Integrating Proteomics into Personalized Medicine for Inflammatory Bowel Disease—Reality or Challenge?
Abstract
1. Introduction
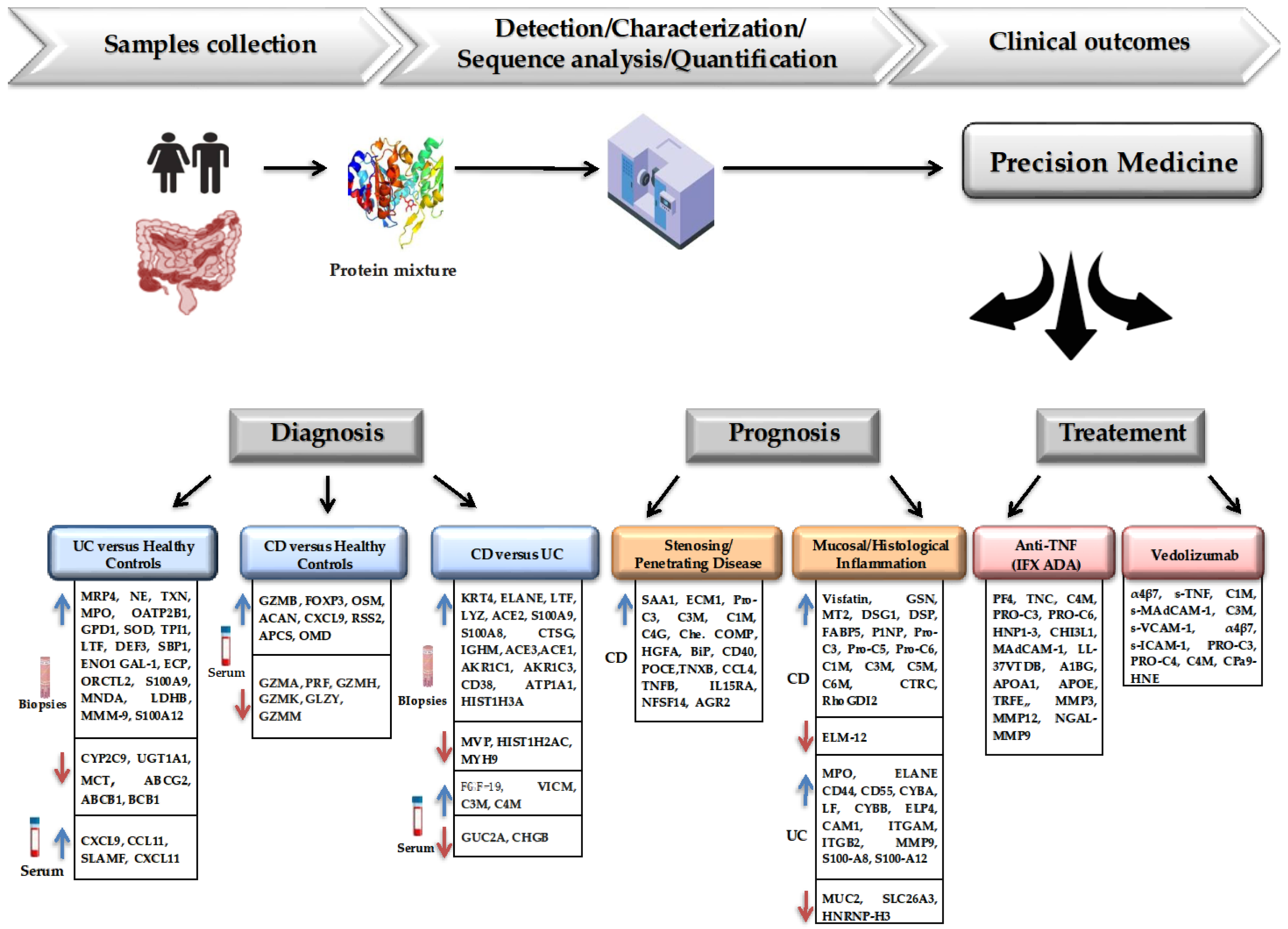
2. The Role of Proteomics in Diagnosis and Susceptibility to IBD
3. The Role of Proteomics as a Prognostic Factor in the Evolution of IBD Patients
4. Application of Proteomics in Optimizing Treatment Approaches for IBD
5. Limitations of Proteomics Study in IBD
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baldan-Martin, M.; Chaparro, M.; Gisbert, J.P. Tissue Proteomic Approaches to Understand the Pathogenesis of Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2021, 27, 1184–1200. [Google Scholar] [CrossRef] [PubMed]
- Cannarozzi, A.L.; Latiano, A.; Massimino, L.; Bossa, F.; Giuliani, F.; Riva, M.; Ungaro, F.; Guerra, M.; Brina, A.L.D.; Biscaglia, G.; et al. Inflammatory bowel disease genomics, transcriptomics, proteomics and metagenomics meet artificial intelligence. United Eur. Gastroenterol. J. 2024, 12, 1461–1480. [Google Scholar] [CrossRef]
- Wang, R.; Li, Z.; Liu, S.; Zhang, D. Global, regional and national burden of inflammatory bowel disease in 204 countries and territories from 1990 to 2019: A systematic analysis based on the Global Burden of Disease Study 2019. BMJ Open 2023, 13, e065186. [Google Scholar] [CrossRef] [PubMed]
- Mak, J.W.Y.; Sun, Y.; Limsrivilai, J.; Abdullah, M.; Kaibullayeva, J.; Balderramo, D.; Vergara, B.I.; Paudel, M.S.; Banerjee, R.; Hilmi, I.; et al. GIVES-21 Consortium. Development of the global inflammatory bowel disease visualization of epidemiology studies in the 21st century (GIVES-21). BMC Med. Res. Methodol. 2023, 23, 129. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, M.; Allin, K.H.; Petralia, F.; Colombel, J.F.; Jess, T. Multiomics to elucidate inflammatory bowel disease risk factors and pathways. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 399–409. [Google Scholar] [CrossRef]
- Agrawal, M.; Spencer, E.A.; Colombel, J.F.; Ungaro, R.C. Approach to the Management of Recently Diagnosed Inflammatory Bowel Disease Patients: A User’s Guide for Adult and Pediatric Gastroenterologists. Gastroenterology 2021, 161, 47–65. [Google Scholar] [CrossRef]
- Atreya, R.; Neurath, M.F. Biomarkers for Personalizing IBD Therapy: The Quest Continues. Clin. Gastroenterol. Hepatol. 2024, 22, 1353–1364. [Google Scholar] [CrossRef]
- Longo, S.; Chieppa, M.; Cossa, L.G.; Spinelli, C.C.; Greco, M.; Maffia, M.; Giudetti, A.M. New Insights into Inflammatory Bowel Diseases from Proteomic and Lipidomic Studies. Proteomes 2020, 8, 18. [Google Scholar] [CrossRef]
- Aldars-García, L.; Marin, A.C.; Chaparro, M.; Gisbert, J.P. The Interplay between Immune System and Microbiota in Inflammatory Bowel Disease: A Narrative Review. Int. J. Mol. Sci. 2021, 22, 3076. [Google Scholar] [CrossRef]
- Ortega Moreno, L.; Sanz-Garcia, A.; Fernández de la Fuente, M.J.; Arroyo Solera, R.; Fernández-Tomé, S.; Marin, A.C.; Mora-Gutierrez, I.; Fernández, P.; Baldan-Martin, M.; Chaparro, M.; et al. Serum adipokines as non-invasive biomarkers in Crohn’s disease. Sci. Rep. 2020, 10, 18027. [Google Scholar] [CrossRef]
- Liu, X.; Peng, T.; Xu, M.; Lin, S.; Hu, B.; Chu, T.; Liu, B.; Xu, Y.; Ding, W.; Li, L.; et al. Spatial multi-omics: Deciphering technological landscape of integration of multi-omics and its applications. J. Hematol. Oncol. 2024, 17, 72. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Guo, S.; Wang, L. Systematic review of metabolomic alterations in ulcerative colitis: Unveiling key metabolic signatures and pathways. Ther. Adv. Gastroenterol. 2024, 17, 17562848241239580. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, S.M.; Sanghi, A.; Wu, S.; Snyder, M.P. A Customizable Analysis Flow in Integrative Multi-Omics. Biomolecules 2020, 10, 1606. [Google Scholar] [CrossRef] [PubMed]
- Borowitz, S.M. The epidemiology of inflammatory bowel disease: Clues to pathogenesis? Front. Pediatr. 2023, 10, 1103713. [Google Scholar] [CrossRef]
- Plevris, N.; Lees, C.W. Disease monitoring in inflammatory bowel disease: Evolving principles and possibilities. Gastroenterology 2022, 162, 1456–1475. [Google Scholar] [CrossRef]
- Nowak, J.K.; Kallab, R.; Satsang, J. Current and emerging biomarkers for ulcerative colitis. Expert Rev. Mol. Diagn. 2023, 23, 1107–1119. [Google Scholar] [CrossRef]
- CORE-IBD Collaborators; Ma, C.; Hanzel, J.; Panaccione, R.; Sandborn, W.J.; D’Haens, G.R.; Ahuja, V.; Atreya, R.; Bernstein, C.N.; Bossuyt, P.; et al. CORE-IBD: A Multidisciplinary International Consensus Initiative to Develop a Core Outcome Set for Randomized Controlled Trials in Inflammatory Bowel Disease. Gastroenterology 2022, 163, 950–964. [Google Scholar]
- Atreya, R.; Neurath, M.F.; Siegmund, B. Personalizing Treatment in IBD: Hype or Reality in 2020? Can We Predict Response to Anti-TNF? Front. Med. 2020, 7, 517. [Google Scholar] [CrossRef]
- Vento-Tormo, R.; Vilarrasa-Blasi, R. Cell-level reference maps for the human body take shape. Nature 2023, 619, 467–468. [Google Scholar] [CrossRef]
- Minea, H.; Singeap, A.M.; Minea, M.; Juncu, S.; Muzica, C.; Sfarti, C.V.; Chiriac, S.; Miftode, I.D.; Stanciu, C.; Trifan, A.; et al. The Contribution of Genetic and Epigenetic Factors: An Emerging Concept in the Assessment and Prognosis of Inflammatory Bowel Diseases. Int. J. Mol. Sci. 2024, 25, 8420. [Google Scholar] [CrossRef]
- Fabian, O.; Bajer, L.; Drastich, P.; Harant, K.; Sticova, E.; Daskova, N.; Modos, I.; Tichanek, F.; Cahova, M. A Current State of Proteomics in Adult and Pediatric Inflammatory Bowel Diseases: A Systematic Search and Review. Int. J. Mol. Sci. 2023, 24, 9386. [Google Scholar] [CrossRef]
- Baldan-Martin, M.; Chaparro, M.; Gisbert, J.P. Systematic Review: Urine Biomarker Discovery for Inflammatory Bowel Disease Diagnosis. Int. J. Mol. Sci. 2023, 24, 10159. [Google Scholar] [CrossRef]
- Gisbert, J.P.; Chaparro, M. Clinical Usefulness of Proteomics in Inflammatory Bowel Disease: A Comprehensive Review. J. Crohns Colitis 2019, 13, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Fiocchi, C. Omics and Multi-Omics in IBD: No Integration, No Breakthroughs. Int. J. Mol. Sci. 2023, 24, 14912. [Google Scholar] [CrossRef]
