Genome-Wide Identification, Phylogeny, and Abiotic Stress Response Analysis of OSCA Family Genes in the Alpine Medicinal Herb Notopterygium franchetii
Abstract
1. Introduction
2. Results
2.1. Identification of OSCA Gene Family and Physicochemical Property Analysis of N. franchetii
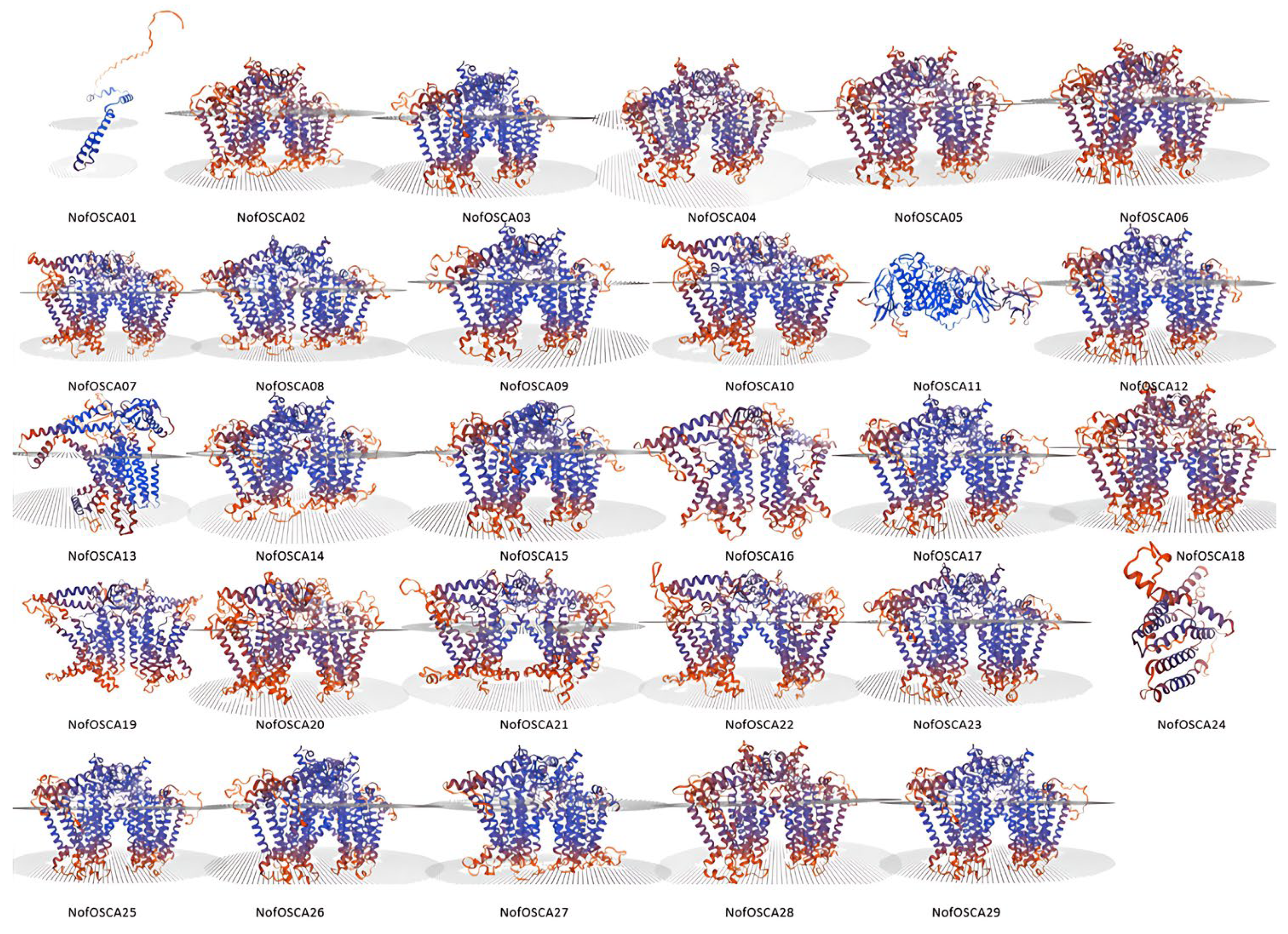
2.2. Secondary and Tertiary Structure Analysis of OSCA gGene Family Proteins in N. franchetii
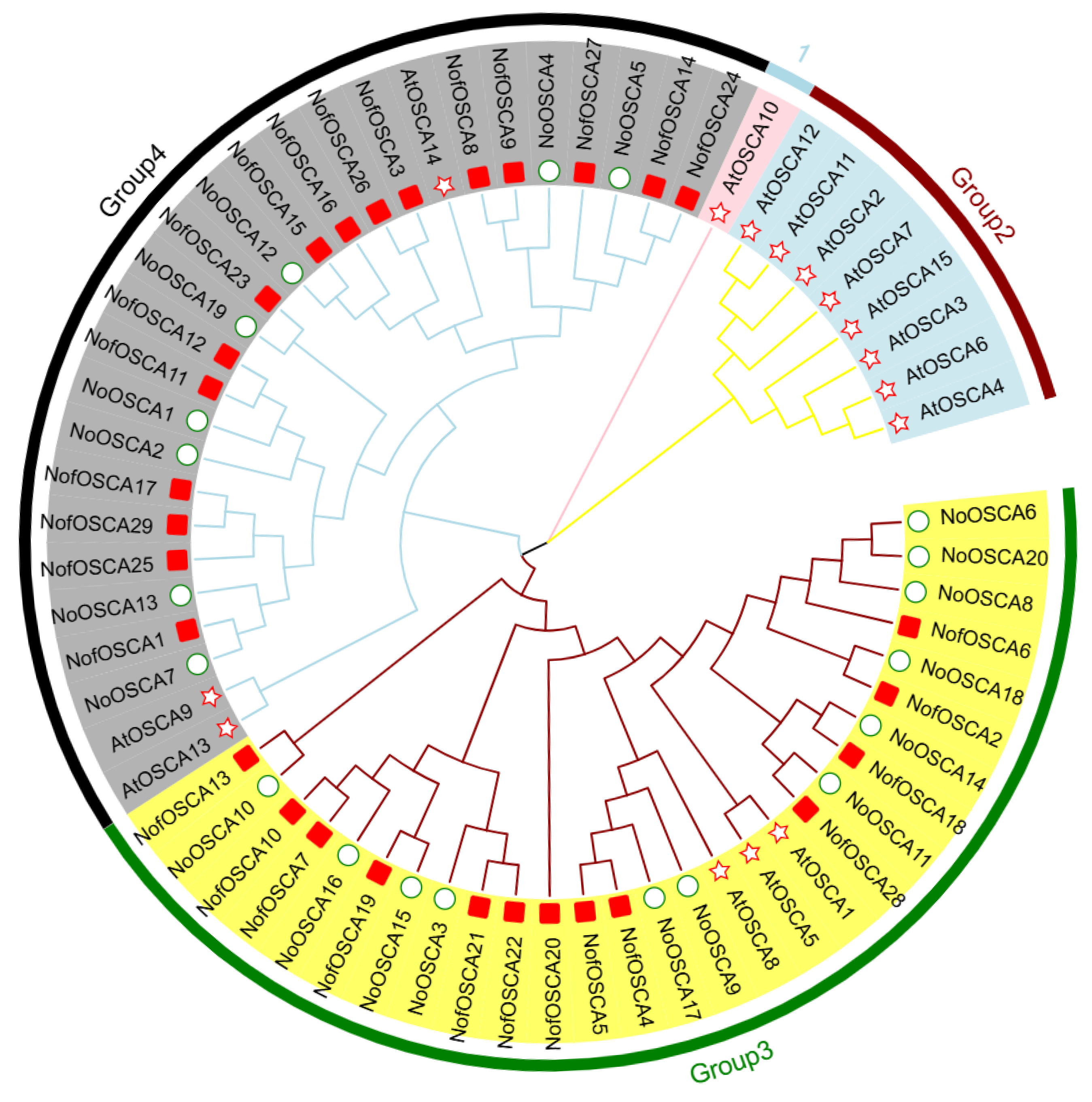
2.3. Phylogenetic Tree Analysis of N. franchetii OSCA Gene Family
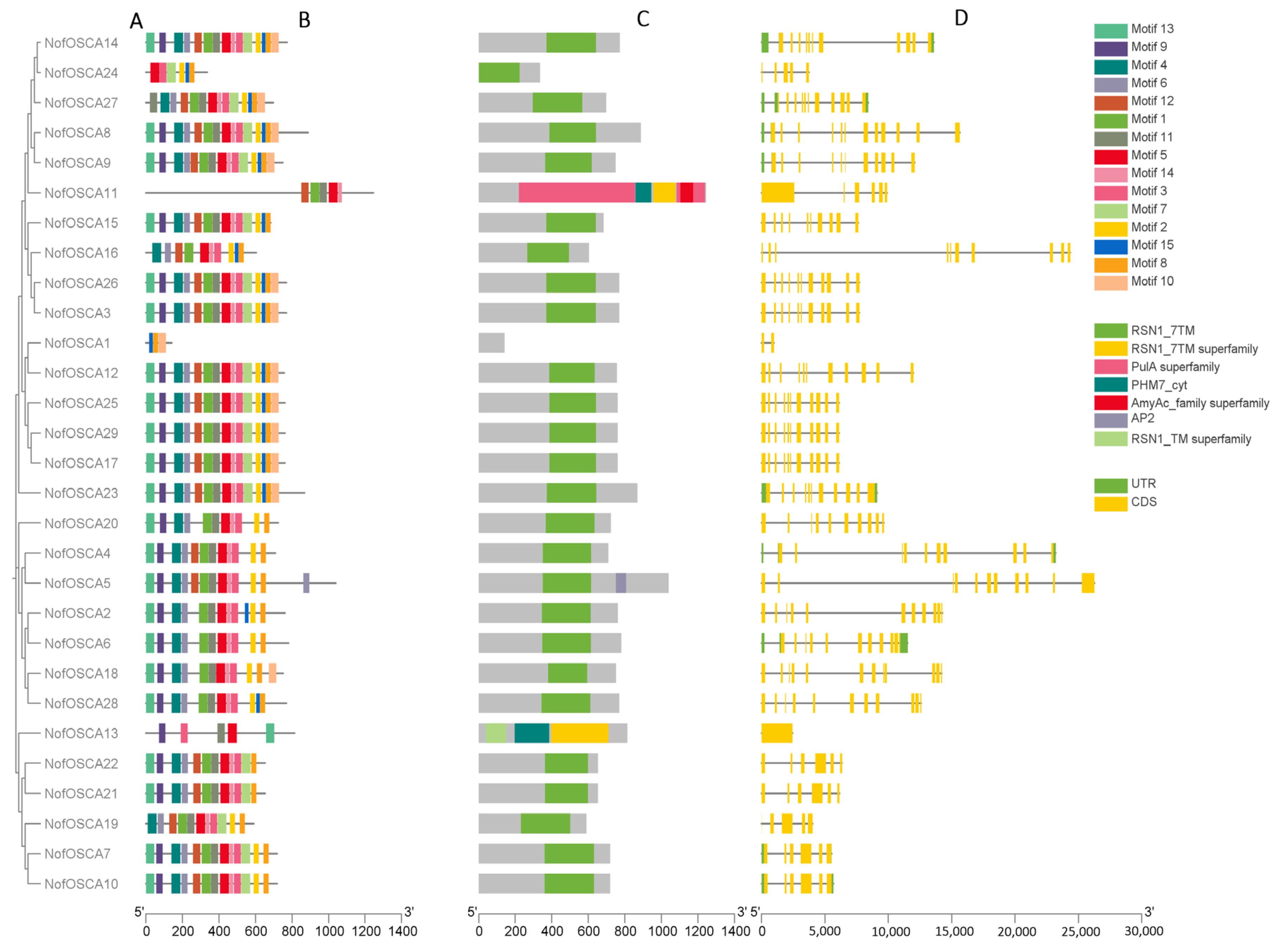
2.4. Conserved Motifs, Conserved Structural Domains, and Gene Structure Analysis of N. franchetii OSCA Gene Family
2.5. Analysis of Cis-Acting Elements of the OSCA Family of N. franchetii
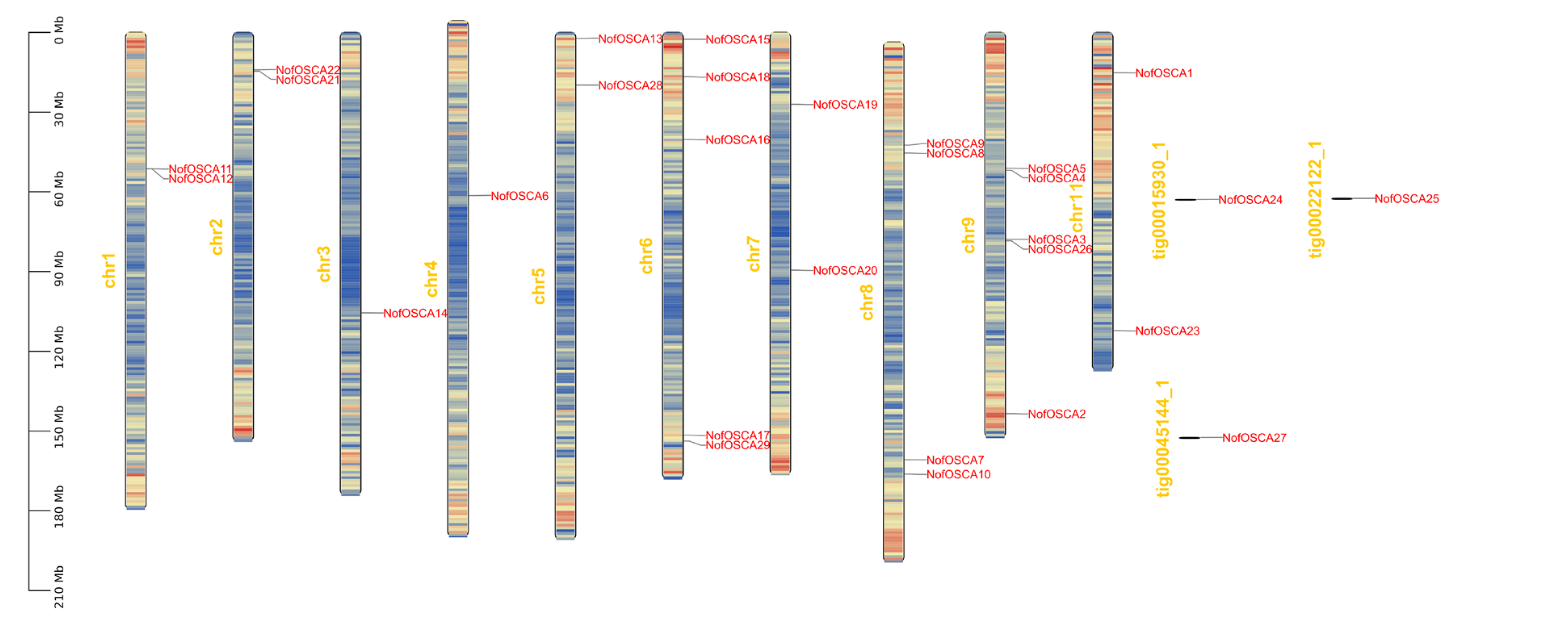
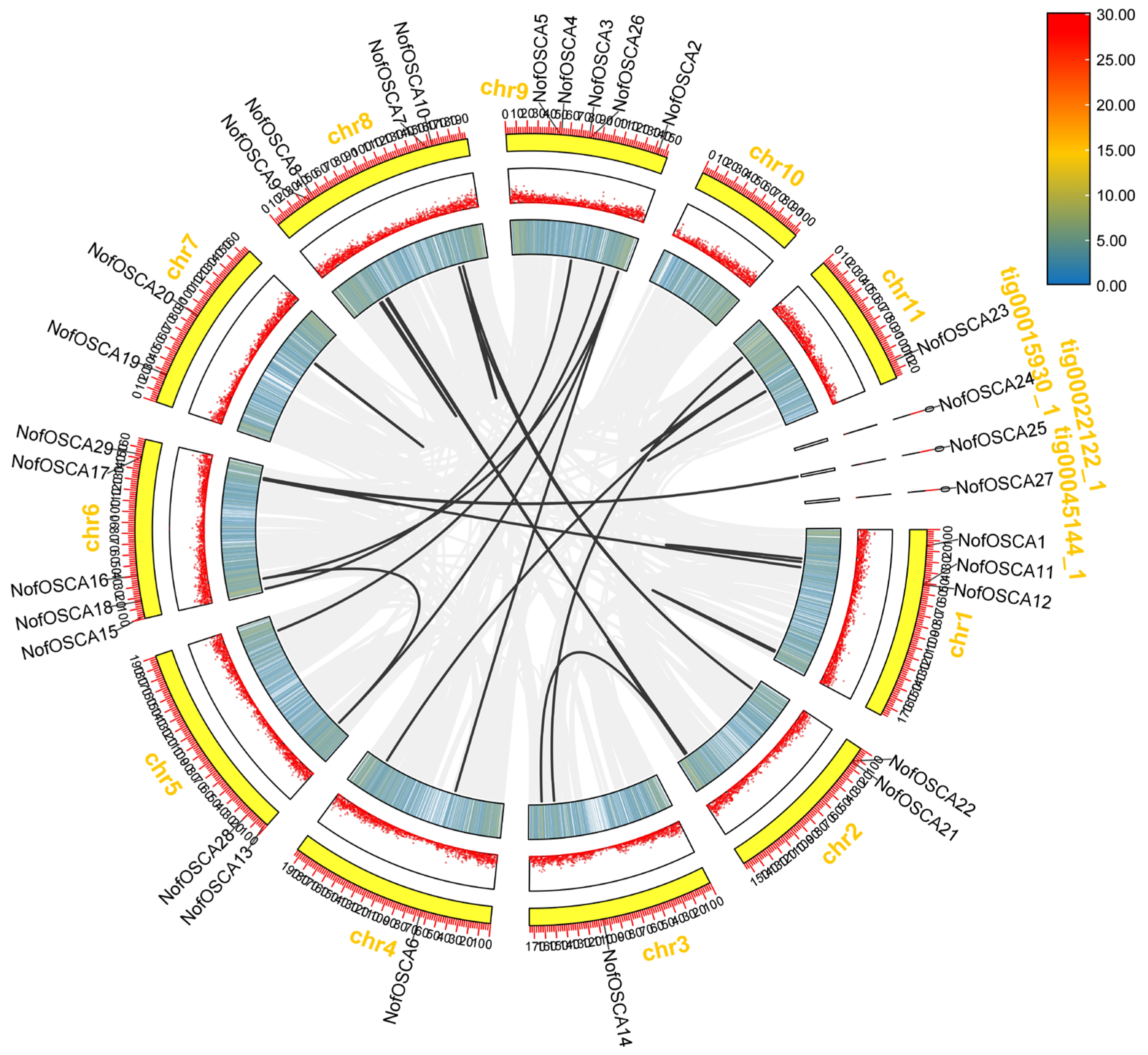
2.6. Chromosomal Localization and Covariance Analysis of the OSCA Gene of N. franche-tii

2.7. Ka/Ks Calculation
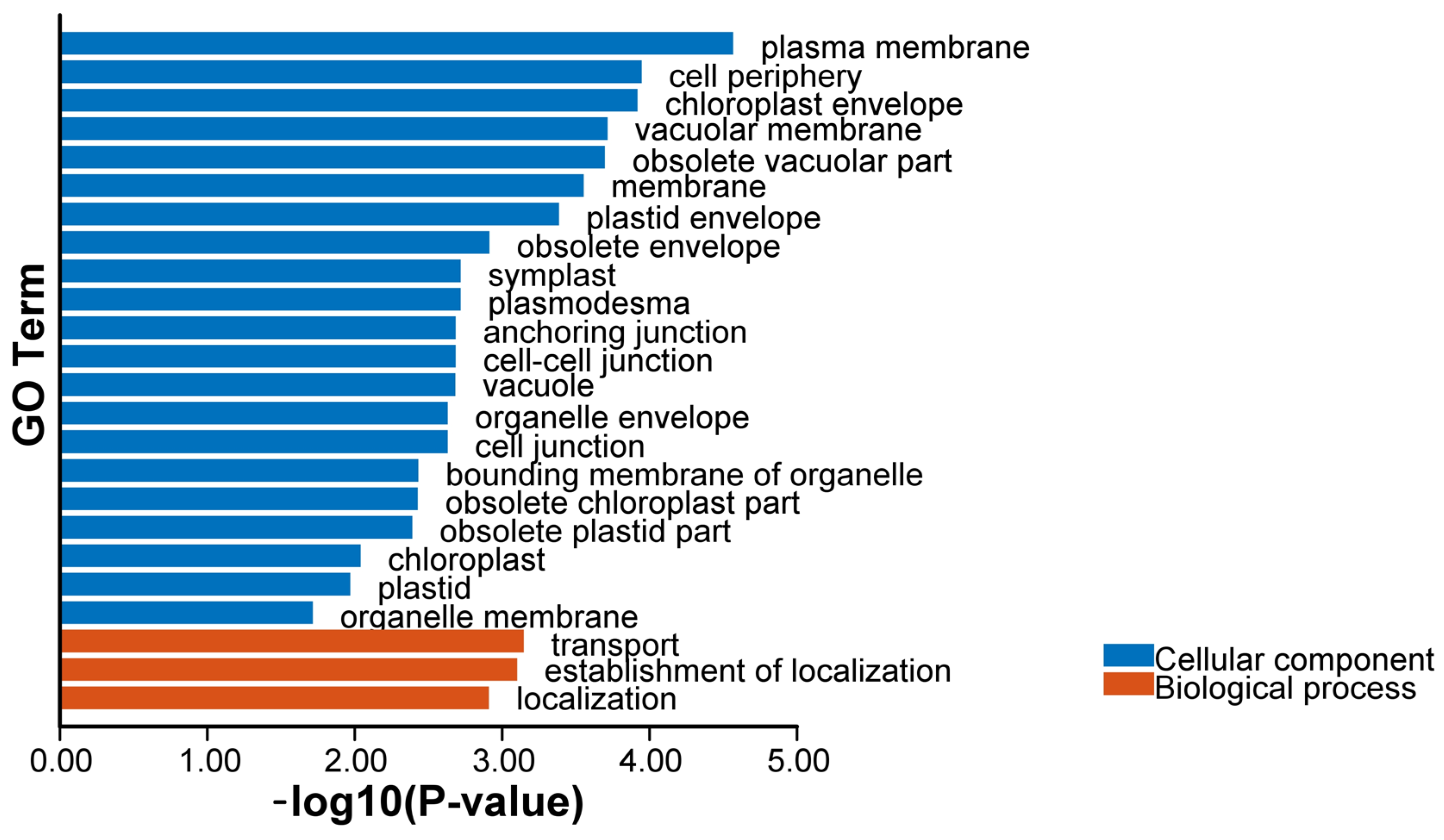
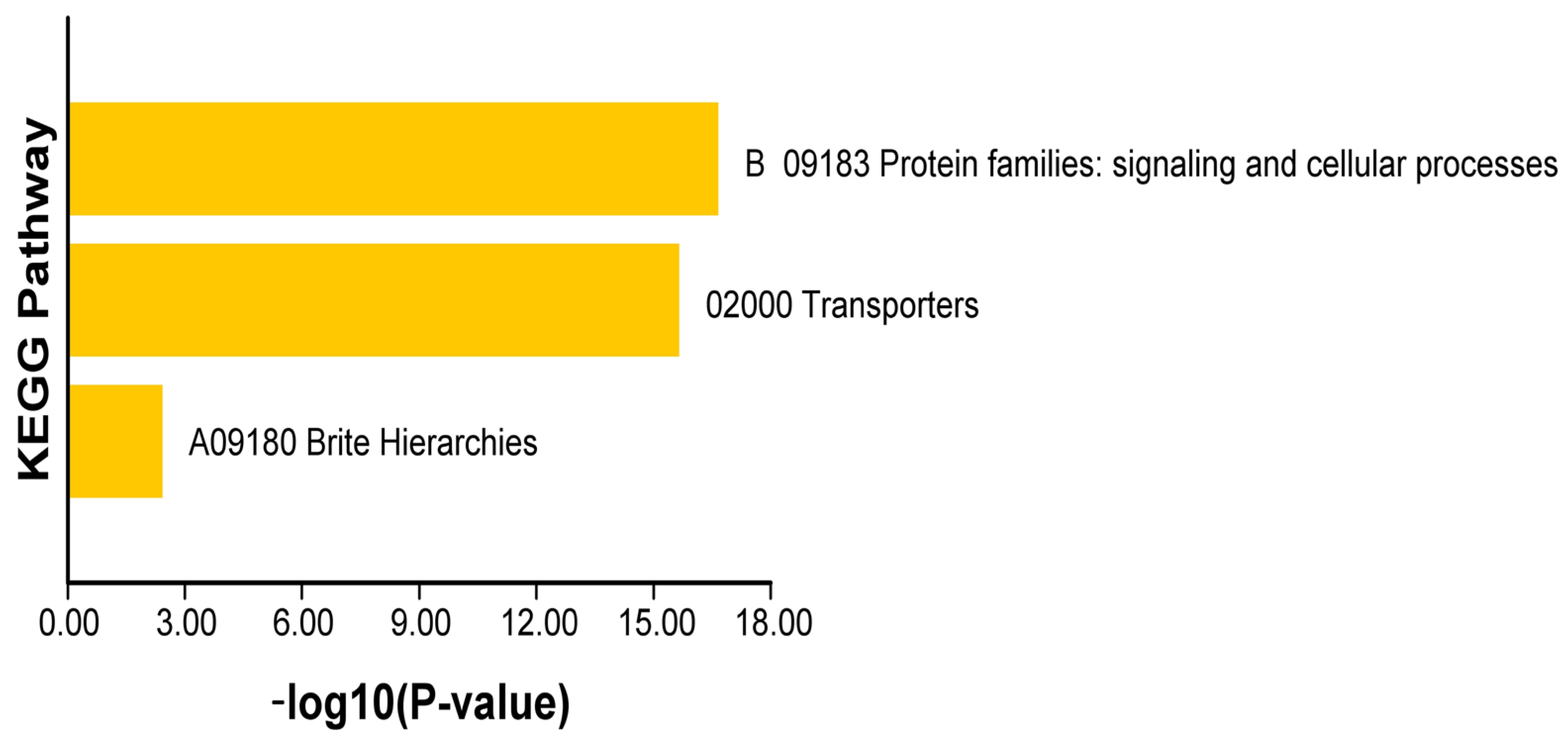
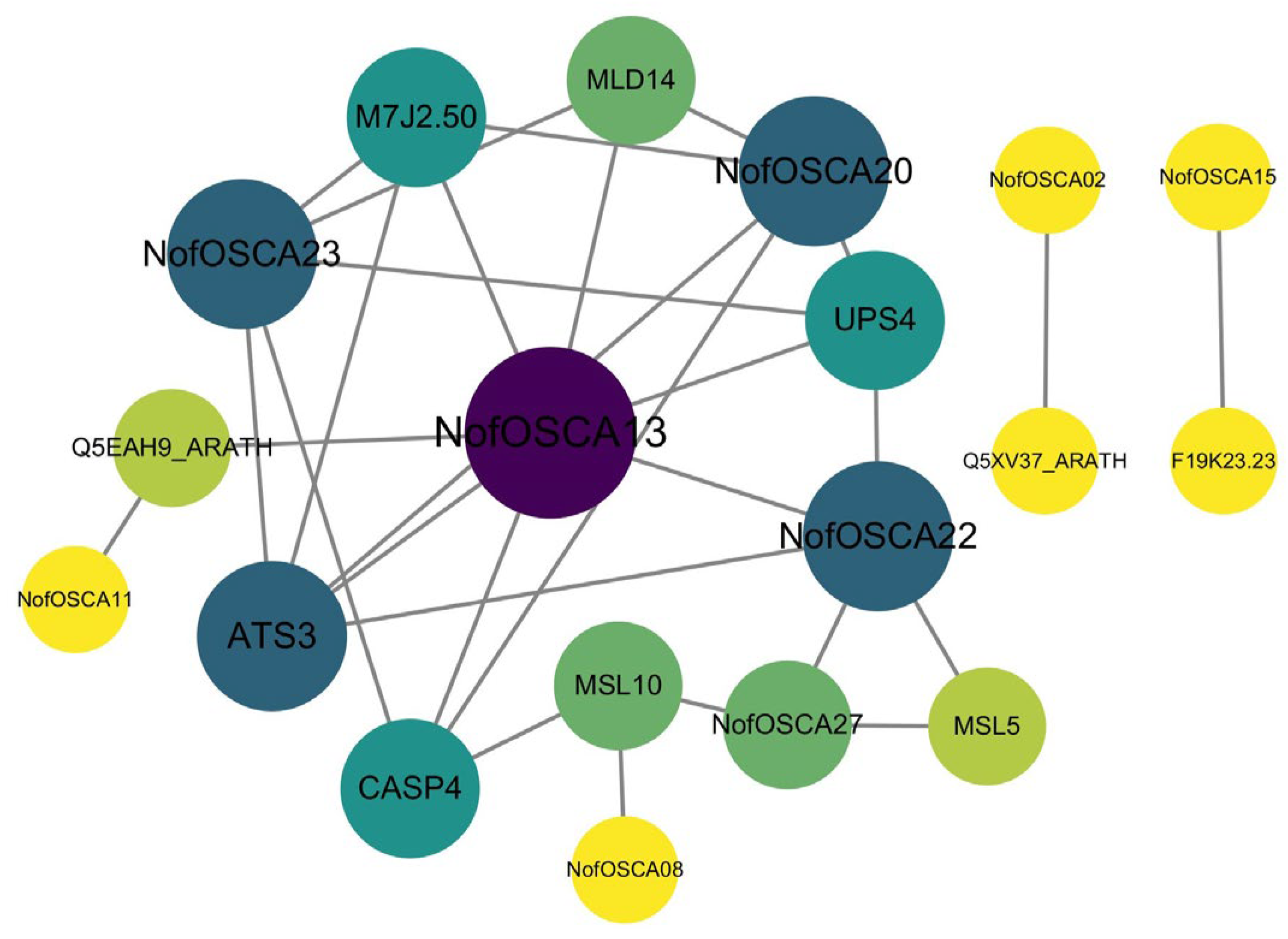
2.8. GO, KEGG Enrichment, and PPI Networks Analysis of N. franchetii
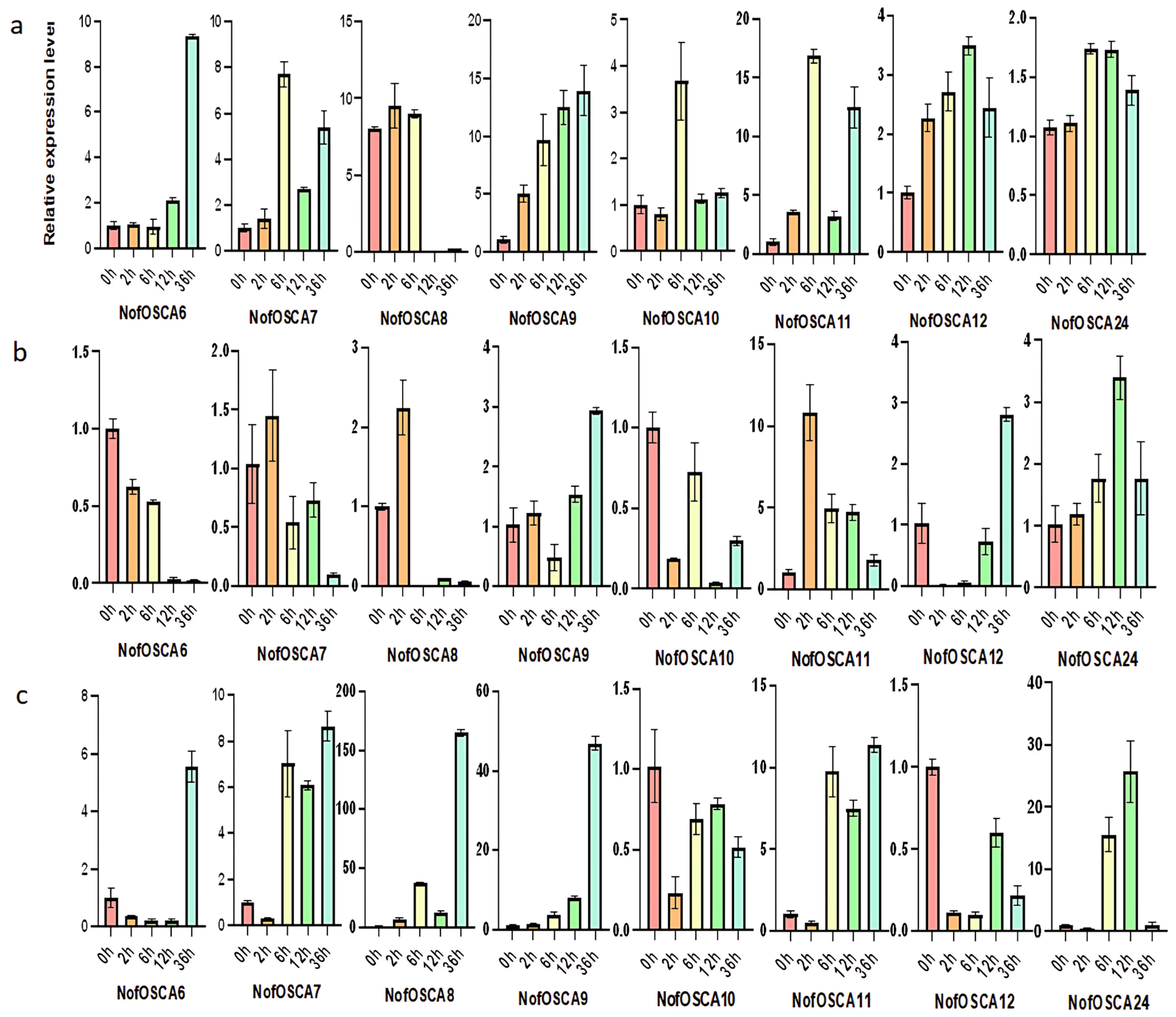
2.9. Expression Analysis of N. franchetii OSCA Gene Under Drought and High-Temperature Stresses
2.10. Expression Analysis of OSCA Gene Expression Among Different Tissues in N. franchetii
3. Discussion
4. Materials and Methods
4.1. Experimental Materials and Treatments
4.2. Identification, Physicochemical Characterization, and Subcellular Localization Prediction of N. franchetii OSCA Family Members
4.3. Conserved Motifs, Conserved Structural Domains, Gene Structure Analysis, and Cis-Acting Element Analysis
4.4. Predictive Analysis of Protein Structure
4.5. Phylogenetic Analysis of the OSCA Family of N. franchetii
4.6. Chromosomal Localization and Covariance Analysis of N. franchetii OSCA Family Members
4.7. Ka/Ks Calculator
4.8. GO, KEGG Enrichment, and PPI Networks Analysis
4.9. Analysis of Stress Response Patterns Based on qRT-PCR
4.10. Expression Pattern Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gao, S.M.; Cong, R.C.; Gao, L.; Zhu, Y.Y.; Meng, Y.; Zhou, Y. Genetic diversity analysis of phenotypic character and SRAP molecular markers in 45 tree peony cultivars. Braz. J. Bot. 2020, 43, 291–302. [Google Scholar] [CrossRef]
- Azietaku, J.T.; Ma, H.; Yu, X.A.; Li, J.; Oppong, M.B.; Cao, J.; An, M.; Chang, Y.X. A review of the ethnopharmacology, phytochemistry and pharmacology of Notopterygium incisum. J. Ethnopharmacol. 2017, 202, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.K.; Zheng, R.L.; Chen, P.; Li, L.C. Phytochemistry and biological profile of the Chinese endemic herb genus Notopterygium. Molecules 2024, 29, 3252. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Liu, M.L.; López-Pujol, J.; Jia, R.W.; Kou, Y.X.; Yue, M.; Guan, T.X.; Li, Z.H. The hybridization origin of the Chinese endemic herb genus Notopterygium (Apiaceae): Evidence from population genomics and ecological niche analysis. Mol. Phylogenet. Evol. 2023, 182, 107736. [Google Scholar] [CrossRef]
