Association of Functional Gene Variants in DYSF–ZNF638, MTSS1 and Ferroptosis-Related Genes with Multiple Sclerosis Severity and Target Gene Expression
Abstract
1. Introduction
2. Results
2.1. Genetic Association Analysis of Selected Variants with MS Disease Severity
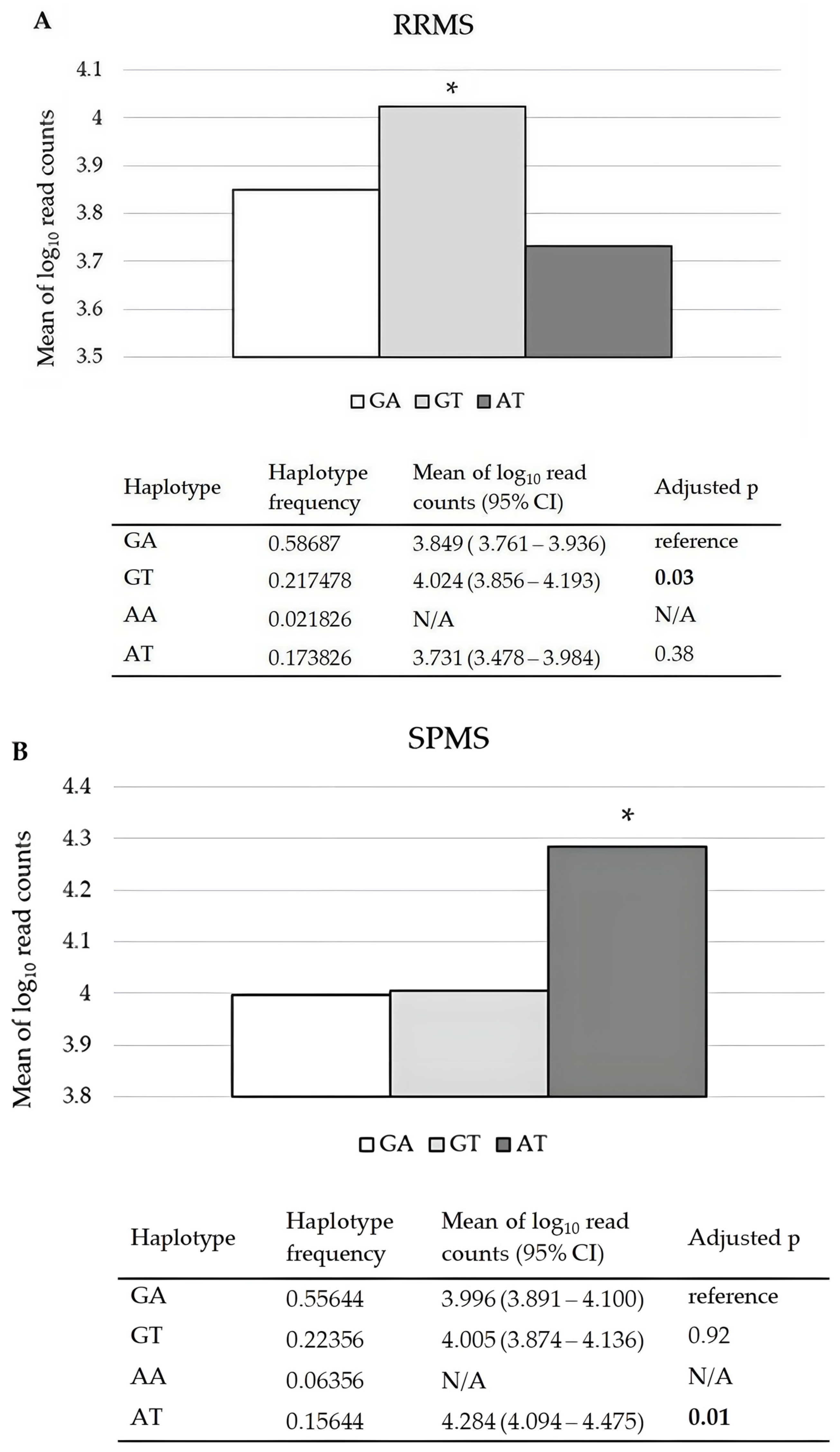
2.2. Effects of CDKN1A rs3176326 and rs3176336, RAB4B-EGLN2 rs111833532, and MAP1B rs62363242 and rs1217817 on Their mRNA Relative Expression Levels in PBMCs of RRMS and SPMS Patients
2.3. Associations of Investigated Gene Variants with MS Neurological Deficit and Severity Parameters (EDSS, MSSS, gARMSS)
2.4. Association of Investigated Gene Variants with Circulatory Molecular Indicators of Processes Associated with Ferroptosis: Lipid Peroxidation (MDA, 4-HNE and HEL), GSH-Related Antioxidant Defense (GSH, GSSG and GPX4) and Iron Metabolism (Free Iron, Transferrin and Ferritin), in MS Patients
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Selection of Genes and Gene Variants
4.3. Genetic Analysis
4.4. Isolation of PBMCs, Extraction of the Total RNA, Targeted RNASeq Library Synthesis and Targeted RNA Sequencing
4.5. Quantification of MDA, 4-HNE, GPX4 and Glutathione in Plasma
4.6. Quantification of Hexanoyl-Lys Adduct (HEL), Iron, Transferrin and Ferritin in Serum
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MS | multiple sclerosis |
| HLA | human leukocyte antigen |
| DEG | differentially expressed genes |
| RR | relapsing–remitting |
| SP | secondary progressive |
| P | progressive |
| CDKN1A | cyclin dependent kinase inhibitor 1A |
| EGLN2 | egl-9 family hypoxia inducible factor 2 |
| MAP1B | microtubule associated protein 1B |
| eQTL | expression quantitative trait locus |
| EDSS | Expanded Disability Status Scale |
| MSSS | Multiple Sclerosis Severity Score |
| gARMSS | Age-related Global MS Severity Score |
| CI | confidence interval |
| N/A | not applicable |
| SE | standard error |
| 4-HNE | 4-Hydroxynonenal |
| HEL | hexanoyl-lys adduct |
| MDA | Malondialdehyde |
References
- International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 2019, 365, eaav7188. [Google Scholar] [CrossRef] [PubMed]
- Jokubaitis, V.G.; Campagna, M.P.; Ibrahim, O.; Stankovich, J.; Kleinova, P.; Matesanz, F.; Hui, D.; Eichau, S.; Slee, M.; Lechner-Scott, J.; et al. Not all roads lead to the immune system: The genetic basis of multiple sclerosis severity. Brain 2023, 146, 2316–2331. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, T.; Moccia, M.; Coetzee, T.; Cohen, J.A.; Correale, J.; Graves, J.; Marrie, R.A.; Montalban, X.; Yong, V.W.; Thompson, A.J.; et al. International Advisory Committee on Clinical Trials in Multiple Sclerosis. Multiple sclerosis progression: Time for a new mechanism-driven framework. Lancet Neurol. 2023, 22, 78–88. [Google Scholar] [CrossRef] [PubMed]