- Alharbi, R.A. Proteomics approach and techniques in identification of reliable biomarkers for diseases. Saudi J. Biol. Sci. 2020, 27, 968–974. [Google Scholar] [CrossRef]
- Naryzhny, S. Inventory of proteoforms as a current challenge of proteomics: Some technical aspects. J. Proteom. 2019, 191, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, Y.; Zhang, T.; Shu, L.; Roepstorff, P.; Yang, F. Quantitative Proteomics Using Isobaric Labeling: A Practical Guide. Genom. Proteom. Bioinform. 2021, 19, 689–706. [Google Scholar] [CrossRef]
- Lee, P.Y.; Saraygord-Afshari, N.; Low, T.Y. The evolution of two-dimensional gel electrophoresis—From proteomics to emerging alternative applications. J. Chromatogr. A 2020, 1615, 460763. [Google Scholar] [CrossRef] [PubMed]
- Assadsangabi, A.; Evans, C.A.; Corfe, B.M.; Lobo, A. Application of Proteomics to Inflammatory Bowel Disease Research: Current Status and Future Perspectives. Gastroenterol. Res. Pract. 2019, 2019, 1426954. [Google Scholar] [CrossRef]
- Deng, J.; Erdjument-Bromage, H.; Neubert, T.A. Quantitative Comparison of Proteomes Using SILAC. Curr. Protoc. Protein Sci. 2019, 95, e74. [Google Scholar] [CrossRef]
- Poljak, A.; Raftery, M.; Polly, P. Evaluating Cellular Viability by iTRAQ Proteomic Profiling. Methods Mol. Biol. 2023, 2644, 193–209. [Google Scholar] [PubMed]
- Fricker, L.D. Quantitative Peptidomics: General Considerations. Methods Mol. Biol. 2024, 758, 89–108. [Google Scholar]
- Beer, L.A.; Liu, P.; Ky, B.; Barnhart, K.T.; Speicher, D.W. Efficient Quantitative Comparisons of Plasma Proteomes Using Label-Free Analysis with MaxQuant. Methods Mol. Biol. 2017, 1619, 339–352. [Google Scholar]
- Anjo, S.I.; Santa, C.; Manadas, B. SWATH-MS as a tool for biomarker discovery: From basic research to clinical applications. Proteomics 2017, 17, 1600278. [Google Scholar] [CrossRef]
- Cecerska-Heryć, E.; Ronkowski, B.; Heryć, R.; Serwin, N.; Grygorcewicz, B.; Roszak, M.; Galant, K.; Dołęgowska, B. Proteomic and lipidomic biomarkers in the diagnosis and progression of inflammatory bowel disease—A review. Proteom. Clin. Appl. 2023, 17, 2200003. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, V.; Nita-Lazar, A. Recent Advancements in Subcellular Proteomics: Growing Impact of Organellar Protein Niches on the Understanding of Cell Biology. J. Proteome Res. 2024, 23, 2700–2722. [Google Scholar] [CrossRef]
- Lucaciu, L.A.; Seicean, R.; Uifălean, A.; Iacobescu, M.; Iuga, C.A.; Seicean, A. Unveiling Distinct Proteomic Signatures in Complicated Crohn’s Disease That Could Predict the Disease Course. Int. J. Mol. Sci. 2023, 24, 16966. [Google Scholar] [CrossRef] [PubMed]
- Fossati, A.; Li, C.; Uliana, F.; Wendt, F.; Frommelt, F.; Sykacek, P.; Heusel, M.; Hallal, M.; Bludau, I.; Capraz, T.; et al. PCprophet: A framework for protein complex prediction and differential analysis using proteomic data. Nat. Methods. 2021, 18, 520–527. [Google Scholar] [CrossRef]
- Pisani, L.F.; Moriggi, M.; Gelfi, C.; Vecchi, M.; Pastorelli, L. Proteomic insights on the metabolism in inflammatory bowel disease. World J. Gastroenterol. 2020, 26, 696–705. [Google Scholar] [CrossRef]
- Alghoul, Z.; Yang, C.; Merlin, D. The Current Status of Molecular Biomarkers for Inflammatory Bowel Disease. Biomedicines 2022, 10, 1492. [Google Scholar] [CrossRef]
- Bennike, T.B. Advances in proteomics: Characterization of the innate immune system after birth and during inflammation. Front. Immunol. 2023, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Mund, A.; Brunner, A.D.; Mann, M. Unbiased spatial proteomics with single-cell resolution in tissues. Mol. Cell 2022, 82, 2335–2349. [Google Scholar] [CrossRef] [PubMed]
- Kalla, R.; Adams, A.T.; Bergemalm, D.; Vatn, S.; Kennedy, N.A.; Ricanek, P.; Lindstrom, J.; Ocklind, A.; Hjelm, F.; Ventham, N.T.; et al. Serum proteomic profiling at diagnosis predicts clinical course, and need for intensification of treatment in inflammatory bowel disease. J. Crohns Colitis 2021, 15, 699–708. [Google Scholar] [CrossRef]
- Torres, J.; Petralia, F.; Sato, T.; Wang, P.; Telesco, S.E.; Choung, R.S.; Strauss, R.; Li, X.J.; Laird, R.M.; Gutierrez, R.L.; et al. Serum Biomarkers Identify Patients Who Will Develop Inflammatory Bowel Diseases Up to 5 Years Before Diagnosis. Gastroenterology 2020, 159, 96–104. [Google Scholar] [CrossRef]
- Bergemalm, D.; Andersson, E.; Hultdin, J.; Eriksson, C.; Rush, S.T.; Kalla, R.; Adams, A.T.; Keita, Å.V.; D’Amato, M.; Gomollon, F.; et al. Systemic Inflammation in Preclinical Ulcerative Colitis. Gastroenterology. 2021, 161, 1526–1539. [Google Scholar] [CrossRef]
- Bennike, T.B.; Carlsen, T.G.; Ellingsen, T.; Bonderup, O.K.; Glerup, H.; Bøgsted, M.; Christiansen, G.; Birkelund, S.; Stensballe, A.; Andersen, V. Neutrophil Extracellular Traps in Ulcerative Colitis: A Proteome Analysis of Intestinal Biopsies. Inflamm. Bowel Dis. 2015, 21, 2052–2067. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, P.; Bruckmueller, H.; Martin, P.; Busch, D.; Haenisch, S.; Muller, J.; Wiechowska-Kozlowska, A.; Partecke, L.I.; Heidecke, C.D.; Cascorbi, I.; et al. Dysregulation of mucosal membrane transporters and drug metabolizing enzymes in ulcerative colitis. J. Pharm. Sci. 2019, 108, 1035–1046. [Google Scholar] [CrossRef]
- Vessby, J.; Wisniewski, J.R.; Lindskog, C.; Eriksson, N.; Gabrysch, K.; Zettl, K.; Wanders, A.; Carlson, M.; Rorsman, F.; Åberg, M. AGPAT1 as a Novel Colonic Biomarker for Discriminating Between Ulcerative Colitis with and Without Primary Sclerosing Cholangitis. Clin. Trans. Gastroenterol. 2022, 13, e00486. [Google Scholar] [CrossRef]
- Grännö, O.; Bergemalm, D.; Salomon, B.; Lindqvist, C.M.; Hedin, C.R.H.; Carlson, M.; Dannenberg, K.; Andersson, E.; Keita, Å.; Magnusson, M.K.; et al. Preclinical Protein Signatures of Crohn’s Disease and Ulcerative Colitis: A Nested Case-Control Study Within Large Population-Based Cohorts. Gastroenterology 2025, 168, 741–753. [Google Scholar] [CrossRef]
- Ning, L.; Li, S.; Gao, J.; Ding, L.; Wang, C.; Chen, W.; Shan, G.; Zhang, F.; Yu, J.; Xu, G. Tenascin-C Is Increased in Inflammatory Bowel Disease and Is Associated with response to Infliximab Therapy. BioMed Res. Int. 2019, 2019, 1475705. [Google Scholar] [CrossRef]