- Yang, J.; Feng, L.; Yue, M.; He, Y.L.; Zhao, G.F.; Li, Z.H. Species delimitation and interspecific relationships of the endangered herb genus Notopterygium inferred from multilocus variations. Mol. Phylogenet. Evol. 2019, 133, 142–151. [Google Scholar] [CrossRef]
- Liu, M.L.; He, Y.L.; López-Pujol, J.; Jia, Y.; Li, Z.H. Complex population evolutionary history of four cold-tolerant Notopterygium herb species in the Qinghai-Tibetan Plateau and adjacent areas. Heredity 2019, 123, 242–263. [Google Scholar] [CrossRef]
- Jia, Y.; Liu, M.L.; Yue, M.; Zhao, Z.; Zhao, G.F.; Li, Z.H. Comparative transcriptome analysis reveals adaptive evolution of Notopterygium incisum and Notopterygium franchetii, two high-alpine herbal species endemic to China. Molecules 2017, 22, 1158. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Jiang, S.Y.; Xu, K.J.; Shi, H.L.; Zhou, Y.; Deng, W.L.; Ding, L.S.; Peng, S.L. Chemical constituents contained in seeds of Notopterygium franchetii. China J. Chin. Meteria Med. 2012, 37, 941–945. [Google Scholar]
- Li, Q.; Dai, Y.Q.; Huang, X.C.; Sun, L.L.; Wang, K.X.; Guo, X.; Xu, D.Q.; Wan, D.G.; An, L.T.; Wang, Z.X.; et al. The chromosome-scale assembly of the Notopterygium incisum genome provides insight into the structural diversity of coumarins. Acta Pharm. Sinica B 2024, 14, 3760–3773. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.Y.; Li, J.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Science China. Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Sytar, O.; Brestic, M.; Zivcak, M.; Olsovska, K.; Kovar, M.; Shao, H.; He, X. Applying hyperspectral imaging to explore natural plant diversity towards improving salt stress tolerance. Sci. Total Environ. 2017, 578, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Wang, T.; Hua, H.L.; Wu, Y.; Su, S.X.; Wei, J.J.; Jiang, S.Y.; Mou, H.L.; Zhu, H.J.; Lv, J.L.; et al. Identification and expression analysis of the OSCA gene family in ginger. J. Agric. Biotechnol. 2024, 32, 1518–1532. [Google Scholar]
- Wu, X.L.; Yuan, J.; Luo, A.X.; Chen, Y.; Fan, Y.J. Drought stress and re-watering increase secondary metabolites and enzyme activity in dendrobium moniliforme. Ind. Crops Prod. 2016, 94, 385–393. [Google Scholar] [CrossRef]
- Zhao, J.G.; Lu, Z.G.; Wang, L.; Jin, B. Plant responses to heat stress: Physiology, transcription, noncoding RNAs, and epigenetics. Int. J. Mol. Sci. 2020, 22, 117. [Google Scholar] [CrossRef]
