Rat Hair Follicle Stem Cell-Derived Exosomes: Isolation, Characterization and Comparative Analysis of Their In Vitro Wound Healing Potential
Abstract
1. Introduction
2. Results
2.1. rHFSC-Derived Exosomes Analysis and Comparison to Secretome
2.2. RT-PCR
2.3. Scanning Electron Microscopy (SEM) with Energy-Dispersive X-Ray Spectroscopy (EDS)
2.4. Total Protein Quantification
2.5. Prestoblue Assay
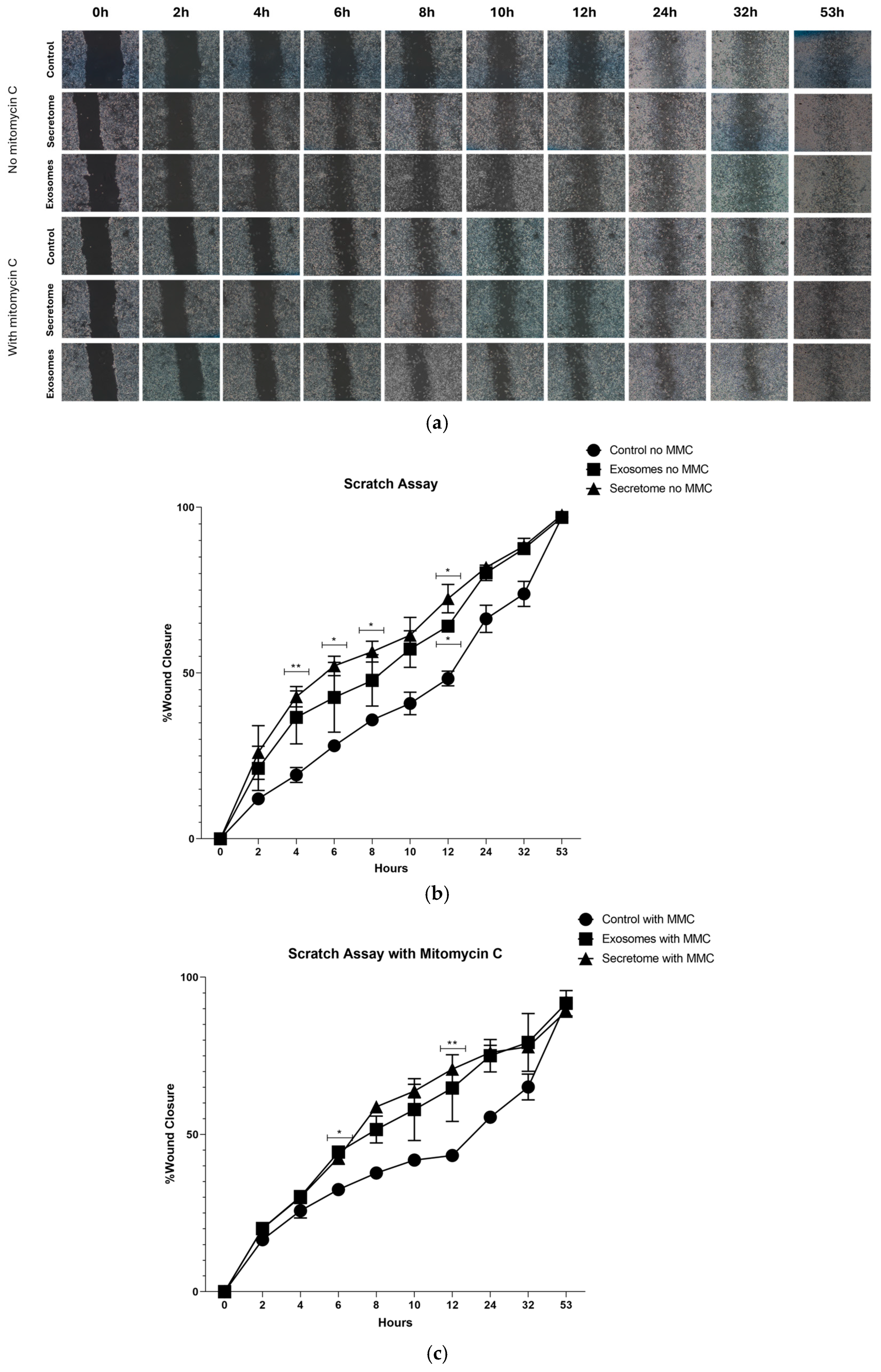
2.6. Scratch Assay
3. Discussion
4. Materials and Methods
4.1. rHFSCs-Derived Secretome and Exosomes Isolation
4.2. rHFSC-Derived Exosomes Analysis
4.3. Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
4.3.1. RNA Isolation and cDNA Synthesis
4.3.2. Quantitative RT-PCR Assay
4.4. Scanning Electron Microscopy (SEM) and Energy Dispersive Spectroscopy (EDS)
4.5. Total Protein Quantification
4.6. Prestoblue Assay
4.7. Scratch Assay
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACTB | Beta-Actin |
| Au | Gold |
| BCA | Bicinchoninic Acid |
| C | Carbon |
| Ca | Calcium |
| CM2D | Conditioned Medium 2D |
| Ct | Threshold cycle |
| DMSO | Dimethyl Sulfoxide |
| EBSD | Electron Backscattered Diffraction |
| ECM | Extracellular Matrix |
| EDS | Energy-Dispersive X-ray Spectroscopy |
| EGF | Epidermal Growth Factor Recombinant Protein |
| EVs | Extracellular Vesicles |
| FBS | Bovine Fetal Serum |
| Fe | Iron |
| GAPDH | Glyceraldehyde 3-phosphate Dehydrogenase |
| G-CSF | Granulocyte Colony-Stimulating Factor |
| GM-CSF | Granulocyte-Macrophage Colony-Stimulating Factor |
| GRO/KC/CINC-1 | Human Growth-Regulated Oncogene/Keratinocyte Chemoattractant/Cytokine-Induced Neutrophil Chemoattractant-1 |
| HMDS | Hexamethyldisilazane |
| IFNγ | Interferon Gamma |
| IL | Interleukin |
| IP-10 | Interferon-Gamma Inducible Protein |
| MCP-1 | Monocyte Chemoattractant Protein-1 |
| MIP | Macrophage Inflammatory Protein |
| MMC | Mitomycin C |
| MSCs | Mesenchymal Stem Cells |
| N | Nitrogen |
| O | Oxygen |
| P | Phosphorus |
| PCR | Polymerase Chain Reaction |
| Pd | Palladium |
| RANTES | Regulated upon Activation, Normal T-Cell Expressed and Presumably Secreted |
| rHFSCs | Rat Hair Follicle Stem Cells |
| RT-PCR | Reverse Transcriptase Polymerase Chain Reaction |
| S | Sulfur |
| SEM | Standard Error of the Mean |
| Si | Silicon |
| TGFβ | Transforming Growth Factor Beta |
| Ti | Titanium |
| TNFα | Tumor Necrosis Factor-Alpha |
| UP | University of Porto |
| VEGF | Vascular Endothelial Growth Factor |
| Z | Zone |
References
- Sousa, P.; Lopes, B.; Sousa, A.C.; Moreira, A.; Coelho, A.; Alvites, R.; Alves, N.; Geuna, S.; Maurício, A.C. Advancements and Insights in Exosome-Based Therapies for Wound Healing: A Comprehensive Systematic Review (2018–June 2023). Biomedicines 2023, 11, 2099. [Google Scholar] [CrossRef]
- Sousa, P.; Lopes, B.; Sousa, A.C.; Coelho, A.; de Sousa Moreira, A.; Rêma, A.; Gonçalves-Maia, M.; Amorim, I.; Alvites, R.; Alves, N.; et al. Isolation, Expansion, and Characterization of Rat Hair Follicle Stem Cells and Their Secretome: Insights into Wound Healing Potential. Biomedicines 2024, 12, 2854. [Google Scholar] [CrossRef] [PubMed]
- Lopes, B.; Sousa, P.; Alvites, R.; Branquinho, M.; Sousa, A.; Mendonça, C.; Atayde, L.M.; Maurício, A.C. The Application of Mesenchymal Stem Cells on Wound Repair and Regeneration. Appl. Sci. 2021, 11, 3000. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, B.; Yang, Y.; Jiang, Q.; Li, T.; Gong, J.; Tang, H.; Zhang, Q. Stem cell-derived exosomes: Emerging therapeutic opportunities for wound healing. Stem Cell Res. Ther. 2023, 14, 107. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, M.T.; Marques, A.P.; Reis, R.L. Using Stem Cells in Skin Regeneration: Possibilities and Reality. Stem Cells Dev. 2012, 21, 1201–1214. [Google Scholar] [CrossRef]
- Rezaie, F.; Momeni-Moghaddam, M.; Naderi-Meshkin, H. Regeneration and Repair of Skin Wounds: Various Strategies for Treatment. Int. J. Low. Extrem. Wounds 2019, 18, 247–261. [Google Scholar] [CrossRef]
- Dean, J.; Hoch, C.; Wollenberg, B.; Navidzadeh, J.; Maheta, B.; Mandava, A.; Knoedler, S.; Sherwani, K.; Baecher, H.; Schmitz, A.; et al. Advancements in bioengineered and autologous skin grafting techniques for skin reconstruction: A comprehensive review. Front. Bioeng. Biotechnol. 2024, 12, 1461328. [Google Scholar] [CrossRef]
- do Amaral, R.J.F.C.; Zayed, N.M.A.; Pascu, E.I.; Cavanagh, B.; Hobbs, C.; Santarella, F.; Simpson, C.R.; Murphy, C.M.; Sridharan, R.; González-Vázquez, A.; et al. Functionalising Collagen-Based Scaffolds With Platelet-Rich Plasma for Enhanced Skin Wound Healing Potential. Front. Bioeng. Biotechnol. 2019, 7, 371. [Google Scholar] [CrossRef]
- Chocarro-Wrona, C.; López-Ruiz, E.; Perán, M.; Gálvez-Martín, P.; Marchal, J.A. Therapeutic strategies for skin regeneration based on biomedical substitutes. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 484–496. [Google Scholar] [CrossRef]
- Zhao, M.; Shi, J.; Cai, W.; Liu, K.; Shen, K.; Li, Z.; Wang, Y.; Hu, D. Advances on Graphene-Based Nanomaterials and Mesenchymal Stem Cell-Derived Exosomes Applied in Cutaneous Wound Healing. Int. J. Nanomed. 2021, 16, 2647–2665. [Google Scholar] [CrossRef]
- Vizoso, F.J.; Eiro, N.; Cid, S.; Schneider, J.; Perez-Fernandez, R. Mesenchymal Stem Cell Secretome: Toward Cell-Free Therapeutic Strategies in Regenerative Medicine. Int. J. Mol. Sci. 2017, 18, 1852. [Google Scholar] [CrossRef]
- Teixeira, F.G.; Salgado, A.J. Mesenchymal stem cells secretome: Current trends and future challenges. Neural Regen. Res. 2020, 15, 75–77. [Google Scholar] [CrossRef] [PubMed]
- Bormann, D.; Gugerell, A.; Ankersmit, H.J.; Mildner, M. Therapeutic Application of Cell Secretomes in Cutaneous Wound Healing. J. Investig. Dermatol. 2023, 143, 893–912. [Google Scholar] [CrossRef] [PubMed]
- Fadilah, N.I.M.; Jailani, M.S.M.A.K.; Hisham, M.A.I.B.; Raj, N.S.; Shamsuddin, S.A.; Ng, M.H.; Fauzi, M.B.; Maarof, M. Cell secretomes for wound healing and tissue regeneration: Next generation acellular based tissue engineered products. J. Tissue Eng. 2022, 13, 20417314221114273. [Google Scholar] [CrossRef] [PubMed]
- Suhandi, C.; Wilar, G.; Elamin, K.M.; Dewayani, A.R.; Ghaliya, S.; Abdullah, A.; Wathoni, N. The Effect of Stem Cell Secretome on the Improvement of Diabetic Wound Recovery: A Systematic Review and Meta-Analysis of In Vivo Studies. Curr. Ther. Res. 2025, 102, 100778. [Google Scholar] [CrossRef]
- Suhandi, C.; Mohammed, A.F.A.; Wilar, G.; El-Rayyes, A.; Wathoni, N. Effectiveness of Mesenchymal Stem Cell Secretome on Wound Healing: A Systematic Review and Meta-analysis. Tissue Eng. Regen. Med. 2023, 20, 1053–1062. [Google Scholar] [CrossRef]
- da Silva, M.M.; Olsson, D.C.; Teixeira, B.L.; Jeremias, T.d.S.; Trentin, A.G. Mesenchymal Stromal Cell Secretome for Therapeutic Application in Skin Wound Healing: A Systematic Review of Preclinical Studies. Cells Tissues Organs 2022, 212, 567–582. [Google Scholar] [CrossRef]
- Kim, J.H.; Green, D.S.; Ju, Y.M.; Harrison, M.; Vaughan, J.W.; Atala, A.; Lee, S.J.; Jackson, J.D.; Nykiforuk, C.; Yoo, J.J. Identification and characterization of stem cell secretome-based recombinant proteins for wound healing applications. Front. Bioeng. Biotechnol. 2022, 10, 954682. [Google Scholar] [CrossRef]