- Starr, A.E.; Deeke, S.A.; Ning, Z.; Chiang, C.K.; Zhan, X.; Mottawea, W.; Singleton, R.; Benchimol, E.I.; Wen, M.; Mack, D.R.; et al. Proteomic analysis of ascending colon biopsies from a paediatric inflammatory bowel disease inception cohort identifies protein biomarkers that differentiate Crohn’s disease from UC. Gut 2017, 66, 1573–1583. [Google Scholar] [CrossRef] [PubMed]
- Wasinger, V.C.; Yau, Y.; Duo, X.; Zeng, M.; Campbell, B.; Shin, S.; Luber, R.; Redmond, D.; Leong, R.W. Low Mass Blood Peptides Discriminative of Inflammatory Bowel Disease (IBD) Severity: A Quantitative Proteomic Perspective. Mol. Cell Proteom. 2016, 15, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Moriggi, M.; Pastorelli, L.; Torretta, E.; Tontini, G.E.; Capitanio, D.; Bogetto, S.F.; Vecchi, M.; Gelfi, C. Contribution of Extracellular Matrix and Signal Mechanotransduction to Epithelial Cell Damage in Inflammatory Bowel Disease Patients: A Proteomic Study. Proteomics 2017, 17, 1700164. [Google Scholar] [CrossRef] [PubMed]
- Vitali, R.; Palone, F.; Armuzzi, A.; Fulci, V.; Negroni, A.; Carissimi, C.; Cucchiara, S.; Stronati, L. Proteomic Analysis Identifies Three Reliable Biomarkers of Intestinal Inflammation in the Stools of Patients With Inflammatory Bowel Disease. J. Crohns Colitis 2023, 17, 92–102. [Google Scholar] [CrossRef]
- Arafah, K.; Kriegsmann, M.; Renner, M.; Lasitschka, F.; Fresnais, M.; Kriegsmann, K.; von Winterfeld, M.; Goepper, B.; Kriegsmann, J.; Casadonte, R.; et al. Microproteomics and immunohistochemistry reveal differences in the aldo-keto-reductase family 1 member C3 in tissue specimens of ulcerative colitis and Crohn’s disease. Proteom. Clin. Appl. 2020, 14, e1900110. [Google Scholar] [CrossRef]
- Louis Sam Titus, A.S.C.; Vanarsa, K.; Soomro, S.; Patel, A.; Prince, J.; Kugathasan, S.; Mohan, C. Resistin, Elastase, and Lactoferrin as Potential Plasma Biomarkers of Pediatric Inflammatory Bowel Disease Based on Comprehensive Proteomic Screens. Mol. Cell Proteom. 2023, 22, 100487. [Google Scholar] [CrossRef]
- Mortensen, J.H.; Godskesen, L.E.; Jensen, M.D.; Van Haaften, W.T.; Klinge, L.G.; Olinga, P.; Dijkstra, G.; Kjeldsen, J.; Karsdal, M.A.; Bay-Jensen, A.C.; et al. Fragments of Citrullinated and MMP-degraded Vimentin and MMP-degraded Type III Collagen Are Novel Serological Biomarkers to Differentiate Crohn’s Disease from Ulcerative Colitis. J. Crohns Colitis 2015, 9, 863–872. [Google Scholar] [CrossRef]
- D’Haens, G.; Kelly, O.; Battat, R.; Silverberg, M.S.; Laharie, D.; Louis, E.; Savarino, E.; Bodini, G.; Yarur, A.; Boland, B.S.; et al. Development and Validation of a Test to Monitor Endoscopic Activity in Patients with Crohn’s Disease Based on Serum Levels of Proteins. Gastroenterology 2020, 158, 515–526. [Google Scholar] [CrossRef]
- Basso, D.; Padoan, A.; D’Incà, R.; Arrigoni, G.; Scapellato, M.L.; Contran, N.; Franchin, C.; Lorenzon, G.; Mescol, C.; Moz, S.; et al. Peptidomic and proteomic analysis of stool for diagnosing IBD and deciphering disease pathogenesis. Clin. Chem. La. Med. 2020, 58, 968–979. [Google Scholar] [CrossRef]
- Pierre, N.; Baiwir, D.; Huynh-Thu, V.A.; Mazzucchelli, G.; Smargiasso, N.; De Pauw, E.; Bouhnik, Y.; Laharie, D.; Colombel, J.F.; Meuwis, M.A.; et al. Discovery of biomarker candidates associated with the risk of short-term and mid/long-term relapse after infliximab withdrawal in Crohn’s patients: A proteomics-based study. Gut 2020, 70, 1450–1457. [Google Scholar] [CrossRef]
- Riaz, T.; Sollid, L.M.; Olsen, I.; de Souza, G.A. Quantitative Proteomics of Gut-Derived Th1 and Th1/Th17 Clones Reveal the Presence of CD28+ NKG2D- Th1 Cytotoxic CD4+ T cells. Mol. Cell Proteom. 2016, 15, 1007–1016. [Google Scholar] [CrossRef]
- Stidham, R.W.; Wu, J.; Shi, J.; Lubman, D.M.; Higgins, P.D. Serum Glycoproteome Profiles for Distinguishing Intestinal Fibrosis from Inflammation in Crohn’s Disease. PLoS ONE 2017, 12, e0170506. [Google Scholar] [CrossRef] [PubMed]
- Townsend, P.; Zhang, Q.; Shapiro, J.; Webb-Robertson, B.J.; Bramer, L.; Schepmoes, A.A.; Weitz, K.K.; Mallette, M.; Moniz, H.; Bright, R.; et al. Serum Proteome Profiles in Stricturing Crohn’s Disease: A Pilot Study. Inflamm. Bowel Dis. 2015, 21, 1935–1941. [Google Scholar] [CrossRef]
- Christensen, B.; Erlich, J.; Gibson, P.R.; Turner, J.R.; Hart, J.; Rubin, D.T. Histologic cure is more strongly associated with clinical outcomes in ileal Crohn’s disease than endoscopic cure. Clin. Gastroenterol. Hepatol. 2020, 18, 2518–2525. [Google Scholar] [CrossRef] [PubMed]
- Gruver, A.M.; Westfall, M.D.; Ackerman, B.L.; Hill, S.; Morrison, R.D.; Bodo, J.; Lai, K.K.; Gemperline, D.C.; His, E.D.; Liebler, D.C.; et al. Proteomic characterizations of endoscopic biopsies of ulcerative colitis are associated with clinically relevant histologic measures of disease severity. J. Clin. Pathol. 2022, 75, 636–642. [Google Scholar] [CrossRef] [PubMed]
- van der Post, S.; Jabbar, K.S.; Birchenough, G.; Arike, L.; Akhtar, N.; Sjovall, H.; Johansson, M.E.V.; Hansson, G.C. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut 2019, 68, 2142–2151. [Google Scholar] [CrossRef]
- Korolkova, O.Y.; Myers, J.N.; Pellom, S.T.; Wang, L.; M’Koma, A.E. Characterization of Serum Cytokine Profile in Predominantly Colonic Inflammatory Bowel Disease to Delineate Ulcerative and Crohn’s Colitides. Clin. Med. Insights Gastroenterol. 2015, 8, 29–44. [Google Scholar] [CrossRef]
- Assadsangabi, A.; Evans, C.A.; Corfe, B.M.; Lobo, A.J. Exploring Predictive Biomarkers of Relapse in Ulcerative Colitis: A Proteomics Approach. Inflamm. Bowel Dis. 2024, 30, 808–819. [Google Scholar] [CrossRef]
- Yau, Y.Y.; Leong, R.W.L.; Pudipeddi, A.; Redmond, D.; Wasinger, V.C. Serological Epithelial Component Proteins Identify Intestinal Complications in Crohn’s Disease. Mol. Cell Proteom. 2017, 16, 1244–1257. [Google Scholar] [CrossRef]
- Wu, J.; Lubman, D.M.; Kugathasan, S.; Denson, L.A.; Hyams, J.S.; Dubinsky, M.C.; Griffiths, A.M.; Baldassano, R.N.; Noe, J.D.; Rabizadeh, S.; et al. Serum Protein Biomarkers of Fibrosis Aid in Risk Stratification of Future Stricturing Complications in Pediatric Crohn’s Disease. Am. J. Gastroenterol. 2019, 114, 777–785. [Google Scholar] [CrossRef]