- Lu, H.D.; Xue, J.Q.; Guo, D.W. Efficacy of planting date adjustment as a cultivation strategy to cope with drought stress and increase rainfed maize yield and water-use efficiency. Agric. Water Manag. 2017, 179, 227–235. [Google Scholar] [CrossRef]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Pandey, M.; Penna, S. Time course of physiological, biochemical, and gene expression changes under short-term salt stress in Brassica juncea L. Crop J. 2016, 5, 219–230. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, L.M.; Zhou, R.Y.; Tang, X.Q.; Bai, Y. Effects of salt stress on photosynthetic characteristics and indexes of adverse circumstances-resistance of Isatis indigotica Fort. seedlings from five areas. J. Nanjing Agric. Univ. 2017, 40, 416–424. [Google Scholar]
- Zhao, S.S.; Zhang, Q.K.; Liu, M.Y.; Zhou, H.P.; Ma, C.L.; Wang, P.P. Regulation of plant responses to salt stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Ding, Y.L.; Yang, S.H. Surviving and thriving: How plants perceive and respond to temperature stress. Dev. Cell 2022, 57, 947–958. [Google Scholar] [CrossRef]
- Li, J.W.; Yang , J.K.; Jia, B.W.; Sun, M.Z.; Liu, Y.; Yin, K.D.; Sun, X.L. Evolution and expression analysis of OSCA gene family in soybean. Chin. J. Oil Crop Sci. 2017, 39, 589–599. [Google Scholar]
- Zhu, Q.L.; Zhao, Y.L.; Lü, H.L.; Luan, F.S.; Gao, P. Identification and characteristic analysis of the CBL family genes in watermelon. North. Hortic. 2017, 15, 18–24. [Google Scholar]
- Yuan, F.; Yang, H.M.; Xue, Y.; Kong, D.D.; Ye, R.; Li, C.J.; Zhang, J.Y.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic -tress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 36–371. [Google Scholar] [CrossRef]
- Pei, S.Y.; Liu, Y.T.; Li, W.K.; Krichilsky, B.; Dai, S.W.; Wang, Y.; Wang, X.; Johnson, D.M.; Crawford, B.M.; Swift, G.B.; et al. OSCA1 is an osmotic specific sensor: A method to distinguish Ca2+-mediated osmotic and ionic perception. New Phytol. 2022, 235, 1665–1678. [Google Scholar] [CrossRef]
- Deng, Z.C.; Tian, D.D.; Song, Q.S.; Guo, C.; Wen, L.C.; Wang, Q.; Chu, Y.M.; Liu, T.; Guo, Y.F. Genome wide identification and expression analysis of the OSCA gene family in response to abiotic stresses in tobacco. Chin. Tob. Sci. 2022, 43, 14–21. [Google Scholar]
- Wang, Z.; Lu, M.; Nan, H.; An, H.M. Identification and expression analysis of OSCA gene family in Rosa roxburahii Tratt. Plant Physiol. Commun. 2022, 58, 350–362. [Google Scholar]
- Li, Y.S.; Yuan, F.; Wen, Z.H.; Li, Y.H.; Wang, F.; Zhu, T.; Zhuo, W.Q.; Jin, X.; Wang, Y.D.; Zhao, H.P.; et al. Genome-wide survey and expression analysis of the OSCA gene family in rice. BMC Plant Biol. 2015, 15, 2–9. [Google Scholar] [CrossRef]
- Han, Y.; Wang, Y.X.; Zhai, Y.J.; Wen, Z.H.; Liu, J.; Xi, C.; Zhao, H.P.; Wang, Y.D.; Han, S.C. OsOSCA1.1 Mediates hyperosmolality and salt stress sensing in Oryza sativa. Biology 2022, 11, 678. [Google Scholar] [CrossRef]
