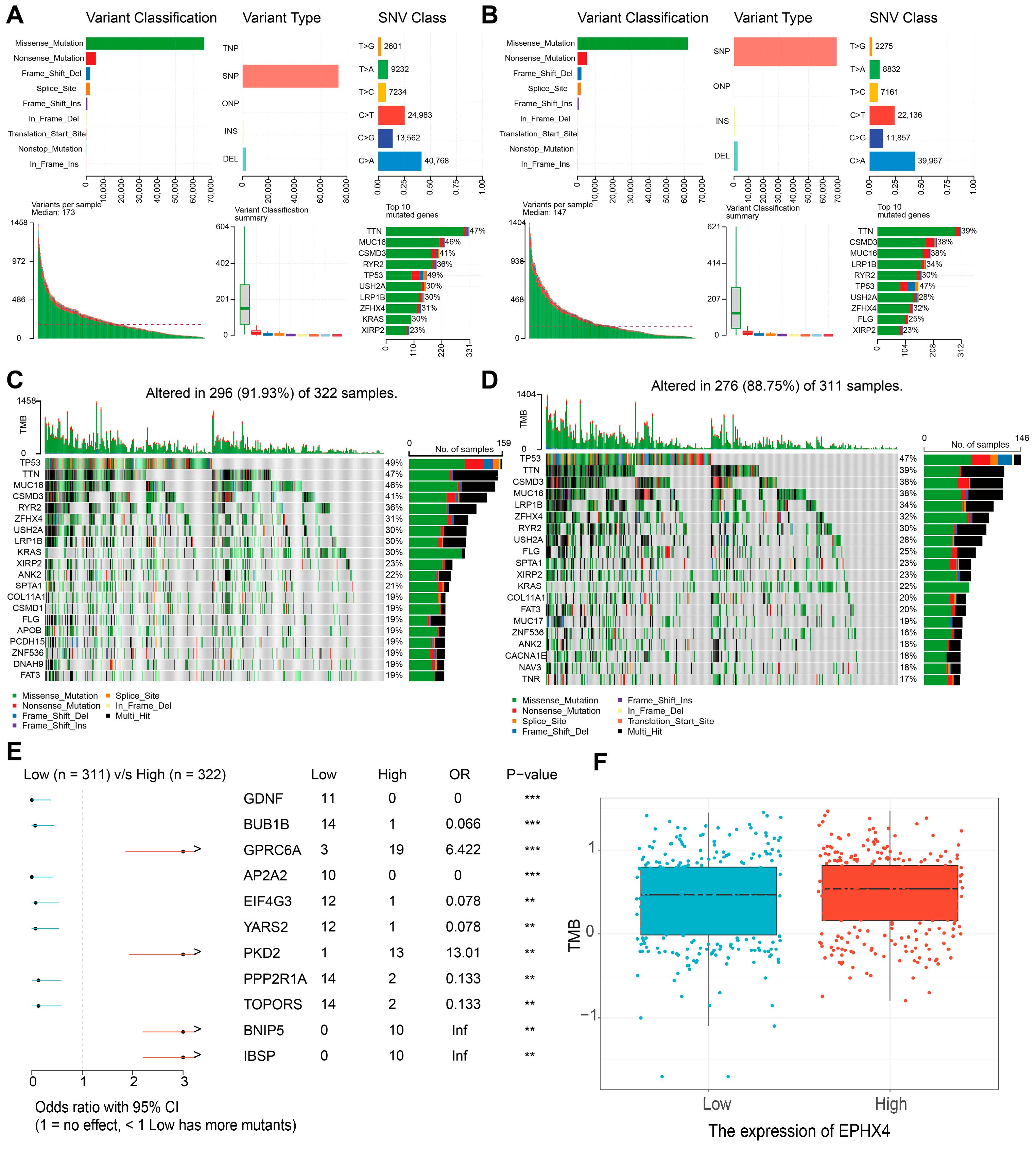
2.6. Relationship Between EPHX4 Expression and Somatic Variants
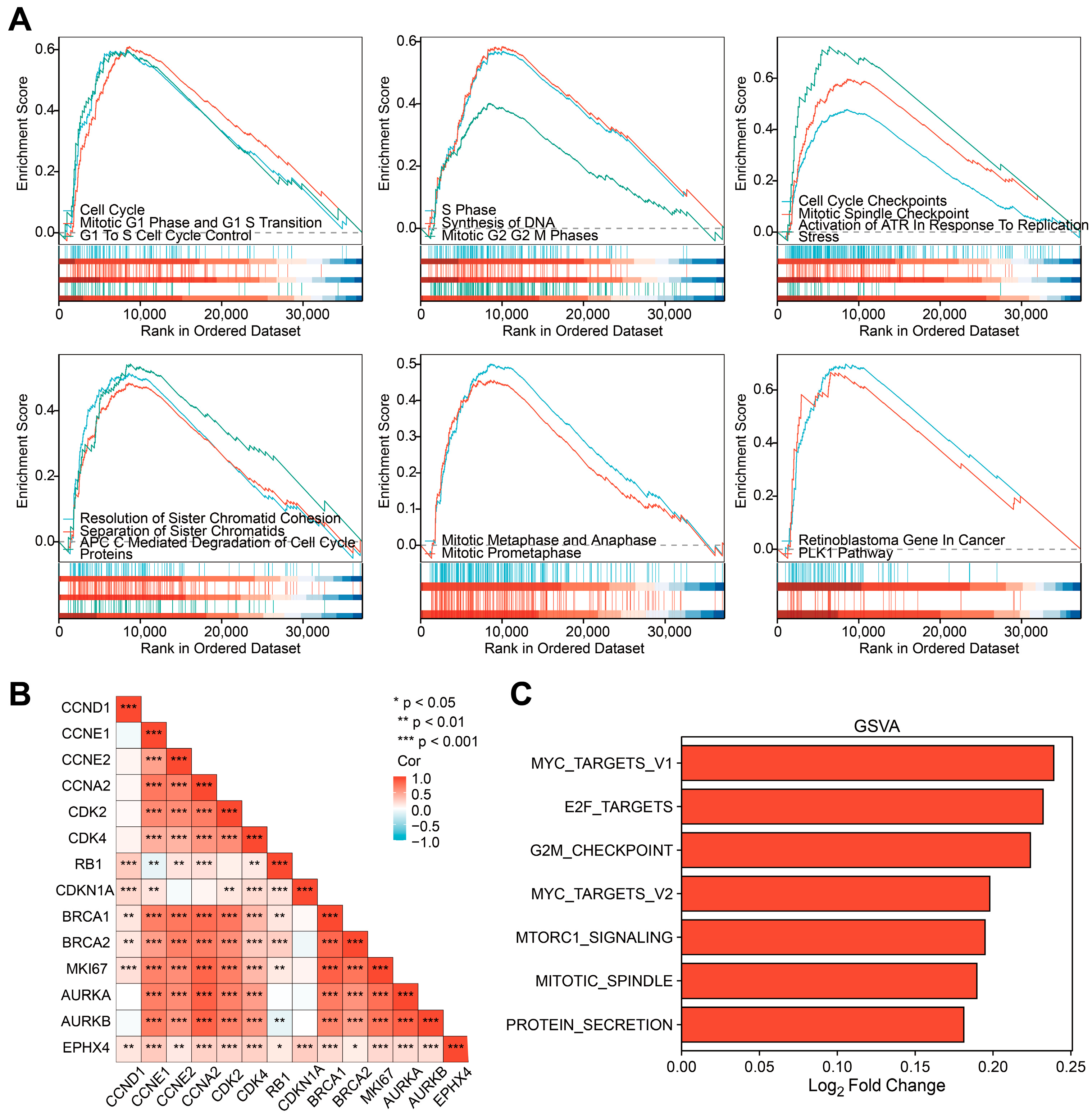
2.7. EPHX4 Might Be Significantly Involved in the Regulation of the Cell Cycle
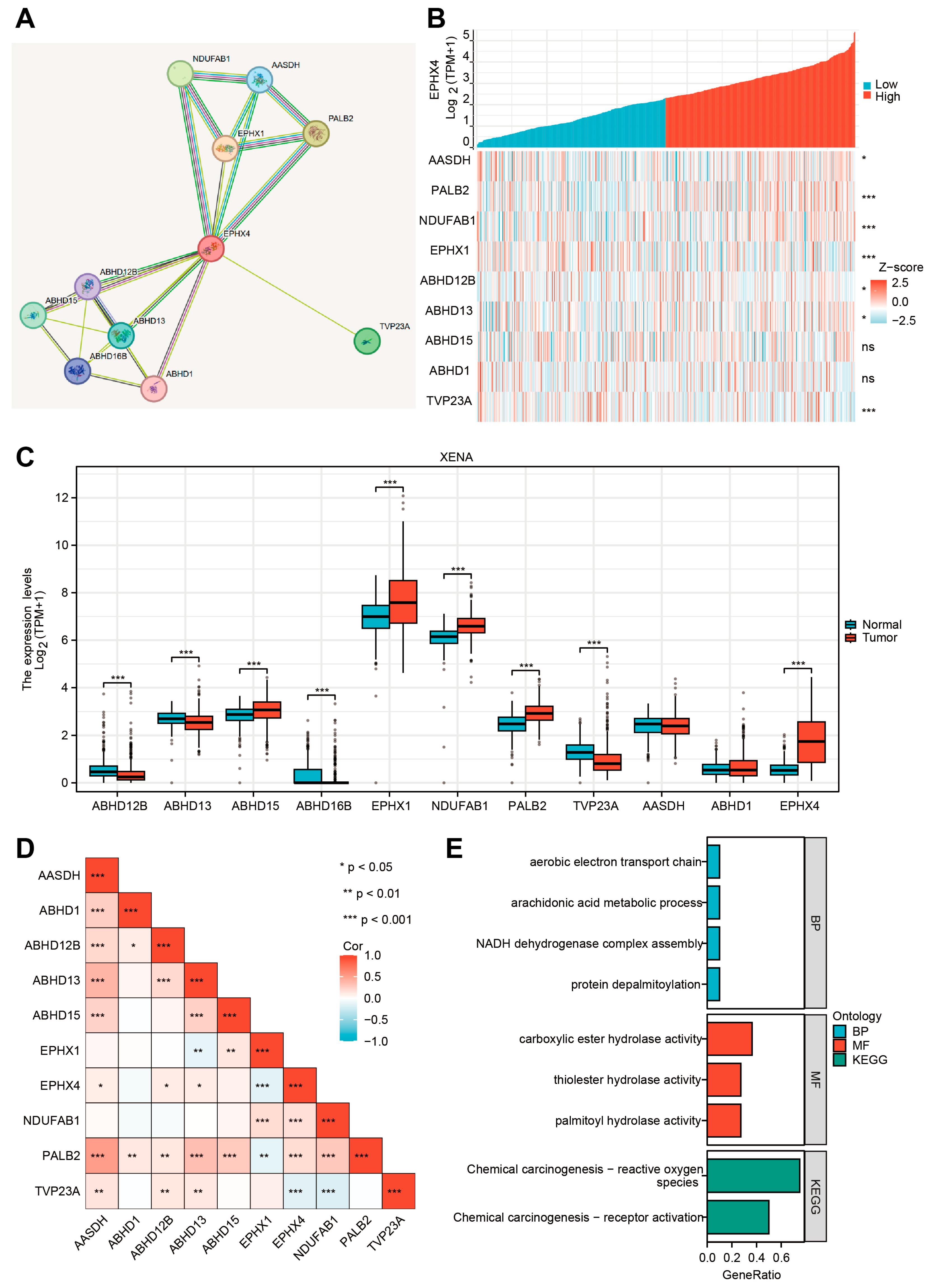
2.8. EPHX4-Related Proteins and Their Functional Enrichment Analyses
3. Discussion
4. Materials and Methods
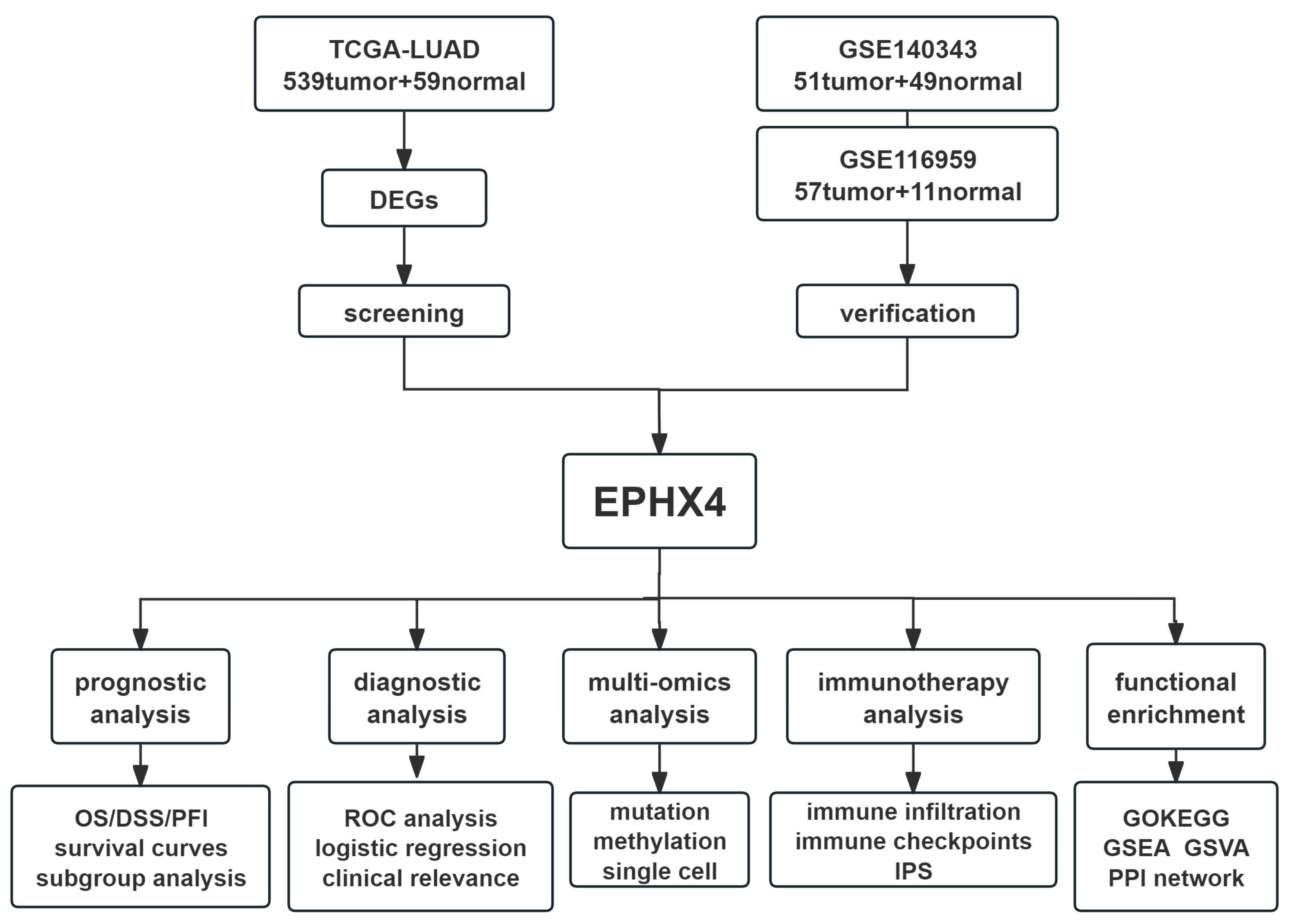
4.1. Flow Chart of the Present Study
4.2. Data Acquisition and Differential Expression Analysis
4.3. Screening of Candidate Genes and Diagnostic Value Analysis
4.4. Prognostic Value Analysis
4.5. Correlation Analysis with Clinical Variables and Prognostic Nomogram Construction
4.6. Immune Cell Infiltration Analysis and Immunotherapy Outcome Prediction
4.7. Gene Mutation and Methylation Analysis
4.8. Functional Enrichment Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adams, S.J.; Stone, E.; Baldwin, D.R.; Vliegenthart, R.; Lee, P.; Fintelmann, F.J. Lung Cancer Screening. Lancet 2023, 401, 390–408. [Google Scholar] [CrossRef] [PubMed]
- Morel, M.; Long, W. FBXL16 Promotes Cell Growth and Drug Resistance in Lung Adenocarcinomas with KRAS Mutation by Stabilizing IRS1 and Upregulating IRS1/AKT Signaling. Mol. Oncol. 2024, 18, 762–777. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, L.; Zhang, H.; Zhao, Y.; Zhang, H.; Wu, S.; Xu, B. A Risk Model Developed Based on Tumor Microenvironment Predicts Overall Survival and Associates with Tumor Immunity of Patients with Lung Adenocarcinoma. Oncogene 2021, 40, 4413–4424. [Google Scholar] [CrossRef]