- Prasai, A.; Jay, J.W.; Jupiter, D.; Wolf, S.E.; El Ayadi, A. Role of Exosomes in Dermal Wound Healing: A Systematic Review. J. Investig. Dermatol. 2022, 142, 662–678.e668. [Google Scholar] [CrossRef]
- Singh, S.; Paul, D.; Nath, V.; A, R. Exosomes: Current knowledge and future perspectives. Tissue Barriers 2024, 12, 2232248. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Zhu, K.; He, W.; Guo, X.; Wang, T.; Gong, S.; Zhu, Z. Exosomes from mesenchymal stem cells: Potential applications in wound healing. Life Sci. 2024, 357, 123066. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Wang, F.; Xu, G.; Shu, F.; Fan, K.; Wang, D. Advancements in engineered exosomes for wound repair: Current research and future perspectives. Front. Bioeng. Biotechnol. 2023, 11, 1301362. [Google Scholar] [CrossRef]
- Lv, H.; Liu, H.; Sun, T.; Wang, H.; Zhang, X.; Xu, W. Exosome derived from stem cell: A promising therapeutics for wound healing. Front. Pharmacol. 2022, 13, 957771. [Google Scholar] [CrossRef]
- Armstrong, J.P.; Holme, M.N.; Stevens, M.M. Re-Engineering Extracellular Vesicles as Smart Nanoscale Therapeutics. ACS Nano 2017, 11, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, X.; Hao, H.; Xu, H.; Shu, J.; Hou, Q.; Wang, M. Exosomes Derived from Umbilical Cord Mesenchymal Stem Cells Treat Cutaneous Nerve Damage and Promote Wound Healing. Front. Cell Neurosci. 2022, 16, 913009. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Quan, H. Adipose-derived stem cells-derived exosomes facilitate cutaneous wound healing by delivering XIST and restoring discoidin domain receptor 2. Cytokine 2022, 158, 155981. [Google Scholar] [CrossRef]
- Zhou, Z.; Zheng, J.; Lin, D.; Xu, R.; Chen, Y.; Hu, X. Exosomes derived from dental pulp stem cells accelerate cutaneous wound healing by enhancing angiogenesis via the Cdc42/p38 MAPK pathway. Int. J. Mol. Med. 2022, 50, 143. [Google Scholar] [CrossRef]
- Tienda-Vázquez, M.A.; Hanel, J.M.; Márquez-Arteaga, E.M.; Salgado-Álvarez, A.P.; Scheckhuber, C.Q.; Alanis-Gómez, J.R.; Espinoza-Silva, J.I.; Ramos-Kuri, M.; Hernández-Rosas, F.; Melchor-Martínez, E.M.; et al. Exosomes: A Promising Strategy for Repair, Regeneration and Treatment of Skin Disorders. Cells 2023, 12, 1625. [Google Scholar] [CrossRef]
- Golchin, A.; Shams, F.; Basiri, A.; Ranjbarvan, P.; Kiani, S.; Sarkhosh-Inanlou, R.; Ardeshirylajimi, A.; Gholizadeh-Ghaleh Aziz, S.; Sadigh, S.; Rasmi, Y. Combination Therapy of Stem Cell-derived Exosomes and Biomaterials in the Wound Healing. Stem Cell Rev. Rep. 2022, 18, 1892–1911. [Google Scholar] [CrossRef]
- Han, X.; Saengow, C.; Ju, L.; Ren, W.; Ewoldt, R.H.; Irudayaraj, J. Exosome-coated oxygen nanobubble-laden hydrogel augments intracellular delivery of exosomes for enhanced wound healing. Nat. Commun. 2024, 15, 3435. [Google Scholar] [CrossRef]
- Narauskaitė, D.; Vydmantaitė, G.; Rusteikaitė, J.; Sampath, R.; Rudaitytė, A.; Stašytė, G.; Aparicio Calvente, M.I.; Jekabsone, A. Extracellular Vesicles in Skin Wound Healing. Pharmaceuticals 2021, 14, 811. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, S.; Shen, Y.; Lu, H.; Zhao, X.; Wang, X.; Wang, Y.; Wang, T.; Liu, B.; Yao, L.; et al. Therapeutic application of mesenchymal stem cell-derived exosomes in skin wound healing. Front. Bioeng. Biotechnol. 2024, 12, 1428793. [Google Scholar] [CrossRef] [PubMed]
- Liew, F.F.; Chew, B.C.; Ooi, J. Wound Healing Properties of Exosomes—A Review and Modelling of Combinatorial Analysis Strategies. Curr. Mol. Med. 2022, 22, 165–191. [Google Scholar] [CrossRef]
- Zhang, Y.; Cui, J.; Cang, Z.; Pei, J.; Zhang, X.; Song, B.; Fan, X.; Ma, X.; Li, Y. Hair follicle stem cells promote epidermal regeneration under expanded condition. Front. Physiol. 2024, 15, 1306011. [Google Scholar] [CrossRef]
- Cui, H.; Fu, L.Q.; Teng, Y.; He, J.J.; Shen, Y.Y.; Bian, Q.; Wang, T.Z.; Wang, M.X.; Pang, X.W.; Lin, Z.W.; et al. Human Hair Follicle Mesenchymal Stem Cell-Derived Exosomes Attenuate UVB-Induced Photoaging via the miR-125b-5p/TGF-β1/Smad Axis. Biomater. Res. 2025, 29, 0121. [Google Scholar] [CrossRef]
- Hu, S.; Zhang, J.; Ji, Q.; Xie, S.; Jiang, J.; Ni, H.; He, X.; Yang, Y.; Wu, M. Exosomes derived from uMSCs promote hair regrowth in alopecia areata through accelerating human hair follicular keratinocyte proliferation and migration. Cell Biol. Int. 2024, 48, 154–161. [Google Scholar] [CrossRef]
- Qiao, Z.; Wang, X.; Zhao, H.; Deng, Y.; Zeng, W.; Yang, K.; Chen, H.; Yan, Q.; Li, C.; Wu, J.; et al. The effectiveness of cell-derived exosome therapy for diabetic wound: A systematic review and meta-analysis. Ageing Res. Rev. 2023, 85, 101858. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Bai, X.; Zhao, B.; Li, Y.; Zhang, Y.; Li, Z.; Wang, X.; Luo, L.; Han, F.; Zhang, J.; et al. Cell-free therapy based on adipose tissue stem cell-derived exosomes promotes wound healing via the PI3K/Akt signaling pathway. Exp. Cell Res. 2018, 370, 333–342. [Google Scholar] [CrossRef]
- Ahmadpour, F.; Rasouli, H.R.; Talebi, S.; Golchin, D.; Esmailinejad, M.R.; Razie, A. Effects of exosomes derived from fibroblast cells on skin wound healing in Wistar rats. Burns 2023, 49, 1372–1381. [Google Scholar] [CrossRef]
- Li, D.; Wu, N. Mechanism and application of exosomes in the wound healing process in diabetes mellitus. Diabetes Res. Clin. Pract. 2022, 187, 109882. [Google Scholar] [CrossRef]
- Wang, K.; Yang, Z.; Zhang, B.; Gong, S.; Wu, Y. Adipose-Derived Stem Cell Exosomes Facilitate Diabetic Wound Healing: Mechanisms and Potential Applications. Int. J. Nanomed. 2024, 19, 6015–6033. [Google Scholar] [CrossRef]
- Li, Y.; Giovannini, S.; Wang, T.; Fang, J.; Li, P.; Shao, C.; Wang, Y.; Agostini, M.; Bove, P.; Mauriello, A.; et al. p63: A crucial player in epithelial stemness regulation. Oncogene 2023, 42, 3371–3384. [Google Scholar] [CrossRef] [PubMed]
- Melino, G.; Memmi, E.M.; Pelicci, P.G.; Bernassola, F. Maintaining epithelial stemness with p63. Sci. Signal. 2015, 8, re9. [Google Scholar] [CrossRef] [PubMed]
- Sidney, L.E.; Branch, M.J.; Dunphy, S.E.; Dua, H.S.; Hopkinson, A. Concise Review: Evidence for CD34 as a Common Marker for Diverse Progenitors. Stem Cells 2014, 32, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Veijouye, S.J.; Yari, A.; Heidari, F.; Sajedi, N.; Moghani, F.G.; Nobakht, M. Bulge Region as a Putative Hair Follicle Stem Cells Niche: A Brief Review. Iran. J. Public Health 2017, 46, 1167–1175. [Google Scholar]
- Kloepper, J.E.; Tiede, S.; Brinckmann, J.; Reinhardt, D.P.; Meyer, W.; Faessler, R.; Paus, R. Immunophenotyping of the human bulge region: The quest to define useful in situ markers for human epithelial hair follicle stem cells and their niche. Exp. Dermatol. 2008, 17, 592–609. [Google Scholar] [CrossRef]
- Xu, J.; Li, Z.; Hou, Y.; Fang, W. Potential mechanisms underlying the Runx2 induced osteogenesis of bone marrow mesenchymal stem cells. Am. J. Transl. Res. 2015, 7, 2527–2535. [Google Scholar]
- Rumiński, S.; Kalaszczyńska, I.; Lewandowska-Szumieł, M. Effect of cAMP Signaling Regulation in Osteogenic Differentiation of Adipose-Derived Mesenchymal Stem Cells. Cells 2020, 9, 1587. [Google Scholar] [CrossRef]
- International Standard ISO 10993-5; Biological Evaluation of Medical Devices—Part 5: Tests for In Vitro Cytotoxicity. ISO: Geneva, Switzerland, 2009.