- Pei, S.; Tao, Q.; Li, W.; Qi, G.; Wang, B.; Wang, Y.; Dai, S.; Shen, Q.; Wang, X.; Wu, X.; et al. Osmosensor-mediated control of Ca2+ spiking in pollen germination. Nature 2024, 629, 1118–1125. [Google Scholar] [CrossRef]
- Bukhari, B.; Guo, C.; Sun, J.; Han, Y.; Lai, X.; Lin, C.; Wang, Y.; Fang, Y. Proteomic, transcriptomic, biochemical and physiological investigations shed light on responses to low temperature stress in two contrasting soybean varieties. Plant Cell Environ. 2025. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, Y.X.; Ni, S.F.; Mu, J.J.; Liu, M.D.; Wang, Y.F.; Zhao, Y.H. Genomewide identification and analysis of the OSCA gene family in barley (Hordeum vulgare L.). J. Genet. 2022, 101, 2–34. [Google Scholar] [CrossRef]
- Batisti, O.; Kudla, J. Analysis of calcium signaling pathways in plants. BBA-Gen. Subj. 2012, 1820, 1283–1293. [Google Scholar] [CrossRef]
- Hedrich, R. Ion channels in plants. Physiol. Rev. 2012, 92, 1777–1811. [Google Scholar] [CrossRef]
- Jezek, M.; Blatt, M.R. The membrane transport system of the guard cell and its integration for stomatal dynamics. Plant Physiol. 2017, 174, 487–519. [Google Scholar] [CrossRef]
- Cao, L.R.; Zhang, P.Y.; Lu, X.M.; Wang, G.R.; Wang, Z.H.; Zhang, Q.J.; Zhang, X.; Wei, X.; Mei, F.J.; Wei, L.; et al. Systematic analysis of the maize OSCA genes revealing ZmOSCA family members involved in osmotic stress and ZmOSCA2.4 confers enhanced drought tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 351. [Google Scholar] [CrossRef]
- Steinhorst, L.; Kudla, J. Calcium—A central regulator of pollen germination and tube growth. Biochim. Et Biophys. Acta-Mol. Cell Res. 2013, 1833, 1573–1581. [Google Scholar] [CrossRef]
- Gu, X.; Wang, P.; Liu, Z.; Wang, L.; Huang, Z.; Zhang, S.; Wu, J. Genome-wide identification and expression analysis of the OSCA gene family in Pyrus bretschneideri. Can. J. Plant Sci. 2018, 98, 918–929. [Google Scholar] [CrossRef]
- Hou, C.C.; Tian, W.; Kleist, T.; He, K.; Garcia, V.; Bai, F.; Hao, Y.L.; Luan, S.; Li, L.G. DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res. 2014, 24, 632–635. [Google Scholar] [CrossRef]
- Waseem, M.; Aslam, M.M.; Shaheen, I. The DUF221 domain-containing (DDP) genes identification and expression analysis in tomato under abiotic and phytohormone stress. GM Crops Food 2021, 12, 586–599. [Google Scholar] [CrossRef]
- Ding, S.C.; Feng, X.; Du, H.W.; Wang, H.W. Genome-wide analysis of maize OSCA family members and their involvement in drought stress. PeerJ 2019, 7, e6765. [Google Scholar] [CrossRef]
- Yin, L.L.; Zhang, M.L.; Wu, R.G.; Chen, X.L.; Liu, F.; Xing, B.L. Genome-wide analysis of OSCA gene family members in Vigna radiata and their involvement in the osmotic response. BMC Plant Biol. 2021, 21, 408. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Y.; Luo, C.Y.; Zhan, K.; Qiu, R.-H.; Huang, C.H.; Xu, X.B.; Jia, D.F. Genome-wide identification of OSCA gene family members and their expression under different abiotic stresses in kiwifruit. J. Fruit Sci. 2024, 41, 436–447. [Google Scholar]