- Bae, S.; Yoon, J.H.; Moon, H.J.; Kim, M.J.; Kim, E.-K. Breast Microcalcifications: Diagnostic Outcomes According to Image-Guided Biopsy Method. Korean J. Radiol. 2015, 16, 996–1005. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef] [PubMed]
- Fretland, A.J.; Omiecinski, C.J. Epoxide Hydrolases: Biochemistry and Molecular Biology. Chem. Biol. Interact. 2000, 129, 41–59. [Google Scholar] [CrossRef]
- Morisseau, C.; Kodani, S.D.; Kamita, S.G.; Yang, J.; Lee, K.S.S.; Hammock, B.D. Relative Importance of Soluble and Microsomal Epoxide Hydrolases for the Hydrolysis of Epoxy-Fatty Acids in Human Tissues. Int. J. Mol. Sci. 2021, 22, 4993. [Google Scholar] [CrossRef]
- Kodani, S.D.; Morisseau, C. Role of Epoxy-Fatty Acids and Epoxide Hydrolases in the Pathology of Neuro-Inflammation. Biochimie 2019, 159, 59–65. [Google Scholar] [CrossRef]
- Gautheron, J.; Jéru, I. The Multifaceted Role of Epoxide Hydrolases in Human Health and Disease. Int. J. Mol. Sci. 2020, 22, 13. [Google Scholar] [CrossRef]
- Decker, M.; Adamska, M.; Cronin, A.; Di Giallonardo, F.; Burgener, J.; Marowsky, A.; Falck, J.R.; Morisseau, C.; Hammock, B.D.; Gruzdev, A.; et al. EH3 (ABHD9): The First Member of a New Epoxide Hydrolase Family with High Activity for Fatty Acid Epoxides. J. Lipid Res. 2012, 53, 2038–2045. [Google Scholar] [CrossRef]