- International Multiple Sclerosis Genetics Consortium; Wellcome Trust Case Control Consortium 2; Sawcer, S.; Hellenthal, G.; Pirinen, M.; Spencer, C.C.; Patsopoulos, N.A.; Moutsianas, L.; Dilthey, A.; Su, Z.; et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011, 476, 214–219. [Google Scholar] [CrossRef]
- Baranzini, S.E.; Wang, J.; Gibson, R.A.; Galwey, N.; Naegelin, Y.; Barkhof, F.; Radue, E.W.; Lindberg, R.L.; Uitdehaag, B.M.; Johnson, M.R.; et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum. Mol. Genet. 2009, 18, 767–778. [Google Scholar] [CrossRef]
- International Multiple Sclerosis Genetics Consortium; MultipleMS Consortium. Locus for severity implicates CNS resilience in progression of multiple sclerosis. Nature 2023, 619, 323–331. [Google Scholar] [CrossRef]
- Campagna, M.P.; Havrdova, E.K.; Horakova, D.; Izquierdo, G.; Matesanz, F.; Eichau, S.; Lechner-Scott, J.; Taylor, B.V.; García-Sanchéz, M.I.; Alcina, A.; et al. No evidence for association between rs10191329 severity locus and longitudinal disease severity in 1813 relapse-onset multiple sclerosis patients from the MSBase registry. Mult. Scler. J. 2024, 30, 1216–1220. [Google Scholar] [CrossRef]
- Kreft, K.L.; Uzochukwu, E.; Loveless, S.; Willis, M.; Wynford-Thomas, R.; Harding, K.E.; Holmans, P.; Lawton, M.; Tallantyre, E.C.; Robertson, N.P. Relevance of Multiple Sclerosis Severity Genotype in Predicting Disease Course: A Real-World Cohort. Ann. Neurol. 2024, 95, 459–470. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Jiang, X. The Chemistry and Biology of Ferroptosis. Cell Chem. Biol. 2020, 27, 365–375. [Google Scholar] [CrossRef]
- Viktorinova, A. Future Perspectives of Oxytosis/Ferroptosis Research in Neurodegeneration Diseases. Cell. Mol. Neurobiol. 2023, 43, 2761–2768. [Google Scholar] [CrossRef]
- Ryan, S.K.; Zelic, M.; Han, Y.; Teeple, E.; Chen, L.; Sadeghi, M.; Shankara, S.; Guo, L.; Li, C.; Pontarelli, F.; et al. Microglia ferroptosis is regulated by SEC24B and contributes to neurodegeneration. Nat. Neurosci. 2023, 26, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Stojkovic, L.; Jovanovic, I.; Dincic, E.; Djordjevic, A.; Kuveljic, J.; Djuric, T.; Stankovic, A.; Vojinovic, S.; Zivkovic, M. Targeted RNAseq Revealed the Gene Expression Signature of Ferroptosis-Related Processes Associated with Disease Severity in Patients with Multiple Sclerosis. Int. J. Mol. Sci. 2024, 25, 3016. [Google Scholar] [CrossRef]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Avila, J.; Domínguez, J.; Díaz-Nido, J. Regulation of microtubule dynamics by microtubule-associated protein expression and phosphorylation during neuronal development. Int. J. Dev. Biol. 1994, 38, 13–25. [Google Scholar]