- Liu, D.D.; Wu, Q.; Jiao, X.Y.; Sun, M.H.; Wang, W.-J. Identification and expression analysis of the OSCA gene family in tea plant. Chin. J. Trop. Crops 2024, 45, 10–22. [Google Scholar]
- Cui, H.L.; Sun, M.Z.; Jia, B.W.; Sun, X.L. Identification of the OSCA gene family in Medicago truncatula and expression analysis under low-temperature stress. Acta Prataculturae Sin. 2024, 33, 111–125. [Google Scholar]
- Tong, K.; Wu, X.W.; He, L.; Qiu, S.Y.; Liu, S.; Cai, L.N.; Rao, S.F.; Chen, J.P. Genome-wide identification and expression profile of OSCA gene family members in Triticum aestivum L. Int. J. Mol. Sci. 2021, 23, 469. [Google Scholar] [CrossRef]
- Li, F.; Liu, J.X.; Guo, X.H.; Yin, L.L.; Zhang, H.L.; Wen, R.Y. Genome-wide survey, characterization, and expression analysis of bZIP transcription factors in Chenopodium quinoa. BMC Plant Biol. 2020, 20, 405. [Google Scholar] [CrossRef]
- Jie, Z.; Chen, X.H.; Pang, D.W.; Duan, A.A.; Li, X.M.; Sun, Y.H.; Yang, M.S. Identification of Populus tomentosa OSCA gene family and its expression analysis under salt stress. Plant Gene Trait 2022, 13, 1–10. [Google Scholar] [CrossRef]
- Qiu, X.W.; Xu, M.Y.; Shao, C.X.; Y, Y.X.; Li, D.; Cheng, L.Y.; Wu, C.J. Identification and expression analysis of OSCA gene family in pepper. Mol. Plant Breed. 2022, 11, 1134. [Google Scholar]
- Liu, X.; Wang, J.W.; Sun, L.F. Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2. Nat. Commun. 2018, 9, 5060–5068. [Google Scholar] [CrossRef]
- Roeber, V.M.; Bajaj, I.; Rohde, M.; Schmülling, T.; Cortleven, A. Light acts as a stressor and influences abiotic and biotic stress responses in plants. Plant Cell Environ. 2021, 44, 645–664. [Google Scholar] [CrossRef]
- González, E.-M. Drought stress tolerance in plants. Int. J. Mol. Sci. 2023, 24, 6562. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015, 58, 29–48. [Google Scholar] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.X.; Zhang, W.X.; Zhang, Y.; Zhang, X.J.; Lang, D.Y.; Zhang, X.H. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2020, 46, 197–212. [Google Scholar] [CrossRef]
- Kaur, A.; Sharma, A.; Madhu; Dixit, S.; Singh, K.; Upadhyay, S.K. OSCA genes in bread wheat: Molecular characterization, expression profiling, and interaction analyses indicated their diverse roles during development and stress response. Int. J. Mol. Sci. 2022, 23, 14867. [Google Scholar] [CrossRef]
- Shan, F.B.; Wu, Y.; Du, R.X.; Yang, Q.F.; Liu, C.H.; Wang, Y.X.; Zhang, C.; Chen, Y. Evolutionary analysis of the OSCA gene family in sunflower (Helianthus annuus L.) and expression analysis under NaCl stress. PeerJ 2023, 11, e15089. [Google Scholar] [CrossRef]