- Ballengee, C.R.; Stidham, R.W.; Liu, C.; Kim, M.O.; Prince, J.; Mondal, K.; Baldassano, R.; Dubinsky, M.; Markowitz, J.; Leleiko, N.; et al. Association Between Plasma Level of Collagen Type III Alpha 1 Chain and Development of Strictures in Pediatric Patients With Crohn’s Disease. Clin. Gastroenterol. Hepatol. 2019, 17, 1799–1806. [Google Scholar] [CrossRef] [PubMed]
- Ungaro, R.C.; Hu, L.; Ji, J.; Nayar, S.; Kugathasan, S.; Denson, L.A.; Hyams, J.; Dubinsky, M.C.; Sands, B.E.; Cho, J.H. Machine learning identifies novel blood protein predictors of penetrating and stricturing complications in newly diagnosed paediatric Crohn’s disease. Aliment. Pharmacol. Ther. 2021, 53, 281–290. [Google Scholar] [CrossRef]
- Vieujean, S.; Hu, S.; Bequet, E.; Salee, C.; Massot, C.; Bletard, N.; Pierre, N.; Quesada-Calv, F.; Baiwir, D.; Mazzucchelli, G.; et al. Potential Role of Epithelial Endoplasmic Reticulum Stress and Anterior Gradient Protein 2 Homologue in Crohn’s Disease Fibrosis. J. Crohns Colitis 2021, 15, 1737–1750. [Google Scholar] [CrossRef] [PubMed]
- van Haaften, W.T.; Mortensen, J.H.; Karsdal, M.A.; Bay-Jensen, A.C.; Dijkstra, G.; Olinga, P. Misbalance in type III collagen formation/degradation as a novel serological biomarker for penetrating (Montreal B3) Crohn’s disease. Aliment. Pharmacol. Ther. 2017, 46, 26–39. [Google Scholar] [CrossRef]
- Bourgonje, A.R.; Alexdottir, M.S.; Otten, A.T.; Loveikyte, R.; Bay-Jensen, A.C.; Pehrsson, M.; van Dullemen, H.M.; Visschedijk, M.C.; Festen, E.A.M.; Weersma, R.K.; et al. Serological biomarkers of type I, III and IV collagen turnover are associated with the presence and future progression of stricturing and penetrating Crohn’s disease. Aliment. Pharmacol. Ther. 2022, 56, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Pehrsson, M.; Domislovic, V.; Alexdottir, M.S.; Brinar, M.; Karsdal, M.A.; Barisic, A.; Krznaric, Z.; Mortensen, J.H. Blood-Based Biomarkers Reflecting Protease 3 and MMP-12 Catalyzed Elastin Degradation as Potential Noninvasive Surrogate Markers of Endoscopic and Clinical Disease in Inflammatory Bowel Disease. J. Clin. Med. 2023, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.O.; Armuzzi, A.; Gisbert, J.P.; Bokemeyer, B.; Peyrin-Biroulet, L.; Nguyen, G.C.; Smyth, M.; Patel, H. Indicators of suboptimal tumor necrosis factor antagonist therapy in inflammatory bowel disease. Dig. Liver Dis. 2017, 49, 1086–1091. [Google Scholar] [CrossRef]
- Adegbola, S.O.; Sahnan, K.; Warusavitarne, J.; Hart, A.; Tozer, P. Anti-TNF Therapy in Crohn’s Disease. Int. J. Mol. Sci. 2018, 19, 2244. [Google Scholar] [CrossRef]
- Cohen, B.L.; Sachar, D.B. Update on anti-tumor necrosis factor agents and other new drugs for inflammatory bowel disease. BMJ 2017, 357, j2505. [Google Scholar] [CrossRef]
- Pierre, N.; Huynh-Thu, V.A.; Marichal, T.; Allez, M.; Bouhnik, Y.; Laharie, D.; Bourreille, A.; Colombel, J.F.; Meuwis, M.A.; Louis, E.; et al. Distinct blood protein profiles associated with the risk of short-term and mid/long-term clinical relapse in patients with Crohn’s disease stopping infliximab: When the remission state hides different types of residual disease activity. Gut 2023, 72, 443–450. [Google Scholar] [CrossRef]
- Winter, D.A.; de Bruyne, P.; van der Woude, J.; Rizopoulos, D.; de Ridder, L.; Samsom, J.; Escher, J.C. Biomarkers predicting the effect of anti-TNF treatment in paediatric and adult inflammatory bowel disease. J. Pediatr. Gastroentero. Nutr. 2024, 79, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Biancheri, P.; Brezski, R.J.; Di Sabatino, A.; Greenplate, A.R.; Soring, K.L.; Corazza, G.R.; Kok, K.B.; Rovedatti, L.; Vossenkämper, A.; Ahmad, N.; et al. Proteolytic cleavage and loss of function of biologic agents that neutralize tumor necrosis factor in the mucosa of patients with inflammatory bowel disease. Gastroenterology 2015, 149, 1564–1574.e3. [Google Scholar] [CrossRef] [PubMed]
- Gazouli, M.; Anagnostopoulos, A.K.; Papadopoulou, A.; Vaiopoulou, A.; Papamichael, K.; Mantzaris, G.; Theodoropoulos, G.E.; Anagnou, N.P.; Tsangaris, G.T. Serum protein profile of Crohn’s disease treated with infliximab. J. Crohns Colitis 2013, 7, e461–e470. [Google Scholar] [CrossRef]
- Gazouli, M.A.; Fillet, M.; Lutteri, L.; Marée, R.; Geurts, P.; de Seny, D.; Malaise, M.; Chapelle, J.P.; Wehenkel, L.; Belaiche, J.; et al. Proteomics for prediction and characterization of response to infliximab in Crohn’s disease: A pilot study. Clin. Biochem. 2008, 41, 960–967. [Google Scholar]
- Soendergaar, M.K.; Strid, H.; Isaksson, S.; Bajor, A.; Lasson, A.; Ung, K.A.; Öhman, L. Response to infliximab therapy in ulcerative colitis is associated with decreased monocyte activation, reduced CCL2 expression and downregulation of Tenascin C. J. Crohns Colitis 2015, 9, 56–65. [Google Scholar]
- Pompili, S.; Latella, G.; Gaudio, E.; Sferra, R.; Vetuschi, A. The Charming World of the Extracellular Matrix: A Dynamic and Protective Network of the Intestinal Wall. Front. Med. 2021, 8, 610189. [Google Scholar] [CrossRef]
- Mortensen, J.H.; Lindholm, M.; Langholm, L.L.; Kjeldsen, J.; Bay-Jensen, A.C.; Karsdal, M.A.; Manon-Jensen, T. The intestinal tissue homeostasis—The role of extracellular matrix remodeling in inflammatory bowel disease. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 977–993. [Google Scholar] [CrossRef]
- van Haaften, W.T.; Mortensen, J.H.; Dige, A.K.; Grønbæk, H.; Hvas, C.L.; Bay-Jensen, A.C.; Karsdal, M.A.; Olinga, P.; Manon-Jensen, T.; Dijkstra, G. Serological Biomarkers of Tissue Turnover Identify Responders to Anti-TNF Therapy in Crohn’s Disease: A Pilot Study. Clin. Transl. Gastroenterol. 2020, 11, e00217. [Google Scholar] [CrossRef]
- Alexdottir, M.S.; Bourgonje, A.R.; Karsdal, M.A.; Pehrsson, M.; Loveikyte, R.; van Dullemen, H.M.; Visschedijk, M.C.; Festen, E.A.M.; Weersma, R.K.; Faber, K.N.; et al. Serological Biomarkers of Intestinal Collagen Turnover Identify Early Response to Infliximab Therapy in Patients With Crohn’s Disease. Front. Med. 2022, 9, 933872. [Google Scholar] [CrossRef]
- Bouhnik, Y.; Carbonnel, F.; Laharie, D.; Stefanescu, C.; Hébuterne, X.; Abitbol, V.; Nachury, M.; Brixi, H.; Bourreille, A.; Picon, L.; et al. Efficacy of adalimumab in patients with Crohn’s disease and symptomatic small bowel stricture: A multicentre, prospective, observational cohort (CREOLE) study. Gut 2018, 67, 53–60. [Google Scholar] [CrossRef]