- Black, M.M.; Slaughter, T.; Fischer, I. Microtubule-associated protein 1b (MAP1b) is concentrated in the distal region of growing axons. J. Neurosci. 1994, 14, 857–870. [Google Scholar] [CrossRef]
- Wu, H.Y.; Dawson, M.R.; Reynolds, R.; Hardy, R.J. Expression of QKI proteins and MAP1B identifies actively myelinating oligodendrocytes in adult rat brain. Mol. Cell. Neurosci. 2001, 17, 292–302. [Google Scholar] [CrossRef]
- Bouquet, C.; Soares, S.; von Boxberg, Y.; Ravaille-Veron, M.; Propst, F.; Nothias, F. Microtubule-associated protein 1B controls directionality of growth cone migration and axonal branching in regeneration of adult dorsal root ganglia neurons. J. Neurosci. 2004, 24, 7204–7213. [Google Scholar] [CrossRef]
- Meixner, A.; Haverkamp, S.; Wässle, H.; Führer, S.; Thalhammer, J.; Kropf, N.; Bittner, R.E.; Lassmann, H.; Wiche, G.; Propst, F. MAP1B is required for axon guidance and Is involved in the development of the central and peripheral nervous system. J. Cell Biol. 2000, 151, 1169–1178. [Google Scholar] [CrossRef]
- Allen, E.; Ding, J.; Wang, W.; Pramanik, S.; Chou, J.; Yau, V.; Yang, Y. Gigaxonin-controlled degradation of MAP1B light chain is critical to neuronal survival. Nature 2005, 438, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Uchida, Y. Overexpression of full-length but not N-terminal truncated isoform of microtubule-associated protein (MAP) 1B accelerates apoptosis of cultured cortical neurons. J. Biol. Chem. 2003, 278, 366–371. [Google Scholar] [CrossRef] [PubMed]
- McArdle, H.J.; Gambling, L.; Kennedy, C. Iron deficiency during pregnancy: The consequences for placental function and fetal outcome. Proc. Nutr. Soc. 2014, 73, 9–15. [Google Scholar] [CrossRef]
- Zierfuss, B.; Wang, Z.; Jackson, A.N.; Moezzi, D.; Yong, V.W. Iron in multiple sclerosis—Neuropathology, immunology, and real-world considerations. Mult. Scler. Relat. Disord. 2023, 78, 104934. [Google Scholar] [CrossRef]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef]
- Kreis, N.N.; Louwen, F.; Yuan, J. The Multifaceted p21 (Cip1/Waf1/CDKN1A) in Cell Differentiation, Migration and Cancer Therapy. Cancers 2019, 11, 1220. [Google Scholar] [CrossRef]
- van den Bosch, A.M.R.; van der Poel, M.; Fransen, N.L.; Vincenten, M.C.J.; Bobeldijk, A.M.; Jongejan, A.; Engelenburg, H.J.; Moerland, P.D.; Smolders, J.; Huitinga, I.; et al. Profiling of microglia nodules in multiple sclerosis reveals propensity for lesion formation. Nat. Commun. 2024, 15, 1667. [Google Scholar] [CrossRef]
- Distéfano-Gagné, F.; Bitarafan, S.; Lacroix, S.; Gosselin, D. Roles and regulation of microglia activity in multiple sclerosis: Insights from animal models. Nat. Rev. Neurosci. 2023, 24, 397–415. [Google Scholar] [CrossRef]