- Del Campo, E.M.; Casano, L.M.; Barreno, E. Evolutionary implications of intron-exon distribution and the properties and sequences of the RPL10A gene in eukaryotes. Mol. Phylogenetics Evol. 2013, 66, 857–867. [Google Scholar] [CrossRef]
- Li, J.Q.; Luo, S.L.; Zhang, S.L.; Zhang, W.Y.; Zhang, G.B. Genome-wide identification of the OSCA gene family in pepper and expression analysis under different stress conditions. Plant Sci. J. 2022, 40, 187–196. [Google Scholar]
- Wang, W.X.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Michel, B.E.; Kaufmann, M.R. The osmotic potential of polyethylene glycol 6000. Plant Physiol. 1973, 51, 914–916. [Google Scholar] [CrossRef]
- Garcia-Hernandez, M.; Berardini, T.; Chen, G.H.; Crist, D.; Doyle, A.; Huala, E.; Knee, E.; Lambrecht, M.; Miller, N.; Mueller, L.A.; et al. TAIR: A resource for integrated Arabidopsis data. Funct. Integr. Genom. 2002, 2, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.N.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. CABIOS 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.L.; Lin, Y.S.; Lin, C.Y.; Chung, Y.C.; Chung, Y.F. CUDA ClustalW: An efficient parallel algorithm for progressive multiple sequence alignment on Multi-GPUs. Comput. Biol. Chem. 2015, 58, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.H.; Lercher, M.J.; Hu, S.N.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- He, Z.L.; Zhang, H.K.; Gao, S.L.; Lercher, M.J.; Chen, W.H.; Hu, S.N. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Zhang, H.K.; Gao, S.H.; Lercher, M.J.; Hu, S.N.; Chen, W.H. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Rocha, D.J.P.G.; Castro, T.L.P.; Aguiar, E.R.G.R.; Pacheco, L.G.C. Gene expression analysis in Bacteria by RT-qPCR. Methods Mol. Biol. 2020, 2065, 119–137. [Google Scholar]
- Seifert, E. OriginPro 9.1: Scientific data analysis and graphing software—Software review. J. Chem. Inf. Model. 2014, 54, 1552–1553. [Google Scholar] [CrossRef]
- Mitteer, D.-R.; Greer, B.-D. Using GraphPad prism’s heat maps for efficient, fine-grained analyses of single-case data. Behav. Anal. Pract. 2022, 15, 505–514. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]


| Gene ID | Sequence ID | Number of Amino Acids | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathi-City | Transmembrane Domain | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| NofOSCA 01 | evm.model.Chr01.3154 | 141 | 15,918.53 | 9.37 | 38.64 | 104.47 | −0.077 | 0 | chloroplast |
| NofOSCA 02 | evm.model.Chr01.5771 | 761 | 86,236.02 | 9.02 | 44 | 100.96 | 0.22 | 10 | plasma membrane |
| NofOSCA 03 | evm.model.Chr01.8116 | 769 | 87,453.56 | 9.23 | 39.54 | 103.2 | 0.159 | 8 | plasma membrane |
| NofOSCA 04 | evm.model.Chr01.8913 | 709 | 80,504.33 | 9.14 | 42.24 | 107.38 | 0.329 | 9 | plasma membrane |