- Ponsioen, C.Y.; de Groof, E.J.; Eshuis, E.J.; Gardenbroek, T.J.; Bossuyt, P.M.M.; Hart, A.; Warusavitarne, J.; Buskens, C.J.; van Bodegraven, A.A.; Brink, M.A.; et al. Laparoscopic ileocaecal resection versus infliximab for terminal ileitis in Crohn’s disease: A randomised controlled, open-label, multicentre trial. Lancet Gastroenterol. Hepatol. 2017, 2, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Pu, D.; Wang, D.; Zhang, M.; Zhou, C.; Zhang, Z.; Feng, B. Proteomic Analysis of Potential Targets for Non-Response to Infliximab in Patients With Ulcerative Colitis. Front. Pharmacol. 2022, 13, 905133. [Google Scholar] [CrossRef] [PubMed]
- Soendergaard, C.; Seidelin, J.B.; Steenholdt, C.; Nielsen, O.H. Putative biomarkers of vedolizumab resistance and underlying inflammatory pathways involved in IBD. BMJ Open Gastroenterol. 2018, 5, e000208. [Google Scholar] [CrossRef]
- Li, L.; Chen, R.; Zhang, Y.; Zhou, G.; Chen, B.; Zeng, Z.; Chen, M.; Zhang, S. A Novel Model Based on Serum Biomarkers to Predict Primary Non-Response to Infliximab in Crohn’s Disease. Front. Immunol. 2021, 12, 646673. [Google Scholar] [CrossRef]
- Ringold, R.; Martens, E.; Ferrante, M.; Van Assche, G.; Opdenakker, G.; Dukler, A.; Vermeire, S. The Ulcerative Colitis Response Index for Detection of Mucosal Healing in Patients Treated With Anti-tumour Necrosis Factor. J. Crohns Colitis 2020, 14, 176–184. [Google Scholar]
- Battat, R.; Dulai, P.S.; Vande Casteele, N.; Evans, E.; Hester, K.D.; Webster, E.; Jain, A.; Proudfoot, J.A.; Mairalles, A.; Neill, J.; et al. Biomarkers Are Associated With Clinical and Endoscopic Outcomes With Vedolizumab Treatment in Ulcerative Colitis. Inflamm. Bowel Dis. 2019, 25, 410–420. [Google Scholar] [CrossRef]
- Holmer, A.K.; Battat, R.; Dulai, P.S.; Vande Casteele, N.; Nguyen, N.; Jain, A.; Miralles, A.; Neill, J.; Le, H.; Singh, S.; et al. Biomarkers are associated with clinical and endoscopic outcomes with vedolizumab treatment in Crohn’s disease. Ther. Adv. Gastroenterol. 2020, 13, 1756284820971214. [Google Scholar] [CrossRef]
- Alexdottir, M.S.; Bourgonje, A.R.; Karsdal, M.A.; Pehrsson, M.; Loveikyte, R.; van Dullemen, H.M.; Visschedijk, M.C.; Festen, E.A.M.; Weersma, R.K.; Faber, K.N.; et al. Serological Biomarkers of Extracellular Matrix Turnover and Neutrophil Activity Are Associated with Long-Term Use of Vedolizumab in Patients with Crohn’s Disease. Int. J. Mol. Sci. 2022, 23, 8137. [Google Scholar] [CrossRef] [PubMed]
- Heier, C.R.; Fiorillo, A.A.; Chaisson, E.; Gordish-Dressman, H.; Hathout, Y.; Damsker, J.M.; Hoffman, E.P.; Conklin, L.S. Identification of Pathway-Specific Serum Biomarkers of Response to Glucocorticoid and Infliximab Treatment in Children with Inflammatory Bowel Disease. Clin. Transl. Gastroenterol. 2016, 7, e192. [Google Scholar] [CrossRef]
- Boden, E.K.; Shows, D.M.; Chiorean, M.V.; Lord, J.D. Identification of Candidate Biomarkers Associated with Response to Vedolizumab in Inflammatory Bowel Disease. Dig. Dis. Sci. 2018, 63, 2419–2429. [Google Scholar] [CrossRef]
- Vandereyken, K.; Sifrim, A.; Thienpont, B.; Voet, T. Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet. 2023, 24, 494–515. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Fan, Q.; Li, Z.; Goll, R.; Florholmen, J. Evaluation of anti-TNF therapeutic response in patients with inflammatory bowel disease: Current and novel biomarkers. EBioMedicine 2021, 66, 103329. [Google Scholar] [CrossRef] [PubMed]
- Elhag, D.A.; Kumar, M.; Saadaoui, M.; Akobeng, A.K.; Al-Mudahka, F.; Elawad, M.; Al Khodor, S. Inflammatory Bowel Disease Treatments and Predictive Biomarkers of Therapeutic Response. Int. J. Mol. Sci. 2022, 23, 6966. [Google Scholar] [CrossRef] [PubMed]
- Cartlidge, P.; Ho, G.T.; Kalla, R. Decoding the proteomic landscape of inflammatory bowel disease. J. Crohns Colitis 2025, 19, jjaf008. [Google Scholar] [CrossRef]
- Centanni, L.; Cicerone, C.; Fanizzi, F.; D’Amico, F.; Furfaro, F.; Zilli, A.; Parigi, T.L.; Peyrin-Biroulet, L.; Danese, S.; Allocca, M. Advancing Therapeutic Targets in IBD: Emerging Goals and Precision Medicine Approaches. Pharmaceuticals 2025, 18, 78. [Google Scholar] [CrossRef]
- Kumar, M.; Murugesan, S.; Ibrahim, N.; Elawad, M.; Al Khodor, S. Predictive biomarkers for anti-TNF alpha therapy in IBD patients. J. Transl. Med. 2024, 22, 284. [Google Scholar] [CrossRef]
| Type of Technology | Principle | Advantages | Disadvantages | References |
|---|---|---|---|---|
| 2DE | Proteins are fractionated and separated based on their isoelectric point and molecular weight on polyacrylamide gels. | Low cost. High resolution. Able to analyze complex structures, including protein isoforms and post-translational modifications. | Biological variation—low reproducibility. Laborious. Protein identification requires additional MS. | [26,28] |
| 2D-DIGE | Uses spectrally distinct fluorescent dyes for sample labeling, allowing comparative analysis of up to three proteins on a single gel. | Higher sensitivity, accuracy, and reproducibility. Increased detection rate compared to 2DE. | Laborious. Protein identification requires additional MS. | [26] |
| SILAC | Normal and heavy stable isotope labeling by amino acids in cell culture enables precise quantification of protein abundance. | Simple. Accurate and reproducible quantification. Suitable for dynamic studies of protein turnover. | Limited to cell culture systems or model organisms. Not applicable to complex biological samples. Samples must be grown in custom media to incorporate stable isotopes during growth. | [30] |
| SILAMi | Isotopically labeled microorganisms are used to trace protein synthesis in microbial communities. | Allows study of microbial dynamics and interactions. Suitable for intestinal microbiome-related proteomics. | Requires specific growth conditions for labeled microorganisms. Limited application for clinical investigations. | [29] |
| ICAT | Using a reagent that contains a reactive group towards thiol groups, a linker to incorporate stable isotopes (2H/1H), and an affinity tag to isolate isotope-labeled proteins/peptides (chemical labeling in vitro). | High accuracy for quantitative proteomic analysis of cells and tissues. | Requires chromatographic separation techniques. | [28] |