- dos Santos, L.S.M.; Trombetta-Lima, M.; Eggen, B.J.L.; Demaria, M. Cellular senescence in brain aging and neurodegeneration. Ageing Res. Rev. 2024, 93, 102141. [Google Scholar] [CrossRef]
- Rivellini, C.; Porrello, E.; Dina, G.; Mrakic-Sposta, S.; Vezzoli, A.; Bacigaluppi, M.; Gullotta, G.S.; Chaabane, L.; Leocani, L.; Marenna, S.; et al. JAB1 deletion in oligodendrocytes causes senescence-induced inflammation and neurodegeneration in mice. J. Clin. Investig. 2022, 132, e145071. [Google Scholar] [CrossRef]
- Karlebach, G.; Steinhaus, R.; Danis, D.; Devoucoux, M.; Anczuków, O.; Sheynkman, G.; Seelow, D.; Robinson, P.N. Alternative splicing is coupled to gene expression in a subset of variably expressed genes. npj Genom. Med. 2024, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Richardson, T.G.; Shihab, H.A.; Hemani, G.; Zheng, J.; Hannon, E.; Mill, J.; Carnero-Montoro, E.; Bell, J.T.; Lyttleton, O.; McArdle, W.L.; et al. Collapsed methylation quantitative trait loci analysis for low frequency and rare variants. Hum. Mol. Genet. 2016, 25, 4339–4349. [Google Scholar] [CrossRef]
- Gaunt, T.R.; Shihab, H.A.; Hemani, G.; Min, J.L.; Woodward, G.; Lyttleton, O.; Zheng, J.; Duggirala, A.; McArdle, W.L.; Ho, K.; et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016, 17, 61. [Google Scholar] [CrossRef]
- Kular, L.; Liu, Y.; Ruhrmann, S.; Zheleznyakova, G.; Marabita, F.; Gomez-Cabrero, D.; James, T.; Ewing, E.; Lindén, M.; Górnikiewicz, B.; et al. DNA methylation as a mediator of HLA-DRB1*15:01 and a protective variant in multiple sclerosis. Nat. Commun. 2018, 9, 2397. [Google Scholar] [CrossRef]
- Xavier, A.; Maltby, V.E.; Ewing, E.; Campagna, M.P.; Burnard, S.M.; Tegner, J.N.; Slee, M.; Butzkueven, H.; Kockum, I.; Kular, L.; et al. DNA Methylation Signatures of Multiple Sclerosis Occur Independently of Known Genetic Risk and Are Primarily Attributed to B Cells and Monocytes. Int. J. Mol. Sci. 2023, 24, 12576. [Google Scholar] [CrossRef]
- Kwong, A.; Boughton, A.P.; Wang, M.; VandeHaar, P.; Boehnke, M.; Abecasis, G.; Kang, H.M. FIVEx: An interactive eQTL browser across public datasets. Bioinformatics 2022, 38, 559–561. [Google Scholar] [CrossRef]
- Manouchehrinia, A.; Westerlind, H.; Kingwell, E.; Zhu, F.; Carruthers, R.; Ramanujam, R.; Ban, M.; Glaser, A.; Sawcer, S.; Tremlett, H.; et al. Age Related Multiple Sclerosis Severity Score: Disability ranked by age. Mult. Scler. J. 2017, 23, 1938–1946. [Google Scholar] [CrossRef]
- Tang, D.; Chen, X.; Kang, R.; Kroemer, G. Ferroptosis: Molecular mechanisms and health implications. Cell Res. 2021, 31, 107–125. [Google Scholar] [CrossRef]