- Wong, R.S.-Y.; Tan, T.; Pang, A.S.-R.; Srinivasan, D.K. The role of cytokines in wound healing: From mechanistic insights to therapeutic applications. Explor. Immunol. 2025, 5, 1003183. [Google Scholar] [CrossRef]
- Nirenjen, S.; Narayanan, J.; Tamilanban, T.; Subramaniyan, V.; Chitra, V.; Fuloria, N.K.; Wong, L.S.; Ramachawolran, G.; Sekar, M.; Gupta, G.; et al. Exploring the contribution of pro-inflammatory cytokines to impaired wound healing in diabetes. Front. Immunol. 2023, 14, 1216321. [Google Scholar] [CrossRef]
- Nosenko, M.A.; Ambaryan, S.G.; Drutskaya, M.S. Proinflammatory Cytokines and Skin Wound Healing in Mice. Mol. Biol. 2019, 53, 653–664. [Google Scholar] [CrossRef]
- Nurkesh, A.; Jaguparov, A.; Jimi, S.; Saparov, A. Recent Advances in the Controlled Release of Growth Factors and Cytokines for Improving Cutaneous Wound Healing. Front. Cell Dev. Biol. 2020, 8, 638. [Google Scholar] [CrossRef]
- Schaffrick, L.; Ding, J.; Kwan, P.; Tredget, E. The dynamic changes of monocytes and cytokines during wound healing post-burn injury. Cytokine 2023, 168, 156231. [Google Scholar] [CrossRef] [PubMed]
- Papait, A.; Ragni, E.; Cargnoni, A.; Vertua, E.; Romele, P.; Masserdotti, A.; Perucca Orfei, C.; Signoroni, P.B.; Magatti, M.; Silini, A.R.; et al. Comparison of EV-free fraction, EVs, and total secretome of amniotic mesenchymal stromal cells for their immunomodulatory potential: A translational perspective. Front. Immunol. 2022, 13, 960909. [Google Scholar] [CrossRef]
- Claudinot, S.; Sakabe, J.-I.; Oshima, H.; Gonneau, C.; Mitsiadis, T.; Littman, D.; Bonfanti, P.; Martens, G.; Nicolas, M.; Rochat, A.; et al. Tp63-expressing adult epithelial stem cells cross lineages boundaries revealing latent hairy skin competence. Nat. Commun. 2020, 11, 5645. [Google Scholar] [CrossRef]
- Lee, B.-W.; Oh, J.-W.; Seo, M.; Hwang, J.-S.; Yoon, B. Expression of p63 and its association with cell proliferation at different stages of murine hair follicle cycle. J. Biomed. Transl. Res. 2018, 19, 10–15. [Google Scholar] [CrossRef]
- Ohyama, M.; Terunuma, A.; Tock, C.L.; Radonovich, M.F.; Pise-Masison, C.A.; Hopping, S.B.; Brady, J.N.; Udey, M.C.; Vogel, J.C. Characterization and isolation of stem cell–enriched human hair follicle bulge cells. J. Clin. Investig. 2006, 116, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Hoogduijn, M.J.; Gorjup, E.; Genever, P.G. Comparative characterization of hair follicle dermal stem cells and bone marrow mesenchymal stem cells. Stem Cells Dev. 2006, 15, 49–60. [Google Scholar] [CrossRef]
- Komori, T. Whole Aspect of Runx2 Functions in Skeletal Development. Int. J. Mol. Sci. 2022, 23, 5776. [Google Scholar] [CrossRef]
- Komori, T. Regulation of Skeletal Development and Maintenance by Runx2 and Sp7. Int. J. Mol. Sci. 2024, 25, 10102. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, Y.; Qin, H.; Tong, S.; Sun, Q.; Wang, T.; Zhang, H.; Cui, M.; Guo, S. Osteogenically-induced exosomes stimulate osteogenesis of human adipose-derived stem cells. Cell Tissue Bank. 2021, 22, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Ievlev, V.; Lynch, T.J.; Freischlag, K.W.; Gries, C.B.; Shah, A.; Pai, A.C.; Ahlers, B.A.; Park, S.; Engelhardt, J.F.; Parekh, K.R. Krt14 and Krt15 differentially regulate regenerative properties and differentiation potential of airway basal cells. JCI Insight 2023, 8, e162041. [Google Scholar] [CrossRef] [PubMed]
- Quan, R.; Zheng, X.; Ni, Y.; Xie, S.; Li, C. Culture and characterization of rat hair follicle stem cells. Cytotechnology 2016, 68, 621–628. [Google Scholar] [CrossRef][Green Version]
- Guda, P.R.; Sharma, A.; Anthony, A.J.; ElMasry, M.S.; Couse, A.D.; Ghatak, P.D.; Das, A.; Timsina, L.; Trinidad, J.C.; Roy, S.; et al. Nanoscopic and Functional Characterization of Keratinocyte-Originating Exosomes in the Wound Fluid of Non-Diabetic and Diabetic Chronic Wound Patients. Nano Today 2023, 52, 101954. [Google Scholar] [CrossRef]
- Garcia-Martin, R.; Brandao, B.B.; Thomou, T.; Altindis, E.; Kahn, C.R. Tissue differences in the exosomal/small extracellular vesicle proteome and their potential as indicators of altered tissue metabolism. Cell Rep. 2022, 38, 110277. [Google Scholar] [CrossRef]