| iTRAQ | Proteins are digested into peptides, which are labeled with isobaric tags. Quantification is achieved by measuring reporter ion intensities during mass spectrometry analysis (chemical labeling in vitro). | Increased multiplexing capability. Enables relative and absolute quantification of proteins in multiple samples. | Complex sample preparation. Requires advanced MS expertise. | [31,32] |
| TMT | Proteins are divided into peptides and labeled with TMT reagents that release reporter ions during MS2 for quantification (chemical labeling in vitro). | Enables multiplexing of up to 16 samples. Suitable for comparative proteomics. | Expensive reagents. Potential interference between reporter ions at high multiplexing levels. Quantitative precision is dependent on the reproducibility of sample preparation. | [32] |
| LFQ | Peptides are quantified based on MS signal intensity or spectral counting without additional labeling. | Simple sample preparation. Supports high-throughput proteomic analysis. Dynamically detects differential protein expression. | Less accurate than the labeling methods (TMT). Reduced repeatability. The stability of experimental operation is demanding. Low reproducibility. Additional time needed for MS analysis. | [33] |
| LC-MS/MS | Chemical compounds are separated by liquid chromatography and analyzed by mass spectrometry to identify and quantify proteins. | High sensitivity and versatility. Capable of analyzing a wide range of biomolecules with high resolution. | Incomplete protein digestion. Difficulties in chromatographic separation of peptides. Requires advanced instrumentation and expertise. High operational costs. | [25] |
| MALDI-TOF-MS | Proteins or peptides are ionized using a laser, and their mass-to-charge ratios are analyzed in a time-of-flight mass spectrometer. | Rapid analysis of biomolecules. Minimal sample preparation. Suitable for high-throughput proteomic studies. | Limited sensitivity for low-abundance proteins. Lower resolution compared to other MS techniques. | [27] |
| SWATH-MS | A data-independent acquisition method where all precursor ions within a defined mass range are fragmented systematically and analyzed simultaneously. | Low-cost. High sensitivity and comprehensive coverage. Allows quantification of thousands of complex proteomes in a single run. | Requires advanced instrumentation and data analysis software. Computationally intensive and time-consuming for processing large datasets. | [34] |
| Disease | Number of Patients | Sample | Proteomic Model | Main Findings | References | |
|---|---|---|---|---|---|---|
| UC | 72 patients versus 140 controls | Plasma | MMP10, MCP-1, CXCL9, CCL11, SLAMF1, CXCL11 | Increased | The model was upregulated before the onset of UC compared to healthy controls (AUC = 0.92, p < 0.05). | [45] |
| UC | 10 patients versus 10 controls | Colonic tissue | LTF, NE, ECP, MMP-9, MPO, MNDA, CatG, S100-A9, Gal-10, S100-A12, DEF3 | Increased | High abundance in neutrophils (on average 42.2 times, p < 0.005) is associated with NETs formation in UC compared to controls. The severity of histological inflammation correlates with LTF (r = 0.91) and S100-A9 (r = 0.82, p < 0.001). | [46] |
| UC | 10 patients versus 10 controls | Colonic tissue | MRP4, ORCTL2, OATP2B1 | Increased | Significant modification of the expression profile of metabolizing enzymes and protein transporters in inflamed tissue in UC patients compared to controls. | [47] |
| ABCB1, MCT1, ABCG2, | Decreased | |||||
| UC | 55 patients versus 7 controls | Colonic tissue | CD47, NDUFAF4, AGPAT1, LSM-7, TMEM192 | Increased | Five proteins are differentially expressed in UC compared to controls. AGPAT1 is a potential colonic biomarker for distinguishing PSC-UC from UC. | [48] |
| UC | 102 patients versus 156 controls | Serum | TAMBP, SIRT2, SCAMP3, CD5, ADAM8, GZMB, MMP-10, CXCL9, CDKN1A, CCL11, ABL1, TNFRSF6B | Increased | The protein panel has a superior ability to discriminate between UC patients and the control group (AUC = 0.95; 95% CI: 0.92–0.99). | [49] |
| CD | 54 patients versus 156 controls | Serum | CXCL9, IL-6, MMP-10, CCL20, MDK, CXCL17 | Increased | Increased ability to differentiate CD patients from controls (AUC = 0.85; 95% CI: 0.78–0.93). | [49] |
| DNER, GPNMB, CX3CL1 | Decreased | |||||
| IBD | 328 patients versus 224 controls | Serum | MMP-12, OSM, CXCL1, IL-8, IL-17A, CXCL9, GrB, MMP-10, CXCL11, HGF | Increased | The model differentiates IBD from the control group (accuracy = 0.798, 95% CI: 0.764–0.832; sensitivity = 0.831, 95% CI: 0.791–0.872; specificity = 0.748, 95% CI: 0.690–0.805). | [43] |
| GAS6, ITGAV | Decreased | |||||
| IBD | 24 patients versus 9 controls | Colonic tissue | CHI3L1, PNP, OLFM4, LCN2, MMP9, NAMPT, NNMT, PARP9, PARP14, NFKB2, CD38, S100A12, | Increased | The model differentiates IBD patients from the control group. | [50] |
| ITLN1, NNT, NT5C3A, NADK2 | Decreased | |||||
| IBD | 60 patients (30 UC, 30 CD) versus 39 controls | Colonic tissue | FABP5, UGDH, Visfatin, LRPPRC, PPA1 | Increased | The model proved superior accuracy to distinguish IBD patients from healthy controls (AUC = 1.0, 95% CI: 0.99–1.0; precision = 0.95, 95% CI: 0.86–1.0; sensitivity = 1.0, 95% CI: 0.83–1.0; specificity = 0.93, 95% CI: 0.78–0.99). | [51] |
| HADHB, LAP3, LTAH4, MT2, B2M, TRFC, SL25A1, ECH1, HNRNPH3 | Increased | The panel of 12 proteins differentiated patients with CD from UC (AUC = 0.95; 95% CI: 0.86–1.0; accuracy = 0.80; sensitivity = 1.0, 95% CI: 0.78–1.0; specificity = 0.93, 95% CI: 0.68–1). | ||||
| SEC61A1, SND1, TF | Decreased | |||||
| IBD | 83 patients (27 UC, 56 CD) versus 12 controls | Serum | GUC2A, CHGB | Increased | UC could be differentiated from CD by elevated levels of GUC2A (AUC = 0.80, specificity = 0.89, sensitivity = 0.67 p = 0.0006) and CHGB (AUC = 0.70, specificity = 0.78, sensitivity = 0.67, p = 0.008). | [52] |
| IBD | 43 patients (22 UC versus 21 CD) | Colonic tissue | ATP1A1, HIST | Increased | The variability of protein signatures in intestinal epithelial cells distinguishes between CD and UC. | [53] |