- Stojkovic, L.; Djordjevic, A.; Stefanovic, M.; Stankovic, A.; Dincic, E.; Djuric, T.; Zivkovic, M. Circulatory Indicators of Lipid Peroxidation, the Driver of Ferroptosis, Reflect Differences between Relapsing-Remitting and Progressive Multiple Sclerosis. Int. J. Mol. Sci. 2024, 25, 11024. [Google Scholar] [CrossRef]
- Van San, E.; Debruyne, A.C.; Veeckmans, G.; Tyurina, Y.Y.; Tyurin, V.A.; Zheng, H.; Choi, S.M.; Augustyns, K.; van Loo, G.; Michalke, B.; et al. Ferroptosis contributes to multiple sclerosis and its pharmacological targeting suppresses experimental disease progression. Cell Death Differ. 2023, 30, 2092–2103. [Google Scholar] [CrossRef]
- Perluigi, M.; Coccia, R.; Butterfield, D.A. 4-Hydroxy-2-nonenal, a reactive product of lipid peroxidation, and neurodegenerative diseases: A toxic combination illuminated by redox proteomics studies. Antioxid. Redox Signal. 2012, 17, 1590–1609. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, L.; Velmeshev, D.; Holmqvist, S.; Kaufmann, M.; Werneburg, S.; Jung, D.; Vistnes, S.; Stockley, J.H.; Young, A.; Steindel, M.; et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 2019, 573, 75–82. [Google Scholar] [CrossRef]
- Absinta, M.; Lassmann, H.; Trapp, B.D. Mechanisms underlying progression in multiple sclerosis. Curr. Opin. Neurol. 2020, 33, 277–285. [Google Scholar] [CrossRef]
- Gasperi, C.; Wiltgen, T.; McGinnis, J.; Cerri, S.; Moridi, T.; Ouellette, R.; Pukaj, A.; Voon, C.; Bafligil, C.; Lauerer, M.; et al. A Genetic Risk Variant for Multiple Sclerosis Severity is Associated with Brain Atrophy. Ann. Neurol. 2023, 94, 1080–1085. [Google Scholar] [CrossRef]
- Schmiedel, B.J.; Singh, D.; Madrigal, A.; Valdovino-Gonzalez, A.G.; White, B.M.; Zapardiel-Gonzalo, J.; Ha, B.; Altay, G.; Greenbaum, J.A.; McVicker, G.; et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 2018, 175, 1701–1715.e16. [Google Scholar] [CrossRef]
- Belinky, F.; Nativ, N.; Stelzer, G.; Zimmerman, S.; Iny Stein, T.; Safran, M.; Lancet, D. PathCards: Multi-source consolidation of human biological pathways. Database 2015, 2015, bav006. [Google Scholar] [CrossRef]
- Wang, P.; Xie, K.; Wang, C.; Bi, J. Oxidative stress induced by lipid peroxidation is related with inflammation of demyelination and neurodegeneration in multiple sclerosis. Eur. Neurol. 2014, 72, 249–254. [Google Scholar] [CrossRef]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Polman, C.H.; Reingold, S.C.; Banwell, B.; Clanet, M.; Cohen, J.A.; Filippi, M.; Fujihara, K.; Havrdova, E.; Hutchinson, M.; Kappos, L.; et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 2011, 69, 292–302. [Google Scholar] [CrossRef]
- Polman, C.H.; Reingold, S.C.; Edan, G.; Filippi, M.; Hartung, H.P.; Kappos, L.; Lublin, F.D.; Metz, L.M.; McFarland, H.F.; O’Connor, P.W.; et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann. Neurol. 2005, 58, 840–846. [Google Scholar] [CrossRef]