- Cao, L.; Ba, Y.; Chen, F.; Li, D.; Zhang, S.; Zhang, H. The Prognostic Significance of Epoxide Hydrolases in Colorectal Cancer. Biochem. Biophys. Rep. 2025, 41, 101912. [Google Scholar] [CrossRef]
- Shen, N.; Gao, G.; Lu, X.; Jin, J.; Lin, L.; Qian, M.; Qin, Y. Comprehensive Analysis of the Immune Implication of EPHX4 Gene in Laryngeal Squamous Cell Carcinoma. Braz. J. Otorhinolaryngol. 2024, 90, 101411. [Google Scholar] [CrossRef]
- Tuppurainen, H.; Laurila, N.; Nätynki, M.; Eshraghi, L.; Tervasmäki, A.; Erichsen, L.; Sørensen, C.S.; Pylkäs, K.; Winqvist, R.; Peltoketo, H. PALB2-Mutated Human Mammary Cells Display a Broad Spectrum of Morphological and Functional Abnormalities Induced by Increased TGFβ Signaling. Cell. Mol. Life Sci. 2024, 81, 173. [Google Scholar] [CrossRef]
- Triepels, R.; Smeitink, J.; Loeffen, J.; Smeets, R.; Buskens, C.; Trijbels, F.; van den Heuvel, L. The Human Nuclear-Encoded Acyl Carrier Subunit (NDUFAB1) of the Mitochondrial Complex I in Human Pathology. J. Inherit. Metab. Dis. 1999, 22, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Stein, I.S.; Gottfried, A.; Zimmermann, J.; von Mollard, G.F. TVP23 Interacts Genetically with the Yeast SNARE VTI1 and Functions in Retrograde Transport from the Early Endosome to the Late Golgi. Biochem. J. 2009, 419, 229–236. [Google Scholar] [CrossRef]