| MYH9, MVP, HIST1H2AC | Decreased | |||||
| IBD | 117 patients (57 UC, 60 CD) versus 31 controls | Feces | GSN, RhoGDI2 | Increased | The two proteins have a much better performance to differentiate CD patients from the control group GSN, (AUC = 0.998, sensitivity = 0.91, specificity = 1.0, p = 0.0009; RhoGDI2 (AUC = 1.0, sensitivity = 1.0, specificity = 1.0, p = 0.0004) compared to FC (AUC = 0.824, sensitivity = 0.86, specificity = 0.68). | [54] |
| RhoGDI2 | Increased | This protein has maximum precision (AUC = 1, sensitivity = 1.0, specificity = 1.0, p = 0.0009) for discriminating UC patients from healthy controls. | ||||
| IBD | 24 patients (12 UC, 12 CD) versus 9 controls | Colonic tissue | ALDOB, FABP2, ACE1, ACE2, S100A8, S100A9, MPO, LTF | Increased | Upregulation of protein expression was observed in CD compared to UC. | [50] |
| IBD | 121 patients (71 CD, 60 UC) versus 10 controls | Colonic tissue | KRT4, ELANE, S100A9, S100A8, CTSG, LTF, LYZ, IGHM, AKR1C3, AKR1C1 | Increased | Each of these proteins is expressed at levels at least three times higher in CD patients compared to UC. AKR1C3 and AKR1C1 were expressed exclusively in CD patients. | [55] |
| IBD | 76 patients (30 UC, 30 CD) versus 16 controls | Plasma | Resistin, Elastase | Increased | Circulating resistin is significantly increased in UC (AUC = 0.82, sensitivity = 0.77, specificity = 0.88) and CD (AUC = 0.77, sensitivity = 0.70, specificity = 0.88) (p < 0.01). Increased levels of elastase are detected in UC (AUC = 0.74, sensitivity = 0.57, specificity = 0.94). | [56] |
| IBD | 193 patients (118 UC, 75 CD) versus 32 controls | Serum | VICM, C3M, C4M | Increased | The combinations of proteins used for discrimination between CD from UC, and UC from martors are VICM, C3M, C4M (AUC = 0.86, specificity = 0.90, sensitivity = 0.75, accuracy = 0.79) and VICM, C3M (AUC = 0.98, specificity = 0.94, sensitivity = 0.96, accuracy = 0.95), respectively. | [57] |
| Disease | Number of Patients | Sample | Main Findings | References |
|---|---|---|---|---|
| UC | 10 patients versus 10 controls | Colonic tissue | Strong correlation between the severity of inflammatory lesions and the presence of specific tissue proteins (S100-A8, r = 0.84; S100-A12, r = 0.91; LF, r = 0.82). | [41] |
| UC | 64 patients versus 47 controls | Colonic tissue | MUC2 and SLC26A3 were significantly reduced in non-inflamed intestinal segments (p < 0.0001). The reduction in mucus-associated SLC26A3 levels was particularly pronounced in individuals in remission. | [66] |
| UC | 19 patients | Colonic tissue | Neutrophil-related proteins (MPO, ELANE, CD44, CD55, CYBA, CYBB, CAM1, ITGAM, ITGB2, and MMP9) correlate with GS scores (sensitivity: 72.7%, specificity: 100%) and RHI index (sensitivity: 75%, specificity: 81.8%) in active UC. | [65] |
| UC | 51 patients versus 17 controls | Colonic tissue | Up-regulation of the tissue proteins TRX (AUC = 0.91, 95% CI: 0.79–100; sensitivity = 86%, specificity = 85%, accuracy = 85%) and IGHA (AUC = 0.89, 95% CI: 0.75–1.00; sensitivity = 71%, specificity = 85%, accuracy = 80%) is predictive for early recurrence. | [68] |
| CD | 64 patients | Serum | A combination of three proteins (DSG1, DSP, and FABP5) released from transmural intestinal lesions is predictive of complications in CD (AUC = 0.777, sensitivity = 70.0%, specificity = 72.5%, p = 0.007). | [69] |
| CD | 20 patients | Serum | A panel of five serum glycoproteins (COMP, HGFA, POCE, Che, TNXB) showed 20% to 80% higher abundance in CD patients with stenotic complications compared to those with the inflammatory phenotype. | [62] |
| CD | 102 patients | Serum | Linked to elevated serum levels of lymphocyte-expressed proteins (LAG3, SH2B3, SIT1; HR: 2.2–4.5) and decreased concentrations of anti-inflammatory effectors (IL-10, HSD11B1; HR: 0.2–0.3) and cell junction proteins (CDSN, CNTNAP2, CXADR, ITGA11; HR: 0.4) is associated with long-term risk of relapse (p < 0.05). | [60] |
| CD | 30 patients versus 15 controls | Serum | The combination of three proteins (WDR3, LRG1, and SAA1) predicts progression in CD patients with a stricturing or penetrating phenotype (AUC = 0.737). | [37] |
| CD | 589 patients | Serum | The EHI includes 13 serum proteins (ANG1, ANG2, CRP, SAA1, IL7, EMMPRIN, MMP1, MMP2, MMP3, MMP9, TGFA, CEACAM1, and VCAM1) for predicting remission in CD patients (AUC = 0.962; sensitivity = 0.971; specificity = 0.690). | [58] |
| CD | 116 patients | Serum | ECM1 (HR = 3.41, 95% CI: 1.33–8.42; p < 0.001), IgA ASCA (HR = 4.99, CI 95%: 1.50–16.68) and CBir (HR = 5.19, CI 95%: 1.83–14.74) represents a predictive biomarkers for the development of colonic strictures in pediatric CD patients. | [70] |
| CD | 161 patients versus 40 controls | Plasma | Elevated COL3A1 and anti-CSF2 concentrations at the time of diagnosis are predictive for the occurrence of stenotic complications in pediatric patients (AUC = 0.8, CI 95%, 0.71–0.89; sensitivity = 0.7, CI 95%, 0.55–0.83; specificity = 0.83, CI 95%, 0.67–0.93). | [71] |
| CD | 265 patients | Serum | A model including five proteins (NFSF14, CCL4, IL15RA, TNFB, and CD40) expressed in ileal T cells and peripheral blood predicted the occurrence of penetrating complications more effectively (AUC = 0.79) compared to serological markers (LnASCA IgA, LnANCA, LnCbir) (AUC = 0.69) and clinical variables (AUC = 0.74). | [72] |
| CD | 73 patients versus 40 controls | Colonic tissue | The development of fibrotic strictures in CD patients is associated with the hypersecretion of BiP and AGR2 in colonic epithelium. | [73] |
| CD | 112 patients versus 24 controls | Serum | Serum levels of collagen formation and degradation products (P1NP, Pro-C3, Pro-C5, Pro-C6, C1M, C3M, C5M, and C6M) are higher in patients with active endoscopic inflammation. Pro-C3 and C3M present the greatest potential for differentiating penetrating vs. non-penetrating CD (AUC = 0.815, p < 0.001) and stricturing disease (AUC = 0.746, p = 0.002). | [74] |
| CD | 101 patients versus 96 controls | Serum | Increased degradation of collagen markers C1M, C3M, and C4G is significantly associated with the development of strictures (HR: 1.71; p < 0.05). Higher baseline concentrations of C1M and C4G were linked to an elevated risk of progression to the penetrating form of the disease (HR: 1.71; 95% CI: 1.05–2.81; p < 0.05). | [75] |