- Lorscheider, J.; Buzzard, K.; Jokubaitis, V.; Spelman, T.; Havrdova, E.; Horakova, D.; Trojano, M.; Izquierdo, G.; Girard, M.; on behalf of the MSBase Study Group; et al. Defining secondary progressive multiple sclerosis. Brain 2016, 139 Pt 9, 2395–2405. [Google Scholar] [CrossRef] [PubMed]
- Lublin, F.D.; Reingold, S.C.; Cohen, J.A.; Cutter, G.R.; Sørensen, P.S.; Thompson, A.J.; Wolinsky, J.S.; Balcer, L.J.; Banwell, B.; Barkhof, F.; et al. Defining the clinical course of multiple sclerosis: The 2013 revisions. Neurology 2014, 83, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Kurtzke, J.F. Rating neurologic impairment in multiple sclerosis: An expanded disability status scale (EDSS). Neurology 1983, 33, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Roxburgh, R.H.; Seaman, S.R.; Masterman, T.; Hensiek, A.E.; Sawcer, S.J.; Vukusic, S.; Achiti, I.; Confavreux, C.; Coustans, M.; le Page, E.; et al. Multiple Sclerosis Severity Score: Using disability and disease duration to rate disease severity. Neurology 2005, 64, 1144–1151. [Google Scholar] [CrossRef]
- The ARRMS Score. Available online: https://aliman.shinyapps.io/ARMSS/ (accessed on 15 January 2024).
- GnomAD. Available online: https://gnomad.broadinstitute.org/ (accessed on 9 January 2024).
- FIVEx. Available online: https://fivex.sph.umich.edu/ (accessed on 9 January 2024).
- GTEx. Available online: https://gtexportal.org (accessed on 9 January 2024).
- RegulomeDB. Available online: https://regulome.stanford.edu/regulome-search (accessed on 9 January 2024).
- The GWAS Catalogue. Available online: https://www.ebi.ac.uk/gwas (accessed on 9 January 2024).
- HaploReg. Available online: https://pubs.broadinstitute.org/ (accessed on 9 January 2024).
- Dupont, W.D.; Plummer, W.D., Jr. Power and sample size calculations: A review and computer program. Control. Clin. Trials 1990, 11, 116–128. [Google Scholar] [CrossRef]
- Tregouet, D.A.; Garelle, V. A new JAVA interface implementation of THESIAS: Testing haplotype effects in association studies. Bioinformatics 2007, 23, 1038–1039. [Google Scholar] [CrossRef]
| RRMS n = 604 | PMS n = 241 | p | |
|---|---|---|---|
| Age, years | 38.86 ± 10.41 | 47.27 ± 9.93 | <0.01 a |
| Sex, f/m, % | 0.61/0.39 | 0.64/0.36 | 0.53 b |
| Disease duration, years | 7.73 ± 5.83 | 14.07 ± 9.44 | <0.01 c |
| EDSS | 2.47 ± 1.32 | 5.59 ± 1.56 | <0.01 c |
| MSSS | 3.91 ± 2.35 | 6.52 ± 2.03 | <0.01 c |
| gARMSS | 4.58 ± 2.23 | 7.17 ± 2.01 | <0.01 c |
| Gene/Locus | Gene Variant | RRMS, n (%) n = 604 | PMS, n (%) n = 241 | p |
|---|---|---|---|---|
| CDKN1A | rs3176326 | |||
| GG | 375 (0.62) | 152 (0.63) | ||
| GA | 193 (0.32) | 77 (0.32) | 0.82 | |
| AA | 36 (0.06) | 12 (0.05) | ||
| allele G/A | 0.78/0.22 | 0.79/0.21 | 0.65 | |