- Samundeshwari, E.L.; Kattaru, S.; Kodavala, S.; Chandrasekhar, C.; Sarma, P. Prominent Expression of COL2A1, ACAN and IHH Genes are Observed in the Differentiation of Human Hematopoietic Stem Cells into Articular Type of Chondrocytes. Stem Cell Rev. Rep. 2024, 20, 1370–1373. [Google Scholar] [CrossRef]
- Levis, H.; Weston, J.; Austin, B.; Larsen, B.; Ginley-Hidinger, M.; Gullbrand, S.E.; Lawrence, B.; Bowles, R.D. Multiplex gene editing to promote cell survival using low-pH clustered regularly interspaced short palindromic repeats activation (CRISPRa) gene perturbation. Cytotherapy 2023, 25, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Shi, Q.; Li, W.; Wu, W.; Zha, Z. ITGB1 promotes the chondrogenic differentiation of human adipose-derived mesenchymal stem cells by activating the ERK signaling. J. Mol. Histol. 2020, 51, 729–739. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Rompolas, P.; Greco, V. Stem cell dynamics in the hair follicle niche. Semin. Cell Dev. Biol. 2014, 25, 34–42. [Google Scholar] [CrossRef]
- Çankirili, N.K.; Altundag, O.; Çelebi-Saltik, B. Skin Stem Cells, Their Niche and Tissue Engineering Approach for Skin Regeneration. In Cell Biology and Translational Medicine, Volume 6: Stem Cells: Their Heterogeneity, Niche and Regenerative Potential; Turksen, K., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 107–126. [Google Scholar]
- Thiagarajan, L.; Abu-Awwad, H.A.M.; Dixon, J.E. Osteogenic Programming of Human Mesenchymal Stem Cells with Highly Efficient Intracellular Delivery of RUNX2. Stem Cells Transl. Med. 2017, 6, 2146–2159. [Google Scholar] [CrossRef]
- Wang, S.; Drummond, M.L.; Guerrero-Juarez, C.F.; Tarapore, E.; MacLean, A.L.; Stabell, A.R.; Wu, S.C.; Gutierrez, G.; That, B.T.; Benavente, C.A.; et al. Single cell transcriptomics of human epidermis identifies basal stem cell transition states. Nat. Commun. 2020, 11, 4239. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.H.; Jeyaraj, M.; Qasim, M.; Kim, J.H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed]
- Soo, C.Y.; Song, Y.; Zheng, Y.; Campbell, E.C.; Riches, A.C.; Gunn-Moore, F.; Powis, S.J. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology 2012, 136, 192–197. [Google Scholar] [CrossRef]
- Franquesa, M.; Hoogduijn, M.J.; Ripoll, E.; Luk, F.; Salih, M.; Betjes, M.G.; Torras, J.; Baan, C.C.; Grinyó, J.M.; Merino, A.M. Update on controls for isolation and quantification methodology of extracellular vesicles derived from adipose tissue mesenchymal stem cells. Front. Immunol. 2014, 5, 525. [Google Scholar] [CrossRef]
- Teng, L.; Maqsood, M.; Zhu, M.; Zhou, Y.; Kang, M.; Zhou, J.; Chen, J. Exosomes Derived from Human Umbilical Cord Mesenchymal Stem Cells Accelerate Diabetic Wound Healing via Promoting M2 Macrophage Polarization, Angiogenesis, and Collagen Deposition. Int. J. Mol. Sci. 2022, 23, 10421. [Google Scholar] [CrossRef]
- Narayanan, K.B.; Bhaskar, R.; Sudhakar, K.; Nam, D.H.; Han, S.S. Polydopamine-Functionalized Bacterial Cellulose as Hydrogel Scaffolds for Skin Tissue Engineering. Gels 2023, 9, 656. [Google Scholar] [CrossRef]
- Rasti, M.; Parniaei, A.H.; Dehghani, L.; Nasr Esfahani, S.; Mirhendi, H.; Yazdani, V.; Azimian Zavareh, V. Enhancing the wound healing process through local injection of exosomes derived from blood serum: An in vitro and in vivo assessment. Regen. Ther. 2024, 26, 281–289. [Google Scholar] [CrossRef]
- Choudhary, V.; Choudhary, M.; Bollag, W.B. Exploring Skin Wound Healing Models and the Impact of Natural Lipids on the Healing Process. Int. J. Mol. Sci. 2024, 25, 3790. [Google Scholar] [CrossRef]
- Zhao, B.; Zhang, Y.; Han, S.; Zhang, W.; Zhou, Q.; Guan, H.; Liu, J.; Shi, J.; Su, L.; Hu, D. Exosomes derived from human amniotic epithelial cells accelerate wound healing and inhibit scar formation. J. Mol. Histol. 2017, 48, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.R.; Wang, C.; Patel, R.; Trujillo, A.; Patel, N.A.; Prather, J.; Gould, L.J.; Wu, M.H. Human Adipose-Derived Stem Cell Conditioned Media and Exosomes Containing MALAT1 Promote Human Dermal Fibroblast Migration and Ischemic Wound Healing. Adv. Wound Care 2018, 7, 299–308. [Google Scholar] [CrossRef]