| IBD | 143 patients (39 UC, 104 CD) versus29 controls | Serum | ELP-3 is specific to UC with an active clinical phenotype (AUC = 0.870; sensitivity = 83.3%; specificity = 76.2%; p < 0.0001), while ELM-12 is significantly elevated in CD patients in endoscopic remission (AUC = 0.73; sensitivity = 94.4%; specificity = 51.2%; p = 0.001). | [76] |
| IBD | 117 patients (60 UC, 57 CD) versus 31 controls | Feces | Three fecal protein markers were significantly correlated with the severity of intestinal inflammation in CD (CTRC: r = 0.64, p < 0.001; GSN: r = 0.82, p < 0.001; RhoGDI2: r = 0.64, p < 0.001) and UC (CTRC: r = 0.76, p < 0.001; GSN: r = 0.75, p < 0.001; RhoGDI2: r = 0.63, p < 0.001). | [54] |
| IBD | 60 patients (30 UC, 30 CD) versus 29 controls | Colonic tissue | Visfatin and MT2 are significantly correlated with the severity of clinical progression in CD (r = 0.5186, p = 0.0025; r = 0.5975, p = 0.007, respectively), while in UC, an opposite relationship was observed with HNRNP-H3 (r = −0.2791, p = 0.035). | [51] |
| Disease | Medication | Proteins | Sample | Main Findings | References |
|---|---|---|---|---|---|
| UC | IFX | TNC, CCL2 | Serum Colonic tissue | A favorable response is associated with downregulation in tissue levels of TNC and serum expression of CCL2. | [85] |
| UC | IFX | ACTBL2, MBL2, BPI, EIF3D, CR1 | Colonic tissue | Potential biomarkers of non-response to IFX therapy. | [92] |
| UC | VDZ | OC | Serum | Expression is increased in responders (sensitivity 85%, specificity 100%). | [93] |
| UC | VDZ | s-α4β7, s-TNF, s-MAdCAM-1, s-VCAM-1, s-ICAM-1 | Serum | An increase in serum s-α4β7 levels accompanied by a decrease in s-MAdCAM-1, s-VCAM-1, s-ICAM-1, and s-TNF concentrations was observed in patients with endoscopic remission. | [94] |
| UC | IFX ADA | NGAL-MMP-9, CHI3L1, CRP, LL-37 | Serum | A significant reduction in serum proteins and neutrophil count (UCRI index) accurately detects MH, after IFX (AUC = 0.83) and ADA (AUC = 0.79). | [95] |
| CD | IFX ADA | C4M | Serum | Patients with elevated baseline serum levels of C4M do not respond to IFX (OR = 39; sensitivity 0.93, specificity 0.75, p = 0.02) or ADA (OR = 26; sensitivity 0.93, specificity 0.67, p = 0.01). | [74] |
| CD | IFX | PF4 | Serum | Higher levels were found in non-responders. | [84] |
| CD | IFX | PRO-C3, PRO-C6, C4M | Serum | C4M discriminates CD patients with a history of surgery into responders versus non-responders before IFX treatment (AUC = 0.84; p = 0.016). PRO-C3 and PRO-C6-used for monitoring therapeutic efficacy (AUC = 0.95; p = 0.004, and AUC = 0.82; p = 0.037). | [89] |
| CD | VDZ | sCD40L | Serum | An increase in serum concentration is predictive of therapeutic non-response (sensitivity 100%, specificity 100%). | [93] |
| CD | IFX | MMP3, CRP, CCL2 | Serum | The combined model measured at week 2 of treatment proves excellent performance (AUC = 0.898) in predicting primary non-response. | [94] |
| CD | VDZ | s-MAdCAM-1, s-VCAM-1, s-ICAM-1, s-α4β7 | Serum | Increased levels of s-ICAM-1 and s-VCAM-1 are predictive of endoscopic remission. In responders, a significant reduction in serum MAdCAM-1 concentration, while s-α4β7 levels are increased. | [96,97] |
| CD | VDZ | C1M, CPa9-HNE, C6Ma3, C3M, C4M, PRO-C3, PRO-C4 | Serum | Serological biomarkers of extracellular matrix turnover and neutrophil activity have significantly increased baseline concentrations in non-responders. | [98] |
| CD | IFX | VTDB,A1BG, C1R, A2GL | Serum | Identification of proteins with increased abundance in infliximab-induced clinical and serological remission compared to baseline samples. | [83] |
| APOA1, CLUS, APOE, APOH, CO4B, TRFE, PLMN | Increased serum expression of these proteins in non-responding patients. | ||||
| IBD | IFX ADA | ITGAV, IL-8, IL-18, EpCAM, SLAMF7 | Serum | The overexpression is predictive of biologic therapy escalation or the necessity for surgical intervention (HR = 3.9; 95% CI: 2.43–6.26). | [43] |
| IBD | IFX | TNC | Colonic tissue | Increased expression of TNC in inflamed intestinal mucosa was associated with a reduced response to IFX therapy. | [50] |
| IBD | IFX | IL-8, HGF, 4E-BP1, MCP-3, MMP-10, OSM, TGF-α | Serum | Non-responding patients—elevated baseline serum concentrations of seven proteins at the initiation of induction therapy. | [81] |
| IBD | IFX ADA | MMP3, MMP12 | Serum | Serum levels of endogenous IgG cleaved by MMP3 and MMP12 were higher in non-responders. | [82] |
| IBD | IFX CS | SERPINA1, CCL23, IGFBP1, IGFBP2, RETNi | Serum | These proteins with inflammatory functions showed significant reductions post-therapy. | [99] |
| IBD | VDZ | α4β7 | Serum | Higher expression of α4β7 on T effector memory cells and NK cells is predictive of a favorable response. | [100] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minea, H.; Singeap, A.-M.; Minea, M.; Juncu, S.; Chiriac, S.A.; Sfarti, C.V.; Stanciu, C.; Trifan, A. Integrating Proteomics into Personalized Medicine for Inflammatory Bowel Disease—Reality or Challenge? Int. J. Mol. Sci. 2025, 26, 4993. https://doi.org/10.3390/ijms26114993
Minea H, Singeap A-M, Minea M, Juncu S, Chiriac SA, Sfarti CV, Stanciu C, Trifan A. Integrating Proteomics into Personalized Medicine for Inflammatory Bowel Disease—Reality or Challenge? International Journal of Molecular Sciences. 2025; 26(11):4993. https://doi.org/10.3390/ijms26114993
Chicago/Turabian StyleMinea, Horia, Ana-Maria Singeap, Manuela Minea, Simona Juncu, Stefan Andrei Chiriac, Catalin Victor Sfarti, Carol Stanciu, and Anca Trifan. 2025. "Integrating Proteomics into Personalized Medicine for Inflammatory Bowel Disease—Reality or Challenge?" International Journal of Molecular Sciences 26, no. 11: 4993. https://doi.org/10.3390/ijms26114993
APA StyleMinea, H., Singeap, A.-M., Minea, M., Juncu, S., Chiriac, S. A., Sfarti, C. V., Stanciu, C., & Trifan, A. (2025). Integrating Proteomics into Personalized Medicine for Inflammatory Bowel Disease—Reality or Challenge? International Journal of Molecular Sciences, 26(11), 4993. https://doi.org/10.3390/ijms26114993