| CDKN1A | rs3176336 | |||
| AA | 187 (0.31) | 70 (0.29) | ||
| AT | 290 (0.48) | 125 (0.52) | 0.51 | |
| TT | 127 (0.21) | 46 (0.19) | ||
| allele A/T | 0.55/0.45 | 0.55/0.45 | 1 | |
| RAB4B-EGLN2 | rs111833532 | |||
| II | 191 (0.32) | 75 (0.31) | ||
| ID | 296(0.49) | 130 (0.54) | 0.25 | |
| DD | 117 (0.19) | 36 (0.15) | ||
| allele I/D | 0.56/0.44 | 0.58/0.42 | 0.46 | |
| DYSF–ZNF638 | rs10191329 * | |||
| CC | 453 (0.75) | 171 (0.71) | 0.27 | |
| CA +AA | 151 (0.25) | 70 (0.29) | ||
| allele C/A | 0.86/0.14 | 0.86/0.14 | 1 | |
| MTSS1 | rs9643199 | |||
| GG | 321 (0.53) | 132 (0.55) | ||
| GA | 225 (0.37) | 91 (0.38) | 0.62 | |
| AA | 58 (0.10) | 18 (0.07) | ||
| allele G/A | 0.72/0.28 | 0.74/0.26 | 0.43 | |
| MAP1B | rs1217817 | |||
| GG | 78 (0.13) | 34 (0.14) | ||
| GA | 242 (0.40) | 101 (0.42) | 0.66 | |
| AA | 284 (0.47) | 106 (0.44) | ||
| allele G/A | 0.33/0.67 | 0.35/0.65 | 0.41 | |
| MAP1B | rs62363242 | |||
| GG | 284 (0.47) | 106 (0.44) | ||
| GA | 254 (0.42) | 111 (0.46) | 0.50 | |
| AA | 66 (0.11) | 24 (0.10) | ||
| allele G/A | 0.68/0.32 | 0.67/0.33 | 0.68 | |
| Females | ||||
| rs62363242 | RRMS, n (%) n = 367 | PMS, n (%) n = 155 | ||
| GG | 187 (0.51) | 64 (0.41) | 0.03 | |
| AG + AA | 179 (0.49) | 91 (0.59) | ||
| allele G/A | 0.71/0.29 | 0.65/0.35 | 0.03 | |
| Predictors | EDSS | MSSS | gARMSS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE Beta | p | Beta | SE Beta | p | Beta | SE Beta | p | ||
| Sex | −0.049 | 0.043 | 0.245 | −0.058 | 0.038 | 0.122 | −0.059 | 0.039 | 0.136 | |
| Disease duration | 0.134 | 0.043 | 0.002 | −0.466 | 0.038 | <0.001 | −0.213 | 0.04 | <0.001 | |
| Age at onset | 0.124 | 0.043 | 0.004 | 0.122 | 0.039 | <0.001 | −0.416 | 0.04 | <0.001 | |
| RAB4B-EGLN2 rs111833532 (II vs. ID + DD) | 0.094 | 0.043 | 0.028 | 0.074 | 0.038 | 0.05 | 0.093 | 0.039 | 0.018 | |
| HLA-DRB1*15:01 rs3135388 A allele-containing genotypes | 0.043 | 0.043 | 0.314 | 0.027 | 0.038 | 0.468 | 0.045 | 0.039 | 0.256 | |
| Predictors | EDSS | MSSS | gARMSS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta | SE Beta | p | Beta | SE Beta | p | Beta | SE Beta | p | |
| Disease duration | 0.181 | 0.113 | 0.116 | −0.491 | 0.11 | <0.001 | −0.234 | 0.093 | 0.014 |
| Age at onset | −0.207 | 0.116 | 0.08 | −0.18 | 0.113 | 0.115 | −0.699 | 0.095 | <0.001 |
| DYSF-ZNF638 rs10191329 CC vs. CA + AA | 0.233 | 0.105 | 0.03 | 0.168 | 0.104 | 0.111 | 0.18 | 0.086 | 0.04 |
| HLA-DRB1*15:01 rs3135388 A allele-containing genotypes | −0.199 | 0.106 | 0.065 | −0.159 | 0.104 | 0.131 | −0.13 | 0.087 | 0.141 |
| Product of Lipid Peroxidation | RAB4B-EGLN2 rs111833532 (PMS) | p | |
|---|---|---|---|
| II | ID + DD | ||
| 4-HNE (pg/mL) | 1299.98 ± 370.21 | 1938.05 ± 1540.60 | 0.04 |
| HLA-DRB1*15:01 (MS patients overall) | |||