| NofOSCA 05 | evm.model.Chr01.8937 | 1039 | 117,120.39 | 5.87 | 45.85 | 95.45 | 0.04 | 8 | nucleus |
| NofOSCA 06 | evm.model.Chr02.1166 | 780 | 88,102.16 | 9.08 | 47.05 | 98.71 | 0.246 | 10 | plasma membrane |
| NofOSCA 07 | evm.model.Chr03.1679 | 719 | 81,000.99 | 9.28 | 29.33 | 103.69 | 0.286 | 9 | plasma membrane |
| NofOSCA 08 | evm.model.Chr03.1823 | 887 | 101,425.11 | 9.22 | 45.8 | 105.64 | 0.173 | 12 | plasma membrane |
| NofOSCA 09 | evm.model.Chr03.1857 | 749 | 85,843.52 | 9.18 | 45.24 | 102.59 | 0.133 | 9 | cell wall |
| NofOSCA 10 | evm.model.Chr03.4191 | 719 | 81,000.99 | 9.28 | 29.33 | 103.69 | 0.286 | 9 | plasma membrane |
| NofOSCA 11 | evm.model.Chr04.1948 | 1244 | 140,334.56 | 5.67 | 50.56 | 89.34 | −0.106 | 2 | chloroplast |
| NofOSCA 12 | evm.model.Chr04.1949 | 756 | 86,055.52 | 8.75 | 40.45 | 108.7 | 0.213 | 10 | plasma membrane |
| NofOSCA 13 | 08_QHevm.model.Chr05.38_R0 | 813 | 91,784.42 | 6.66 | 41.19 | 102.62 | 0.147 | 9 | plasma membrane |
| NofOSCA 14 | evm.model.Chr05.4066 | 772 | 88,535.84 | 8.9 | 45.36 | 101.31 | 0.151 | 8 | plasma membrane |
| NofOSCA 15 | evm.model.Chr06.115 | 683 | 78295.34 | 9.37 | 41.1 | 107.06 | 0.302 | 8 | plasma membrane |
| NofOSCA 16 | evm.model.Chr06.1871 | 603 | 69,265.51 | 8.99 | 41.25 | 97.5 | 0.023 | 6 | plasma membrane |
| NofOSCA 17 | evm.model.Chr06.5147 | 760 | 86,060.78 | 8.89 | 37.78 | 104.33 | 0.242 | 10 | plasma membrane |
| NofOSCA 18 | evm.model.Chr06.860 | 751 | 84,489.39 | 8.45 | 43.01 | 99.68 | 0.254 | 11 | nucleus |
| NofOSCA 19 | evm.model.Chr07.27 | 589 | 67,484.38 | 9.27 | 30.79 | 104.6 | 0.201 | 7 | plasma membrane |
| NofOSCA 20 | evm.model.Chr07.3709 | 723 | 82,372.78 | 9.05 | 46.42 | 108.27 | 0.188 | 11 | plasma membrane |
| NofOSCA 21 | evm.model.Chr09.276 | 652 | 73,488.58 | 9 | 33.51 | 101.49 | 0.175 | 8 | plasma membrane |
| NofOSCA 22 | evm.model.Chr09.307 | 652 | 73,488.58 | 9 | 33.51 | 101.49 | 0.175 | 8 | plasma membrane |
| NofOSCA 23 | evm.model.Chr10.604 | 868 | 99,515.12 | 8.98 | 45.95 | 93.81 | −0.043 | 8 | nucleus |
| NofOSCA 24 | evm.model.Contig1135.1 | 335 | 37,935.67 | 8.46 | 54.76 | 107.7 | 0.267 | 3 | plasma membrane |
| NofOSCA 25 | evm.model.Contig219.2 | 760 | 86,088.83 | 8.89 | 37.78 | 104.58 | 0.245 | 10 | plasma membrane |
| NofOSCA 26 | evm.model.Contig25.2 | 769 | 87,467.58 | 9.23 | 39.29 | 103.2 | 0.159 | 8 | plasma membrane |
| NofOSCA 27 | evm.model.Contig2643.1 | 697 | 79,801.41 | 8.04 | 45.04 | 100.59 | 0.119 | 8 | plasma membrane |
| NofOSCA 28 | evm.model.Contig7.67 | 769 | 87,313.84 | 8.35 | 43.53 | 100.36 | 0.263 | 10 | plasma membrane |
| NofOSCA 29 | evm.model.Contig84.18 | 760 | 86,088.83 | 8.89 | 37.78 | 104.58 | 0.245 | 10 | plasma membrane |
| Protein | Alpha Helix | Extended Strand | Beta Turn | Random Coil |