- Mino-Kenudson, M.; Schalper, K.; Cooper, W.; Dacic, S.; Hirsch, F.R.; Jain, D.; Lopez-Rios, F.; Tsao, M.S.; Yatabe, Y.; Beasley, M.B.; et al. Predictive Biomarkers for Immunotherapy in Lung Cancer: Perspective From the International Association for the Study of Lung Cancer Pathology Committee. J. Thorac. Oncol. 2022, 17, 1335–1354. [Google Scholar] [CrossRef] [PubMed]
- Gibney, G.T.; Weiner, L.M.; Atkins, M.B. Predictive Biomarkers for Checkpoint Inhibitor-Based Immunotherapy. Lancet Oncol. 2016, 17, e542–e551. [Google Scholar] [CrossRef]
- Taranto, D.; Kloosterman, D.J.; Akkari, L. Macrophages and T Cells in Metabolic Disorder-Associated Cancers. Nat. Rev. Cancer 2024, 24, 744–767. [Google Scholar] [CrossRef]
- Huang, S.; Shi, J.; Shen, J.; Fan, X. Metabolic Reprogramming of Neutrophils in the Tumor Microenvironment: Emerging Therapeutic Targets. Cancer Lett. 2025, 612, 217466. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Y.; Huang, J.; Nie, R.-C.; Wu, Q.-N.; Zuo, Z.; Yuan, S.; Yu, K.; Liang, C.-C.; Pan, Y.-Q.; et al. CAF-Macrophage Crosstalk in Tumour Microenvironments Governs the Response to Immune Checkpoint Blockade in Gastric Cancer Peritoneal Metastases. Gut 2025, 74, 350–363. [Google Scholar] [CrossRef]
- Bao, L.; Li, Y.; Hu, X.; Gong, Y.; Chen, J.; Huang, P.; Tan, Z.; Ge, M.; Pan, Z. Targeting SIGLEC15 as an Emerging Immunotherapy for Anaplastic Thyroid Cancer. Int. Immunopharmacol. 2024, 133, 112102. [Google Scholar] [CrossRef] [PubMed]
- Asciolla, J.J.; Wu, X.; Adamopoulos, C.; Gavathiotis, E.; Poulikakos, P.I. Resistance Mechanisms and Therapeutic Strategies of CDK4 and CDK6 Kinase Targeting in Cancer. Nat. Cancer 2025, 6, 24–40. [Google Scholar] [CrossRef]