- Villatoro, A.J.; Alcoholado, C.; Martín-Astorga, M.C.; Fernández, V.; Cifuentes, M.; Becerra, J. Comparative analysis and characterization of soluble factors and exosomes from cultured adipose tissue and bone marrow mesenchymal stem cells in canine species. Vet. Immunol. Immunopathol. 2019, 208, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Villatoro, A.J.; Martín-Astorga, M.d.C.; Alcoholado, C.; Sánchez-Martín, M.d.M.; Becerra, J. Proteomic Analysis of the Secretome and Exosomes of Feline Adipose-Derived Mesenchymal Stem Cells. Animals 2021, 11, 295. [Google Scholar] [CrossRef]
- Shakhakarmi, K.; Seo, J.-E.; Lamichhane, S.; Thapa, C.; Lee, S. EGF, a veteran of wound healing: Highlights on its mode of action, clinical applications with focus on wound treatment, and recent drug delivery strategies. Arch. Pharm. Res. 2023, 46, 299–322. [Google Scholar] [CrossRef]
- Choi, S.M.; Lee, K.-M.; Kim, H.J.; Park, I.K.; Kang, H.J.; Shin, H.-C.; Baek, D.; Choi, Y.; Park, K.H.; Lee, J.W. Effects of structurally stabilized EGF and bFGF on wound healing in type I and type II diabetic mice. Acta Biomater. 2018, 66, 325–334. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, Q.; Liu, J.; Hao, H.; Jiang, C.; Han, W. Granulocyte-Colony Stimulating Factor (G-CSF) Accelerates Wound Healing in Hemorrhagic Shock Rats by Enhancing Angiogenesis and Attenuating Apoptosis. Med. Sci. Monit. 2017, 23, 2644–2653. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.-Y.; Park, I.-H.; Song, Y.-S.; Joo, H.-W.; Lee, Y.; Shin, J.-H.; Kim, K.-S.; Kim, H. Local injection of granulocyte-colony stimulating factor accelerates wound healing in a rat excisional wound model. Tissue Eng. Regen. Med. 2016, 13, 297–303. [Google Scholar] [CrossRef]
- Goswami, A.G.; Basu, S.; Huda, F.; Pant, J.; Ghosh Kar, A.; Banerjee, T.; Shukla, V.K. An appraisal of vascular endothelial growth factor (VEGF): The dynamic molecule of wound healing and its current clinical applications. Growth Factors 2022, 40, 73–88. [Google Scholar] [CrossRef]
- Wilgus, T.A.; DiPietro, L.A. Complex roles for VEGF in dermal wound healing. J. Investig. Dermatol. 2012, 132, 493–494. [Google Scholar] [CrossRef]
- Aslan, C.; Çelebi, N.; Değim, İ.T.; Atak, A.; Özer, Ç. Development of Interleukin-2 Loaded Chitosan-Based Nanogels Using Artificial Neural Networks and Investigating the Effects on Wound Healing in Rats. AAPS PharmSciTech 2017, 18, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- Biglari, S.; Le, T.Y.L.; Tan, R.P.; Wise, S.G.; Zambon, A.; Codolo, G.; De Bernard, M.; Warkiani, M.; Schindeler, A.; Naficy, S.; et al. Simulating Inflammation in a Wound Microenvironment Using a Dermal Wound-on-a-Chip Model. Adv. Healthc. Mater. 2019, 8, 1801307. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, J.K.; Austin, E.; Huang, A.; Mamalis, A.; Jagdeo, J. The IL-4/IL-13 axis in skin fibrosis and scarring: Mechanistic concepts and therapeutic targets. Arch. Dermatol. Res. 2020, 312, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Serezani, A.P.M.; Bozdogan, G.; Sehra, S.; Walsh, D.; Krishnamurthy, P.; Sierra Potchanant, E.A.; Nalepa, G.; Goenka, S.; Turner, M.J.; Spandau, D.F.; et al. IL-4 impairs wound healing potential in the skin by repressing fibronectin expression. J. Allergy Clin. Immunol. 2017, 139, 142–151.e145. [Google Scholar] [CrossRef] [PubMed]
- Coden, M.E.; Berdnikovs, S. Eosinophils in wound healing and epithelial remodeling: Is coagulation a missing link? J. Leukoc. Biol. 2020, 108, 93–103. [Google Scholar] [CrossRef]
- Singampalli, K.L.; Balaji, S.; Wang, X.; Parikh, U.M.; Kaul, A.; Gilley, J.; Birla, R.K.; Bollyky, P.L.; Keswani, S.G. The Role of an IL-10/Hyaluronan Axis in Dermal Wound Healing. Front. Cell Dev. Biol. 2020, 8, 636. [Google Scholar] [CrossRef]
- Short, W.D.; Steen, E.; Kaul, A.; Wang, X.; Olutoye II, O.O.; Vangapandu, H.V.; Templeman, N.; Blum, A.J.; Moles, C.M.; Narmoneva, D.A.; et al. IL-10 promotes endothelial progenitor cell infiltration and wound healing via STAT3. FASEB J. 2022, 36, e22298. [Google Scholar] [CrossRef]