|---|---|---|---|---|
| NofOSCA01 | 39.01 | 10.64 | 2.13 | 48.23 |
| NofOSCA02 | 49.41 | 10.51 | 1.45 | 38.63 |
| NofOSCA03 | 51.76 | 10.01 | 1.43 | 36.8 |
| NofOSCA04 | 57.4 | 11.85 | 1.27 | 29.48 |
| NofOSCA05 | 44.08 | 8.28 | 0.77 | 46.87 |
| NofOSCA06 | 49.23 | 9.62 | 1.15 | 40 |
| NofOSCA07 | 55.77 | 11.27 | 1.25 | 31.71 |
| NofOSCA08 | 50.39 | 10.37 | 1.69 | 37.54 |
| NofOSCA09 | 55.01 | 10.81 | 1.07 | 33.11 |
| NofOSCA10 | 55.77 | 11.27 | 1.25 | 31.71 |
| NofOSCA11 | 25.64 | 18.81 | 4.66 | 50.88 |
| NofOSCA12 | 52.38 | 10.98 | 1.32 | 35.32 |
| NofOSCA13 | 48.95 | 10.58 | 1.35 | 39.11 |
| NofOSCA14 | 50.91 | 10.49 | 1.3 | 37.31 |
| NofOSCA15 | 54.76 | 10.98 | 1.9 | 32.36 |
| NofOSCA16 | 49.92 | 10.78 | 1 | 38.31 |
| NofOSCA17 | 52.11 | 10.26 | 1.71 | 35.92 |
| NofOSCA18 | 51.26 | 10.12 | 1.2 | 37.42 |
| NofOSCA19 | 59.08 | 10.87 | 1.02 | 29.03 |
| NofOSCA20 | 54.5 | 10.93 | 1.24 | 33.33 |
| NofOSCA21 | 55.67 | 11.2 | 1.23 | 31.9 |
| NofOSCA22 | 55.67 | 11.2 | 1.23 | 31.9 |
| NofOSCA23 | 45.74 | 9.1 | 1.04 | 44.12 |
| NofOSCA24 | 53.43 | 8.36 | 1.79 | 36.42 |
| NofOSCA25 | 52.11 | 10.26 | 1.71 | 35.92 |
| NofOSCA26 | 51.5 | 10.79 | 1.17 | 36.54 |
| NofOSCA27 | 51.79 | 11.91 | 1.58 | 34.72 |
| NofOSCA28 | 50.72 | 9.75 | 1.17 | 38.36 |
| NofOSCA29 | 52.11 | 10.26 | 1.71 | 35.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.-Y.; He, X.-J.; Xie, Y.-Z.; Zhou, L.-P.; Meng, X.; Kang, J.; Luo, C.-Y.; Wang, Y.-N.; Li, Z.-H.; Guan, T.-X. Genome-Wide Identification, Phylogeny, and Abiotic Stress Response Analysis of OSCA Family Genes in the Alpine Medicinal Herb Notopterygium franchetii. Int. J. Mol. Sci. 2025, 26, 5043. https://doi.org/10.3390/ijms26115043
Zhang Q-Y, He X-J, Xie Y-Z, Zhou L-P, Meng X, Kang J, Luo C-Y, Wang Y-N, Li Z-H, Guan T-X. Genome-Wide Identification, Phylogeny, and Abiotic Stress Response Analysis of OSCA Family Genes in the Alpine Medicinal Herb Notopterygium franchetii. International Journal of Molecular Sciences. 2025; 26(11):5043. https://doi.org/10.3390/ijms26115043
Chicago/Turabian StyleZhang, Qi-Yue, Xiao-Jing He, Yan-Ze Xie, Li-Ping Zhou, Xin Meng, Jia Kang, Cai-Yun Luo, Yi-Nuo Wang, Zhong-Hu Li, and Tian-Xia Guan. 2025. "Genome-Wide Identification, Phylogeny, and Abiotic Stress Response Analysis of OSCA Family Genes in the Alpine Medicinal Herb Notopterygium franchetii" International Journal of Molecular Sciences 26, no. 11: 5043. https://doi.org/10.3390/ijms26115043
APA StyleZhang, Q.-Y., He, X.-J., Xie, Y.-Z., Zhou, L.-P., Meng, X., Kang, J., Luo, C.-Y., Wang, Y.-N., Li, Z.-H., & Guan, T.-X. (2025). Genome-Wide Identification, Phylogeny, and Abiotic Stress Response Analysis of OSCA Family Genes in the Alpine Medicinal Herb Notopterygium franchetii. International Journal of Molecular Sciences, 26(11), 5043. https://doi.org/10.3390/ijms26115043