- Chou, J.; Quigley, D.A.; Robinson, T.M.; Feng, F.Y.; Ashworth, A. Transcription-Associated Cyclin-Dependent Kinases as Targets and Biomarkers for Cancer Therapy. Cancer Discov. 2020, 10, 351–370. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, S.I.; Pelletier, L.A.; Espada, A.; Gutiérrez, J.; Sanfeliciano, S.M.G.; Rauch, C.T.; Ganado, M.P.; Baquero, C.; Zapatero, E.; Zhang, A.; et al. Crystal Structure of Active CDK4-Cyclin D and Mechanistic Basis for Abemaciclib Efficacy. npj Breast Cancer 2022, 8, 126. [Google Scholar] [CrossRef]
- Zhang, D.; Baldwin, P.; Leal, A.S.; Carapellucci, S.; Sridhar, S.; Liby, K.T. A Nano-Liposome Formulation of the PARP Inhibitor Talazoparib Enhances Treatment Efficacy and Modulates Immune Cell Populations in Mammary Tumors of BRCA-Deficient Mice. Theranostics 2019, 9, 6224–6238. [Google Scholar] [CrossRef]
- Uxa, S.; Castillo-Binder, P.; Kohler, R.; Stangner, K.; Müller, G.A.; Engeland, K. Ki-67 Gene Expression. Cell Death Differ. 2021, 28, 3357–3370. [Google Scholar] [CrossRef]
- Teli, G.; Maji, L.; Pal, R.; Maheshwari, N.; Purawarga Matada, G.S.; Chawla, P.A.; Chawla, V. Recent Advancements in Mechanistic Research, Therapeutic Potential, and Structure-Activity Relationships of Aurora Kinase Inhibitors in Cancer Therapies. Bioorganic Chem. 2025, 154, 107976. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A Tool for Pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for Analysis of Tumor-Infiltrating Immune Cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An Update to the Integrated Cancer Data Analysis Platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Nastou, K.; Koutrouli, M.; Kirsch, R.; Mehryary, F.; Hachilif, R.; Hu, D.; Peluso, M.E.; Huang, Q.; Fang, T.; et al. The STRING Database in 2025: Protein Networks with Directionality of Regulation. Nucleic Acids Res. 2025, 53, D730–D737. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Chen, Y. Integrative Analysis of EPHX4 as a Novel Prognostic and Diagnostic Biomarker in Lung Adenocarcinoma. Int. J. Mol. Sci. 2025, 26, 5095. https://doi.org/10.3390/ijms26115095
Liu P, Chen Y. Integrative Analysis of EPHX4 as a Novel Prognostic and Diagnostic Biomarker in Lung Adenocarcinoma. International Journal of Molecular Sciences. 2025; 26(11):5095. https://doi.org/10.3390/ijms26115095
Chicago/Turabian StyleLiu, Pengze, and Yutong Chen. 2025. "Integrative Analysis of EPHX4 as a Novel Prognostic and Diagnostic Biomarker in Lung Adenocarcinoma" International Journal of Molecular Sciences 26, no. 11: 5095. https://doi.org/10.3390/ijms26115095
APA StyleLiu, P., & Chen, Y. (2025). Integrative Analysis of EPHX4 as a Novel Prognostic and Diagnostic Biomarker in Lung Adenocarcinoma. International Journal of Molecular Sciences, 26(11), 5095. https://doi.org/10.3390/ijms26115095