- Takagi, N.; Kawakami, K.; Kanno, E.; Tanno, H.; Takeda, A.; Ishii, K.; Imai, Y.; Iwakura, Y.; Tachi, M. IL-17A promotes neutrophilic inflammation and disturbs acute wound healing in skin. Exp. Dermatol. 2017, 26, 137–144. [Google Scholar] [CrossRef]
- Ahmed, M.; Huh, J.R. Cutting edge: Interleukin-17a prompts HIF1α for wound healing. Trends Immunol. 2022, 43, 861–863. [Google Scholar] [CrossRef]
- Wood, S.; Jayaraman, V.; Huelsmann, E.J.; Bonish, B.; Burgad, D.; Sivaramakrishnan, G.; Qin, S.; DiPietro, L.A.; Zloza, A.; Zhang, C.; et al. Pro-Inflammatory Chemokine CCL2 (MCP-1) Promotes Healing in Diabetic Wounds by Restoring the Macrophage Response. PLoS ONE 2014, 9, e91574. [Google Scholar] [CrossRef]
- Badr, G.; Badr, B.M.; Mahmoud, M.H.; Mohany, M.; Rabah, D.M.; Garraud, O. Treatment of diabetic mice with undenatured whey protein accelerates the wound healing process by enhancing the expression of MIP-1α, MIP-2, KC, CX3CL1 and TGF-β in wounded tissue. BMC Immunol. 2012, 13, 32. [Google Scholar] [CrossRef]
- Ritsu, M.; Kawakami, K.; Kanno, E.; Tanno, H.; Ishii, K.; Imai, Y.; Maruyama, R.; Tachi, M. Critical role of tumor necrosis factor-α in the early process of wound healing in skin. J. Dermatol. Dermatol. Surg. 2017, 21, 14–19. [Google Scholar] [CrossRef]
- Ashcroft, G.S.; Jeong, M.-J.; Ashworth, J.J.; Hardman, M.; Jin, W.; Moutsopoulos, N.; Wild, T.; McCartney-Francis, N.; Sim, D.; McGrady, G.; et al. Tumor necrosis factor-alpha (TNF-α) is a therapeutic target for impaired cutaneous wound healing. Wound Repair Regen. 2012, 20, 38–49. [Google Scholar] [CrossRef]
- Skoda, M.; Stangret, A.; Szukiewicz, D. Fractalkine and placental growth factor: A duet of inflammation and angiogenesis in cardiovascular disorders. Cytokine Growth Factor Rev. 2018, 39, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Szukiewicz, D. CX3CL1 (Fractalkine)-CX3CR1 Axis in Inflammation-Induced Angiogenesis and Tumorigenesis. Int. J. Mol. Sci. 2024, 25, 4679. [Google Scholar] [CrossRef]
- Yuan, C.; Liao, J.; Zheng, L.; Ding, L.; Teng, X.; Lin, X.; Wang, L. Current knowledge of leptin in wound healing: A collaborative review. Front. Pharmacol. 2022, 13, 968142. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, S.; Ide, S.; Tokuyama, R.; Umeki, H.; Tatehara, S.; Kataoka, S.; Satomura, K. Leptin Promotes Wound Healing in the Skin. PLoS ONE 2015, 10, e0121242. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Yao, P.; Lee, E.; Greenhalgh, D.; Soulika, A.M. Interferon-gamma inhibits healing post scald burn injury. Wound Repair Regen. 2012, 20, 580–591. [Google Scholar] [CrossRef]
- Yates-Binder, C.C.; Rodgers, M.; Jaynes, J.; Wells, A.; Bodnar, R.J.; Turner, T. An IP-10 (CXCL10)-Derived Peptide Inhibits Angiogenesis. PLoS ONE 2012, 7, e40812. [Google Scholar] [CrossRef]
- Korbecki, J.; Maruszewska, A.; Bosiacki, M.; Chlubek, D.; Baranowska-Bosiacka, I. The Potential Importance of CXCL1 in the Physiological State and in Noncancer Diseases of the Cardiovascular System, Respiratory System and Skin. Int. J. Mol. Sci. 2023, 24, 205. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, L.; Zheng, H.; Feng, J.; Wei, S.; Li, J.; Cui, J.; Chen, F. The Stimulation of Macrophages by Systematical Administration of GM-CSF Can Accelerate Adult Wound Healing Process. Int. J. Mol. Sci. 2022, 23, 11287. [Google Scholar] [CrossRef] [PubMed]
- Lichtman, M.K.; Otero-Vinas, M.; Falanga, V. Transforming growth factor beta (TGF-β) isoforms in wound healing and fibrosis. Wound Repair Regen. 2016, 24, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Kiritsi, D.; Nyström, A. The role of TGFβ in wound healing pathologies. Mech. Ageing Dev. 2018, 172, 51–58. [Google Scholar] [CrossRef] [PubMed]
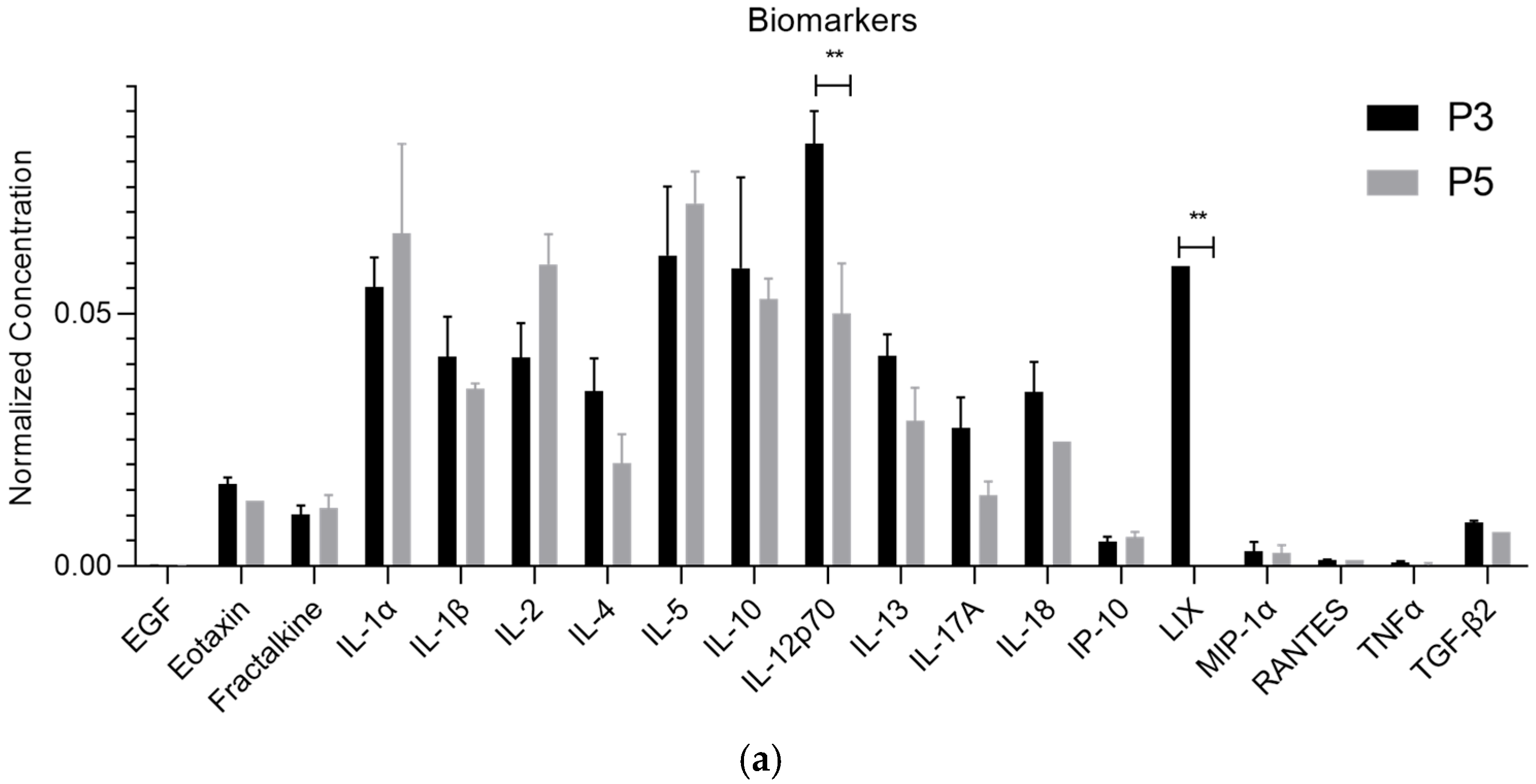
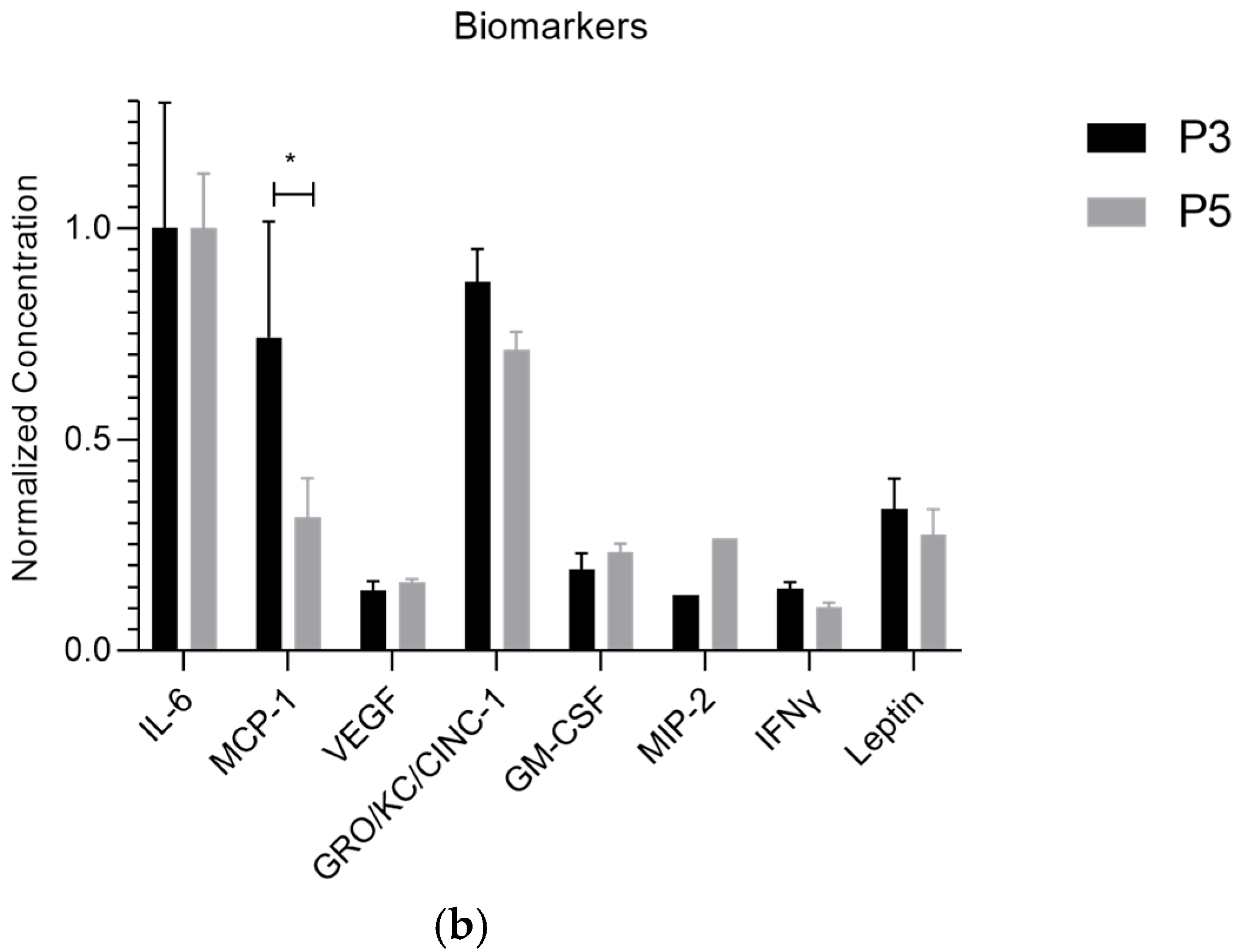
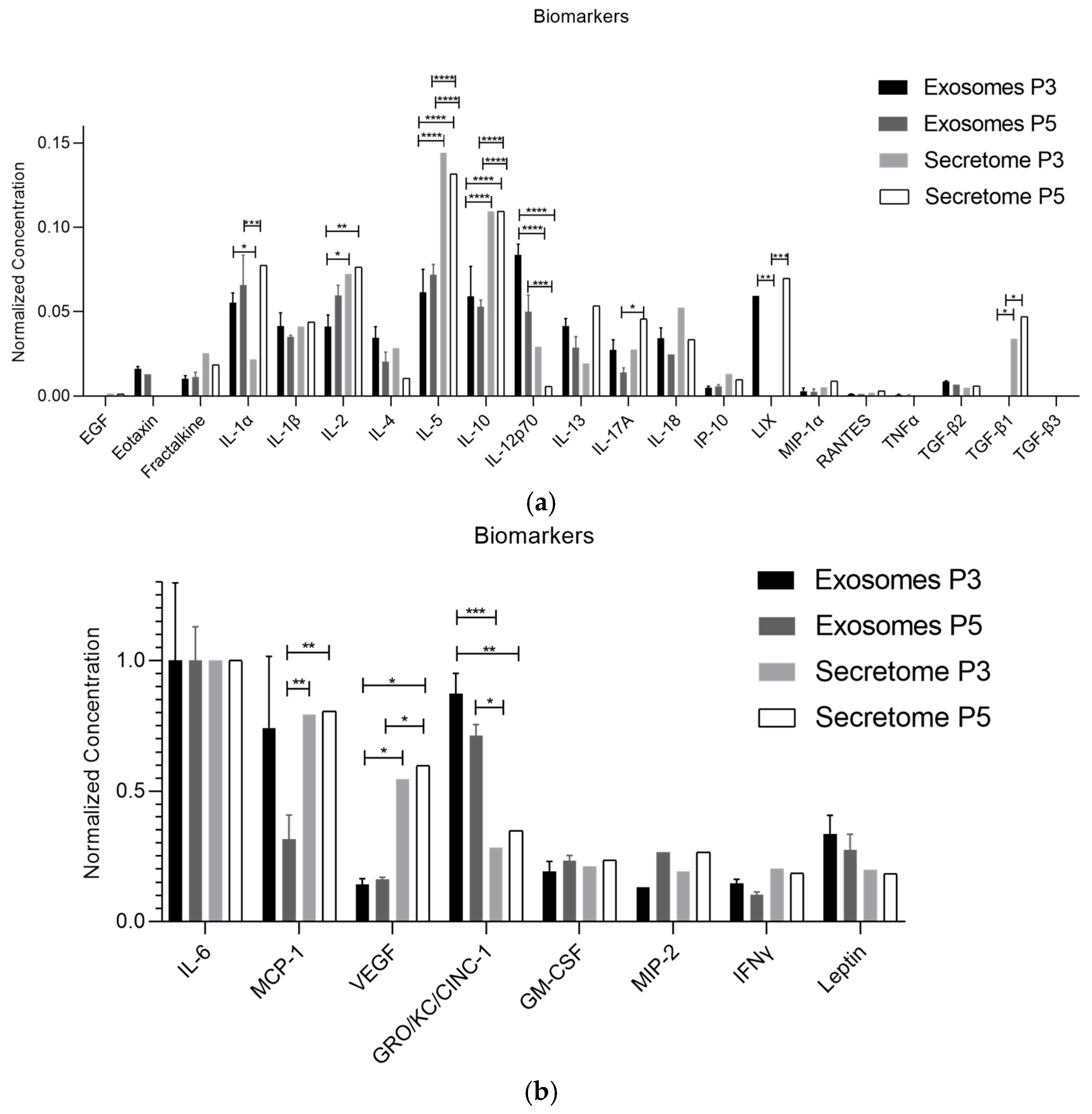
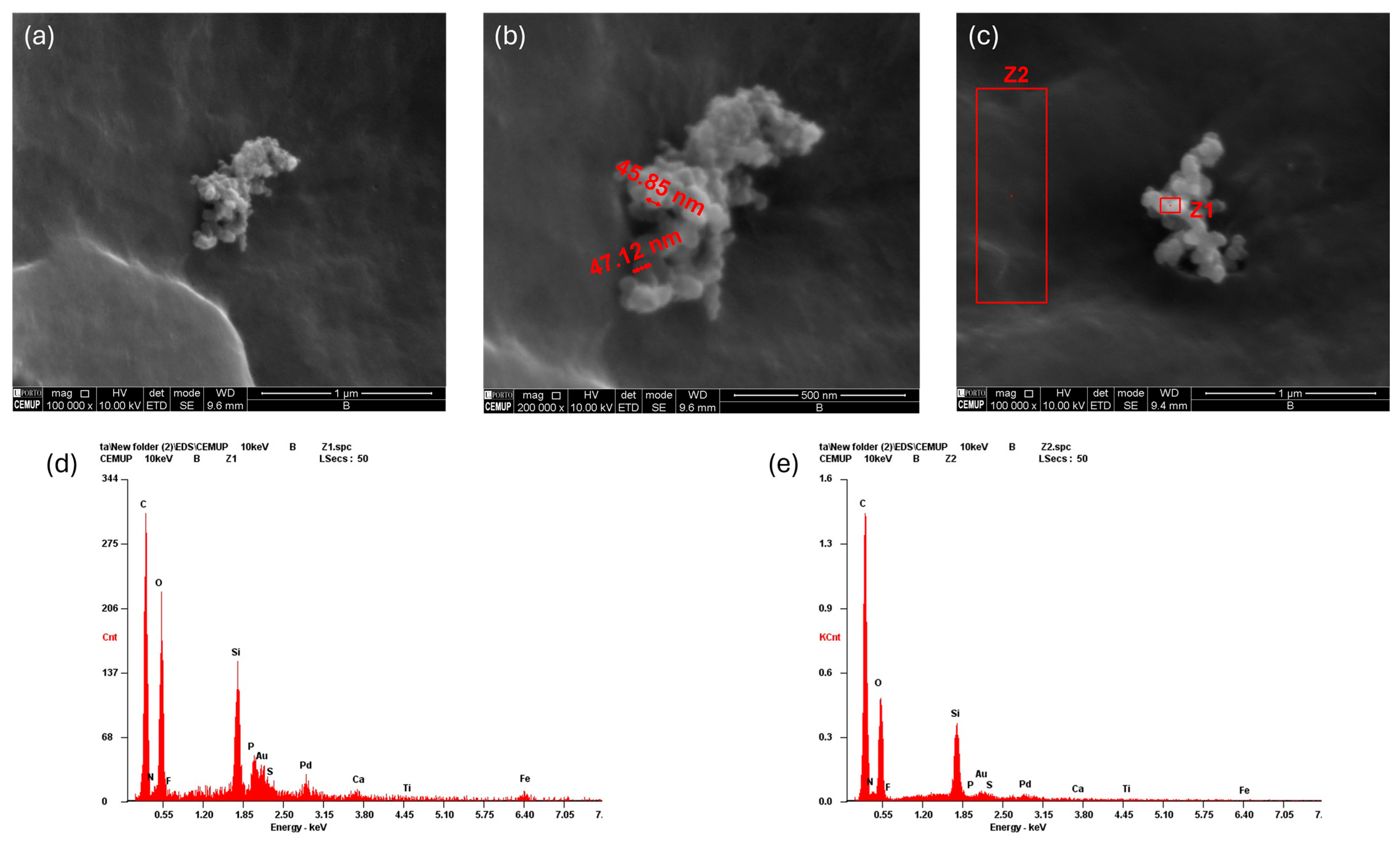
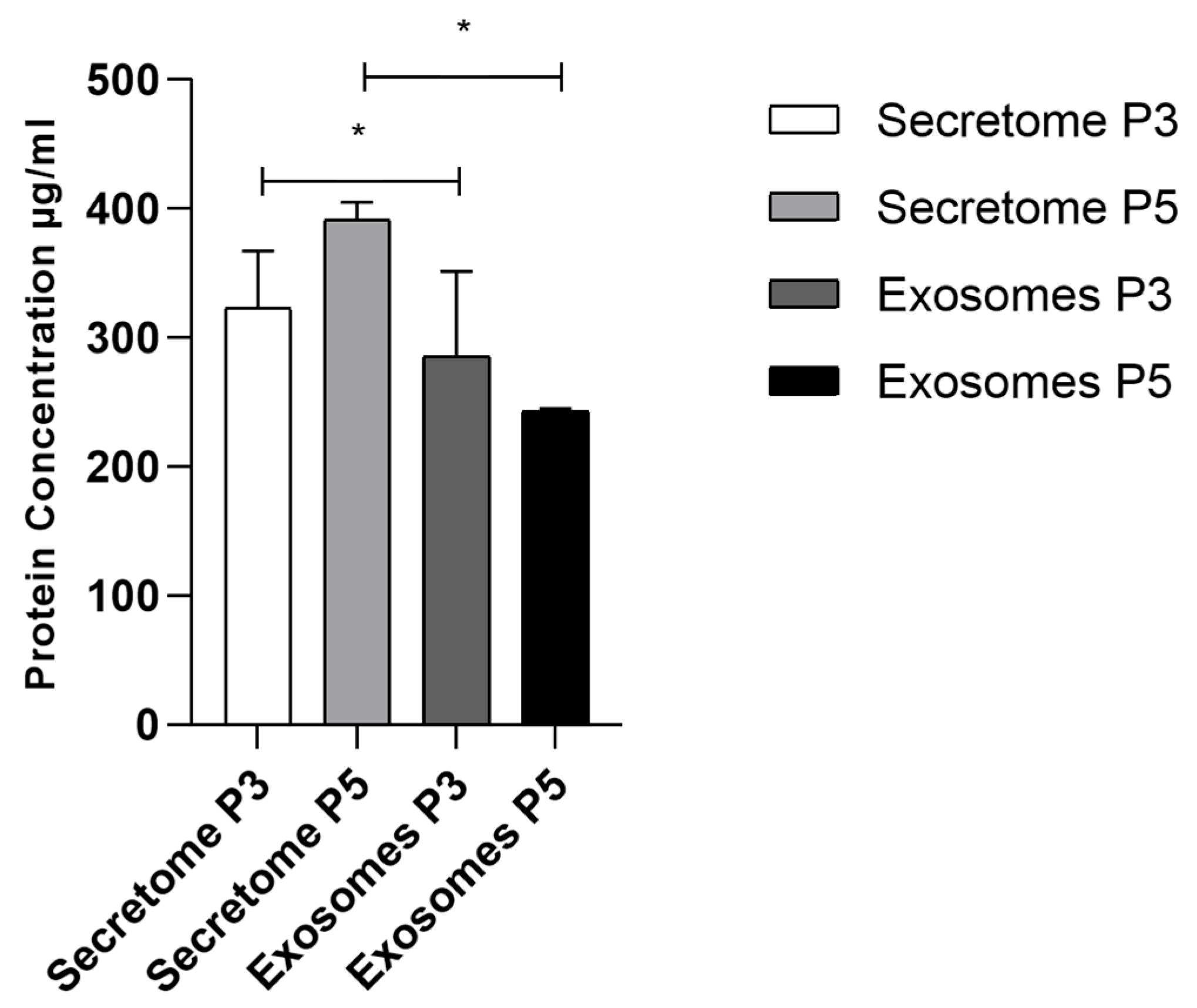
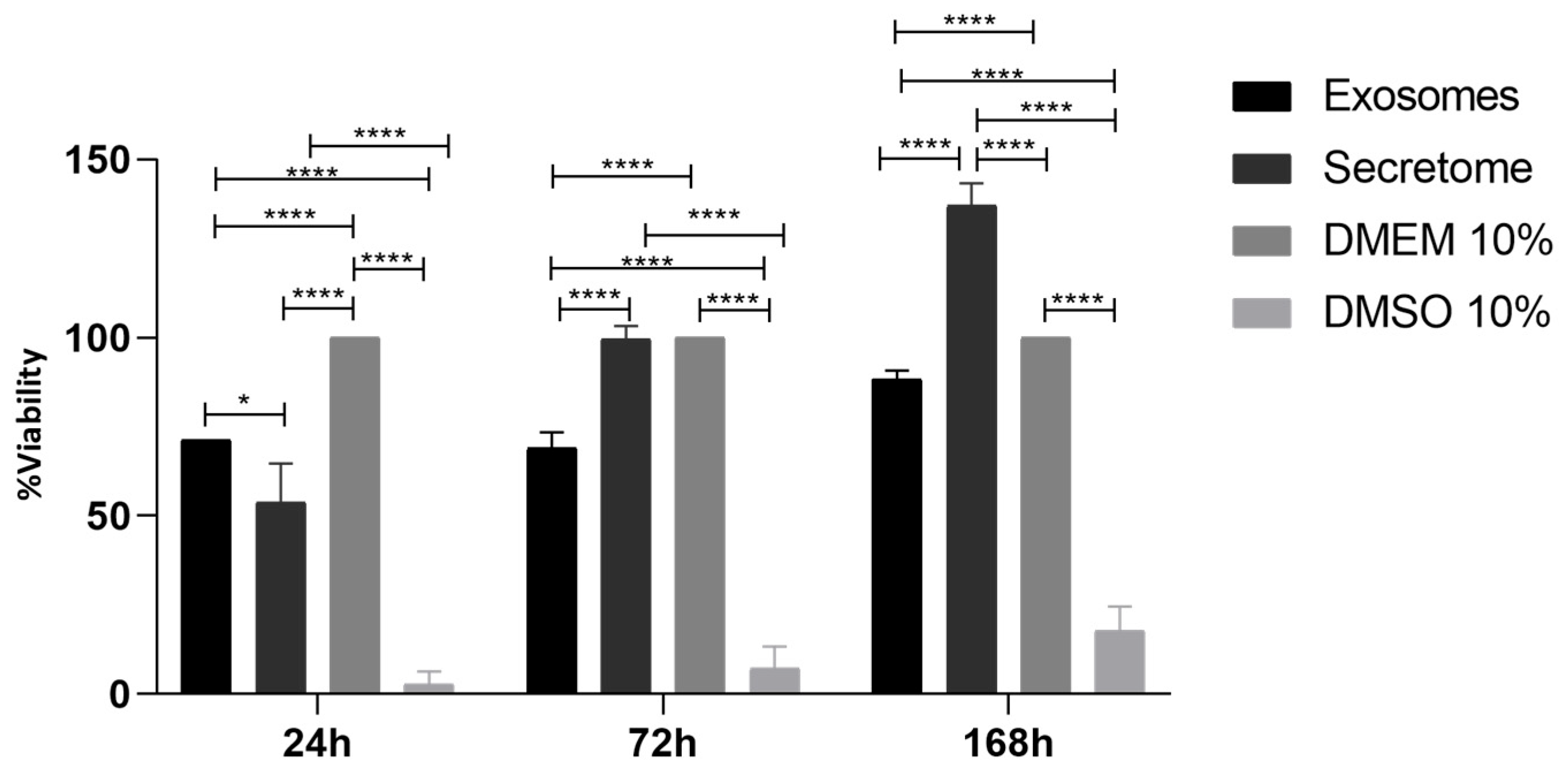
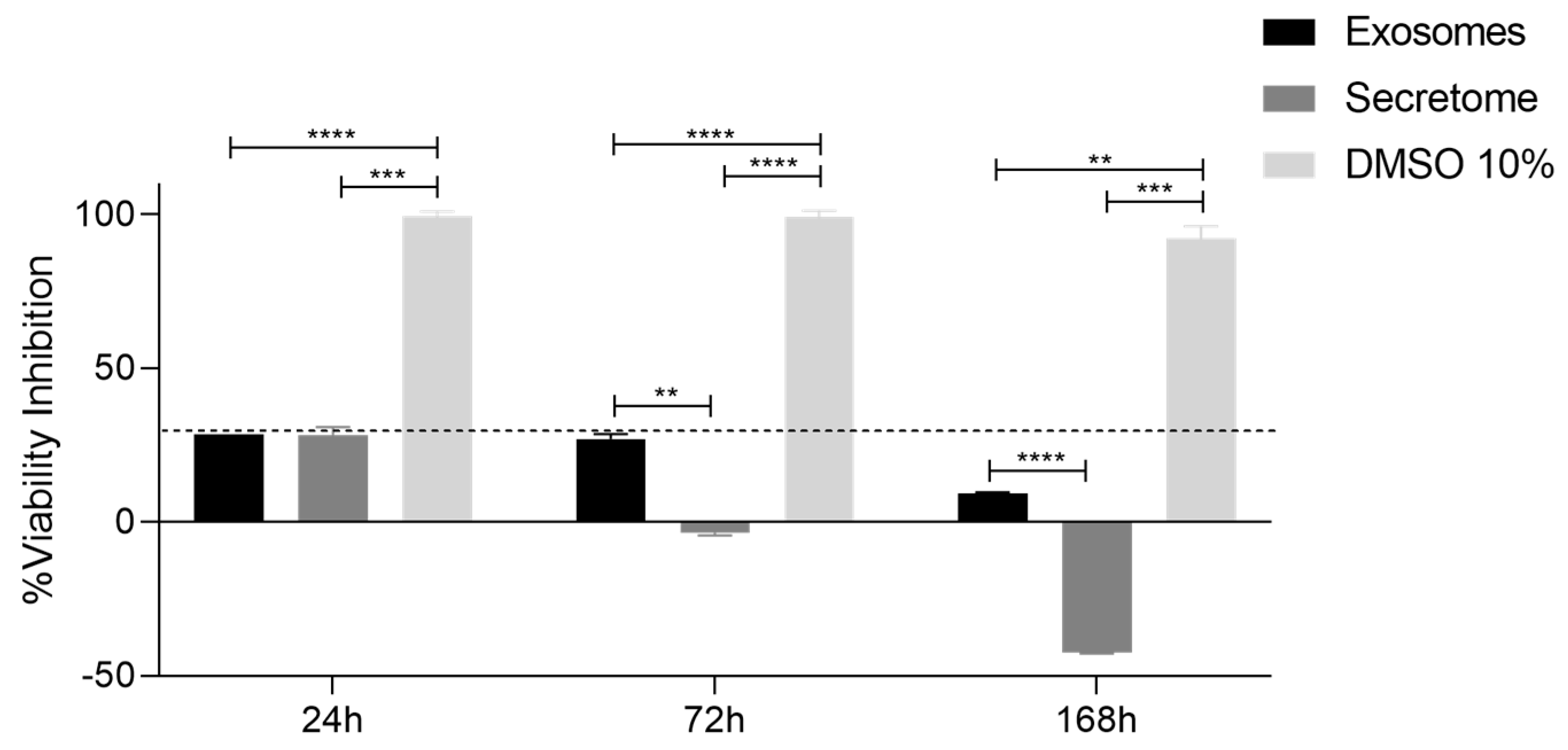
| Biomolecule | Exosomes Mean ± SEM (P3) | Exosomes Mean ± SEM (P5) | Secretome Mean ± SEM (P3) | Secretome Mean ± SEM (P5) |
|---|---|---|---|---|
| EGF | 0.10 ± 0.01 | 0.1 ± 0.02 | 0.16 ± 0.03 | 0.18 ± 0.04 |
| Eotaxin | 1.79 ± 0.14 | 1.65 ± 0.00 | 0.16 ± 0.00 | 0.00 ± 0.00 |