| Without allele A | With allele A | ||
| HEL (nmol/L) | 12.51 ± 4.03 | 13.54 ± 3.63 | 0.018 |
| Iron metabolism | MAP1B rs62363242 (PMS) | ||
| GG | GA + AA | ||
| Iron (µmol/L) | 15.91 ± 4.14 | 13.66 ± 4.78 | 0.03 |
| Transferrin (g/L) | 2.27 ± 0.37 | 2.53 ± 0.44 | 0.03 |
| Ferritin (ng/mL) | 72.25 ± 64.47 | 58.38 ± 73.62 | 0.08 |
| GENE or LOCUS | rs ID Number | Allelic Change | Position | MAF | eQTL | RegulomeDB | GWAS | Variants in LD, n |
|---|---|---|---|---|---|---|---|---|
| CDKN1A | rs3176326 | A/G | Intronic | 0.2 | Yes | 1a | yes | 4 |
| Blood, LCL | CVD | |||||||
| rs3176336 | A/T | Intronic | 0.4 | Yes | 1f | yes | 2 | |
| CD8 T cells, CD4 T cells | CVD | |||||||
| EGLN2 | rs111833532 | TCTG/- | Intronic | 0.45 | Yes | 1f | No | None |
| Blood, T cells, B cells | ||||||||
| MAP1B | rs62363242 | G/A | 2.6 kb 3′ of MAP1B | 0.34 | Yes | 1f | No | 24 |
| Artery, brain, heart | ||||||||
| rs1217817 | A/G | 13 kb 5′ of MAP1B | 0.42 | Yes | 1f | No | 5 | |
| LCL, brain, aorta, CD4+, T cells | ||||||||
| DYSF–ZNF638 | rs10191329 | C/A | 3.9 kb 5′ of DYSF | 0.17 | Yes for | 1f | Yes | 2 |
| ZNF638 in blood, T cells, B cells | MS severity | |||||||
| MTSS1 | rs9643199 | A/G | Intronic | 0.26 | Yes | 4 | Yes | 1 |
| Brain, blood | MS severity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Djuric, T.; Djordjevic, A.; Kuveljic, J.; Stefanovic, M.; Dincic, E.; Kolakovic, A.; Zivkovic, M. Association of Functional Gene Variants in DYSF–ZNF638, MTSS1 and Ferroptosis-Related Genes with Multiple Sclerosis Severity and Target Gene Expression. Int. J. Mol. Sci. 2025, 26, 4986. https://doi.org/10.3390/ijms26114986
Djuric T, Djordjevic A, Kuveljic J, Stefanovic M, Dincic E, Kolakovic A, Zivkovic M. Association of Functional Gene Variants in DYSF–ZNF638, MTSS1 and Ferroptosis-Related Genes with Multiple Sclerosis Severity and Target Gene Expression. International Journal of Molecular Sciences. 2025; 26(11):4986. https://doi.org/10.3390/ijms26114986
Chicago/Turabian StyleDjuric, Tamara, Ana Djordjevic, Jovana Kuveljic, Milan Stefanovic, Evica Dincic, Ana Kolakovic, and Maja Zivkovic. 2025. "Association of Functional Gene Variants in DYSF–ZNF638, MTSS1 and Ferroptosis-Related Genes with Multiple Sclerosis Severity and Target Gene Expression" International Journal of Molecular Sciences 26, no. 11: 4986. https://doi.org/10.3390/ijms26114986
APA StyleDjuric, T., Djordjevic, A., Kuveljic, J., Stefanovic, M., Dincic, E., Kolakovic, A., & Zivkovic, M. (2025). Association of Functional Gene Variants in DYSF–ZNF638, MTSS1 and Ferroptosis-Related Genes with Multiple Sclerosis Severity and Target Gene Expression. International Journal of Molecular Sciences, 26(11), 4986. https://doi.org/10.3390/ijms26114986