| Fractalkine | 1.17 ± 0.19 | 1.47 ± 0.54 | 4.38 ± 0.29 | 3.06 ± 0.15 |
| GM-CSF | 20.10 ± 3.95 | 28.06 ± 4.02 | 35.39 ± 10.69 | 39.75 ± 11.55 |
| GRO/KC/CINC-1 | 90.97 ± 8.13 | 85.26 ± 8.73 | 47.32 ± 8.64 | 57.64 ± 7.24 |
| IFNγ | 15.31 ± 1.68 | 12.31 ± 2.20 | 34.01 ± 2.26 | 30.67 ± 0.73 |
| IL-1α | 5.85 ± 0.61 | 7.97 ± 3.67 | 3.79 ± 2.27 | 12.83 ± 5.50 |
| IL-1β | 4.42 ± 0.83 | 4.29 ± 0.23 | 6.99 ± 0.86 | 7.27 ± 0.63 |
| IL-2 | 4.40 ± 0.71 | 7.24 ± 1.23 | 12.21 ± 1.23 | 12.65 ± 0.58 |
| IL-4 | 3.71 ± 0.68 | 2.54 ± 1.17 | 4.88 ± 1.30 | 2.63 ± 0.00 |
| IL-5 | 6.50 ± 1.42 | 8.68 ± 1.32 | 24.13 ± 2.69 | 21.13 ± 2.46 |
| IL-6 | 104.20 ± 30.96 | 119.66 ± 21.91 | 166.36 ± 50.22 | 165.62 ± 0.00 |
| IL-10 | 6.24 ± 1.87 | 6.42 ± 0.83 | 18.37 ± 1.87 | 18.17 ± 0.88 |
| IL-12p70 | 8.80 ± 0.68 | 6.08 ± 2.04 | 4.34 ± 1.80 | 1.42 ± 0.00 |
| IL-13 | 4.43 ± 0.45 | 3.54 ± 1.34 | 3.39 ± 0.43 | 8.84 ± 0.51 |
| IL-17A | 2.94 ± 0.63 | 1.77 ± 0.58 | 4.77 ± 0.30 | 7.57 ± 1.11 |
| IL-18 | 3.68 ± 0.63 | 3.05 ± 0.00 | 8.89 ± 1.31 | 5.55 ± 1.24 |
| IP-10 | 0.61 ± 0.1 | 0.79 ± 0.21 | 2.33 ± 0.31 | 1.96 ± 0.38 |
| Leptin | 35.04 ± 7.41 | 32.97 ± 12.35 | 32.31 ± 8.97 | 30.49 ± 4.08 |
| LIX | 6.29 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| MCP-1 | 77.34 ± 28.53 | 37.68 ± 19.27 | 131.64 ± 38.26 | 137.21 ± 19.77 |
| MIP-1α | 0.40 ± 0.20 | 0.40 ± 0.34 | 1.70 ± 0.57 | 2.19 ± 0.39 |
| MIP-2 | 13.81 ± 0.00 | 31.83 ± 0.00 | 32.78 ± 5.77 | 44.02 ± 7.97 |
| RANTES | 0.23 ± 0.01 | 0.24 ± 0.00 | 0.51 ± 0.03 | 0.50 ± 0.01 |
| TNFα | 0.17 ± 0.03 | 0.13 ± 0.07 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| VEGF | 14.77 ± 2.43 | 19.47 ± 1.60 | 91.75 ± 3.22 | 98.88 ± 1.06 |
| G-CSF | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| TGF-β1 | 0.00 ± 0.00 | 0.00 ± 0.00 | 5.82 ± 1.07 | 7.14 ± 1.02 |
| TGF-β2 | 0.99 ± 0.07 | 0.90 ± 0.00 | 0.97 ± 0.04 | 0.98 ± 0.00 |
| TGF-β3 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.19 ± 0.00 | 0.00 ± 0.00 |
| Exosomes P3 vs. Exosomes P5 | Exosomes P3 vs. Secretome P3 | Exosomes P3 vs. Secretome P5 | Exosomes P5 vs. Secretome P3 | Exosomes P5 vs. Secretome P5 | Secretome P3 vs. Secretome P5 | |
|---|---|---|---|---|---|---|
| IL-1α | ns | * | ns | *** | ns | **** |
| IL-2 | ns | * | ** | ns | ns | ns |
| IL-5 | ns | **** | **** | **** | **** | ns |
| IL-10 | ns | **** | **** | **** | **** | ns |
| IL-12p70 | ** | **** | **** | ns | *** | ns |
| IL-13 | ns | ns | ns | ns | ns | ** |
| IL-17a | ns | ns | ns | ns | * | ns |
| LIX | ** | ** | ns | ns | *** | ** |
| TGF-β1 | ns | ns | * | ns | * | ns |
| MCP-1 | * | ns | ns | ** | ** | ns |
| VEGF | ns | * | * | ns | * | ns |
| GRO/KC/CINC-1 | ns | *** | ** | * | ns | ns |
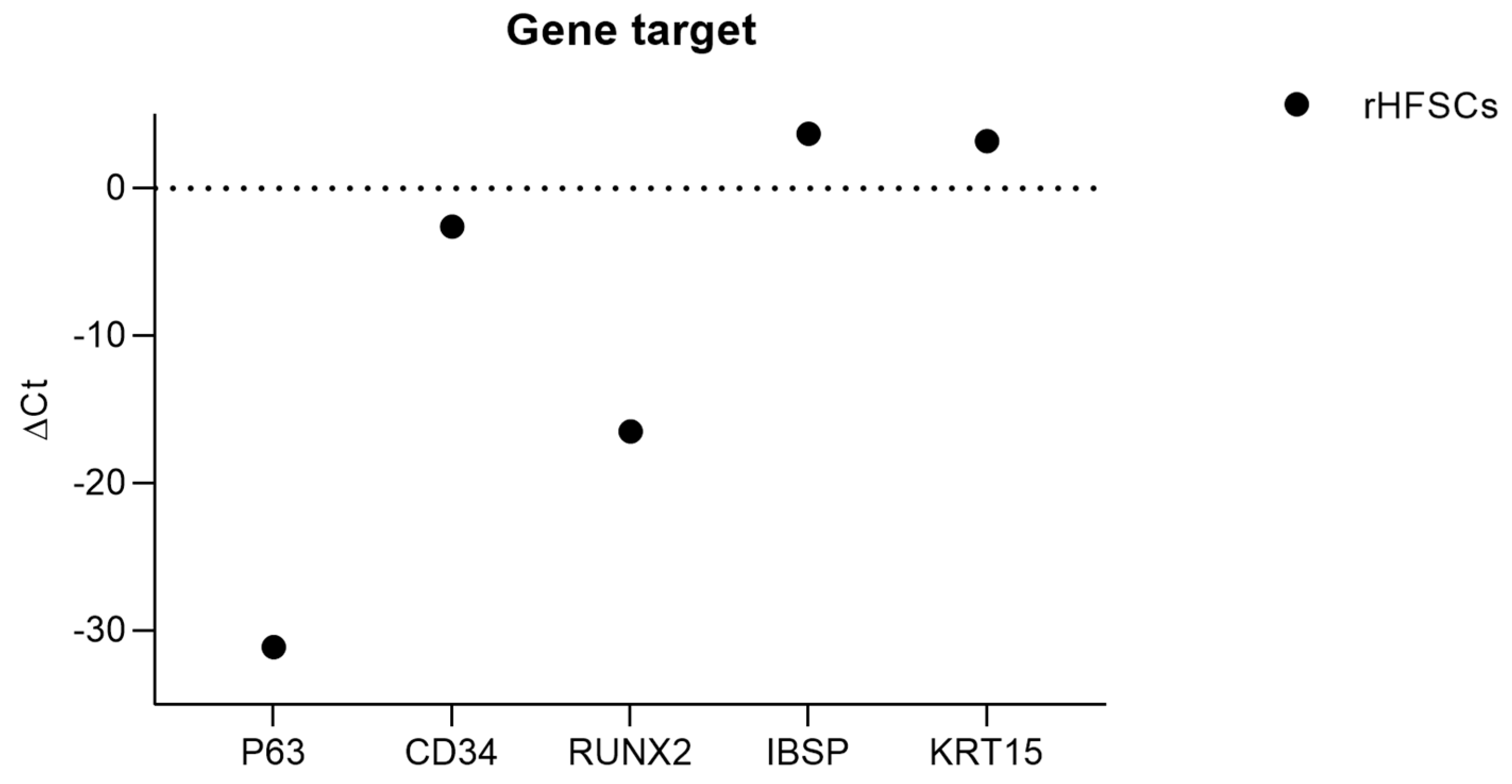
| Target Gene | Ct Average | ΔCt |
|---|---|---|
| KRT14 | nd | nd |
| p63 | 4.92 ± 0.00 | −31.1 |
| CD34 | 33.40 ± 1.41 | −2.6 |
| COL2A1 | nd | nd |
| ITGα6 | nd | nd |
| ACAN | nd | nd |
| ITGβ1 | Nd | nd |
| RUNX2 | 19.45 ± 0.00 | −16.5 |
| KRT10 | nd | nd |
| IBSP | 39.67 ± 0.00 | 3.7 |
| KRT15 | 39.19 ± 0.00 | 3.2 |
| ADIPOQ | nd | nd |
| AAK1 | nd | nd |
| KRT19 | nd | nd |
| Secretome P3 | Secretome P5 | Exosomes P3 | Exosomes P5 | |
|---|---|---|---|---|
| Secretome P3 | ns | * | ns | |
| Secretome P5 | ns | * | ||
| Exosomes P3 | ns |
| 24 h | 72 h | 168 h | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exosomes | Secretome | DMEM 10% | DMSO 10% | Exosomes | Secretome | DMEM 10% | DMSO 10% | Exosomes | Secretome | DMEM 10% | DMSO 10% | |
| Exosomes | * | **** | **** | **** | **** | **** | **** | **** | **** | |||
| Secretome | **** | **** | ns | **** | **** | **** | ||||||
| DMEM 10% | **** | **** | **** | |||||||||
| 24 h | 72 h | 168 h | ||||
|---|---|---|---|---|---|---|
| Secretome | DMSO 10% | Secretome | DMSO 10% | Secretome | DMSO 10% | |
| Exosomes | ns | **** | ** | **** | **** | ** |
| Secretome | *** | **** | *** | |||
| No MMC | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 h | 4 h | 6 h | 8 h | 10 h | 12 h | 24 h | 32 h | 53 h | |
| Control vs. Exosomes | ns | ns | ns | ns | ns | * | ns | ns | ns |
| Control vs. Secretome | ns | ** | * | * | ns | * | ns | ns | ns |
| Exosomes vs. Secretome | ns | ns | ns | ns | ns | ns | ns | ns | ns |
| With MMC | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 h | 4 h | 6 h | 8 h | 10 h | 12 h | 24 h | 32 h | 53 h | |
| Control vs. Exosomes | ns | ns | * | ns | ns | ns | ns | ns | ns |
| Control vs. Secretome | ns | ns | ns | ns | ns | ** | ns | ns | ns |
| Exosomes vs. Secretome | ns | ns | ns | ns | ns | ns | ns | ns | ns |
| Biomarker | Function in Wound Healing |
|---|---|
| EGF | Promotes keratinocyte and fibroblast proliferation, aiding re-epithelialization and collagen synthesis [87,88]. |
| G-CSF | Enhances neutrophil production, supporting debris clearance during the inflammatory phase [89,90]. |
| VEGF | Critical for angiogenesis, ensuring oxygen and nutrient delivery to healing tissues [91,92]. |
| IL-6, IL-1α and IL-1β | Key pro-inflammatory cytokines that regulate inflammation, recruit immune cells and stimulate fibroblasts and keratinocytes [50]. |
| IL-2 and IL-12p70 | Primarily modulate immune responses, indirectly affecting wound healing [93,94]. |
| IL-4 and IL-13 | Promote fibroblast differentiation into myofibroblasts, impacting wound contraction and fibrosis [95,96]. |
| IL-5 and Eotaxin | Mainly recruit eosinophils, with limited direct impact on typical wound healing [97]. |
| IL-10 | Anti-inflammatory cytokine, crucial for resolving inflammation and minimizing scarring [98,99]. |
| IL-17A and IL-18 | Contribute to inflammation and influence keratinocyte activity and angiogenesis [100,101]. |
| RANTES (CCL5), MCP-1 (CCL2), MIP-1α (CCL3) and MIP-2 (CXCL2) | Chemokines that recruit immune cells to the wound site, supporting inflammation and repair [102,103]. |
| TNFα | It stimulates the production of other cytokines and chemokines, activates immune cells and can influence fibroblast and keratinocyte behavior. Drives early inflammation but may impair healing if chronically elevated [104,105]. |
| Fractalkine (CX3CL1) | Aids immune cell recruitment and endothelial interaction [106,107]. |
| Leptin | Supports keratinocyte proliferation, angiogenesis and collagen production [108,109]. |
| IFNγ and IP-10 (CXCL10) | Influence inflammation and ECM remodeling, with prolonged expression potentially impairing healing [110,111]. |
| GRO/KC/CINC-1 (CXCL1) and LIX (CXCL5) | Attract neutrophils during early wound responses [112]. |
| GM-CSF | Promotes differentiation of immune cells, supporting both inflammation and repair [113]. |
| TGFβ1 and TGFβ2 | Stimulate fibroblast proliferation, myofibroblast differentiation and ECM production [114]. |
| TGFβ3 | Encourages regenerative healing with reduced scarring [115]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa, P.; Lopes, B.; Sousa, A.C.; de Sousa Moreira, A.; Rêma, A.; Alvites, R.; Geuna, S.; Alves, N.; Maurício, A.C. Rat Hair Follicle Stem Cell-Derived Exosomes: Isolation, Characterization and Comparative Analysis of Their In Vitro Wound Healing Potential. Int. J. Mol. Sci. 2025, 26, 5081. https://doi.org/10.3390/ijms26115081
Sousa P, Lopes B, Sousa AC, de Sousa Moreira A, Rêma A, Alvites R, Geuna S, Alves N, Maurício AC. Rat Hair Follicle Stem Cell-Derived Exosomes: Isolation, Characterization and Comparative Analysis of Their In Vitro Wound Healing Potential. International Journal of Molecular Sciences. 2025; 26(11):5081. https://doi.org/10.3390/ijms26115081
Chicago/Turabian StyleSousa, Patrícia, Bruna Lopes, Ana Catarina Sousa, Alícia de Sousa Moreira, Alexandra Rêma, Rui Alvites, Stefano Geuna, Nuno Alves, and Ana Colette Maurício. 2025. "Rat Hair Follicle Stem Cell-Derived Exosomes: Isolation, Characterization and Comparative Analysis of Their In Vitro Wound Healing Potential" International Journal of Molecular Sciences 26, no. 11: 5081. https://doi.org/10.3390/ijms26115081
APA StyleSousa, P., Lopes, B., Sousa, A. C., de Sousa Moreira, A., Rêma, A., Alvites, R., Geuna, S., Alves, N., & Maurício, A. C. (2025). Rat Hair Follicle Stem Cell-Derived Exosomes: Isolation, Characterization and Comparative Analysis of Their In Vitro Wound Healing Potential. International Journal of Molecular Sciences, 26(11), 5081. https://doi.org/10.3390/ijms26115081












