RNA Through Time: From the Origin of Life to Therapeutic Frontiers in Transcriptomics and Epitranscriptional Medicine
Abstract
1. RNA in the Origin of Life
- Replication and catalysis: RNA possesses the ability to self-replicate and catalyze chemical reactions, whereas DNA necessitates the presence of proteins for replication and catalysis [6].
- Structural flexibility: The single-stranded nature of RNA allows it to form complex three-dimensional structures, facilitating its ability to perform various functions including catalysis [7].
- Spontaneous synthesis: RNA nucleotides can be synthesized spontaneously in primordial environments, whereas DNA synthesis requires complex chemical pathways [2].
- Versatility: RNA functions as a genetic material, catalyst, and structural component, whereas DNA primarily serves as a repository for genetic information [8].
- Error tolerance: A higher mutation rate of RNA facilitates rapid evolution and adaptation, which is advantageous in primordial environments [4].
- Stability and Complexity: RNA is relatively unstable compared to DNA, and the complexity of forming functional ribozymes in a prebiotic environment is a significant challenge.
- Transition to the DNA/Protein World: The transition from an RNA-based world to one utilizing DNA and proteins raises questions about how such a shift occurred and what mechanisms facilitated it.
- Alternative Theories: Other hypotheses, such as the role of peptide nucleic acids (PNAs) or the involvement of other polymers, also explore possible pathways for the origin of life.
2. Technical Improvements in RNA Extraction
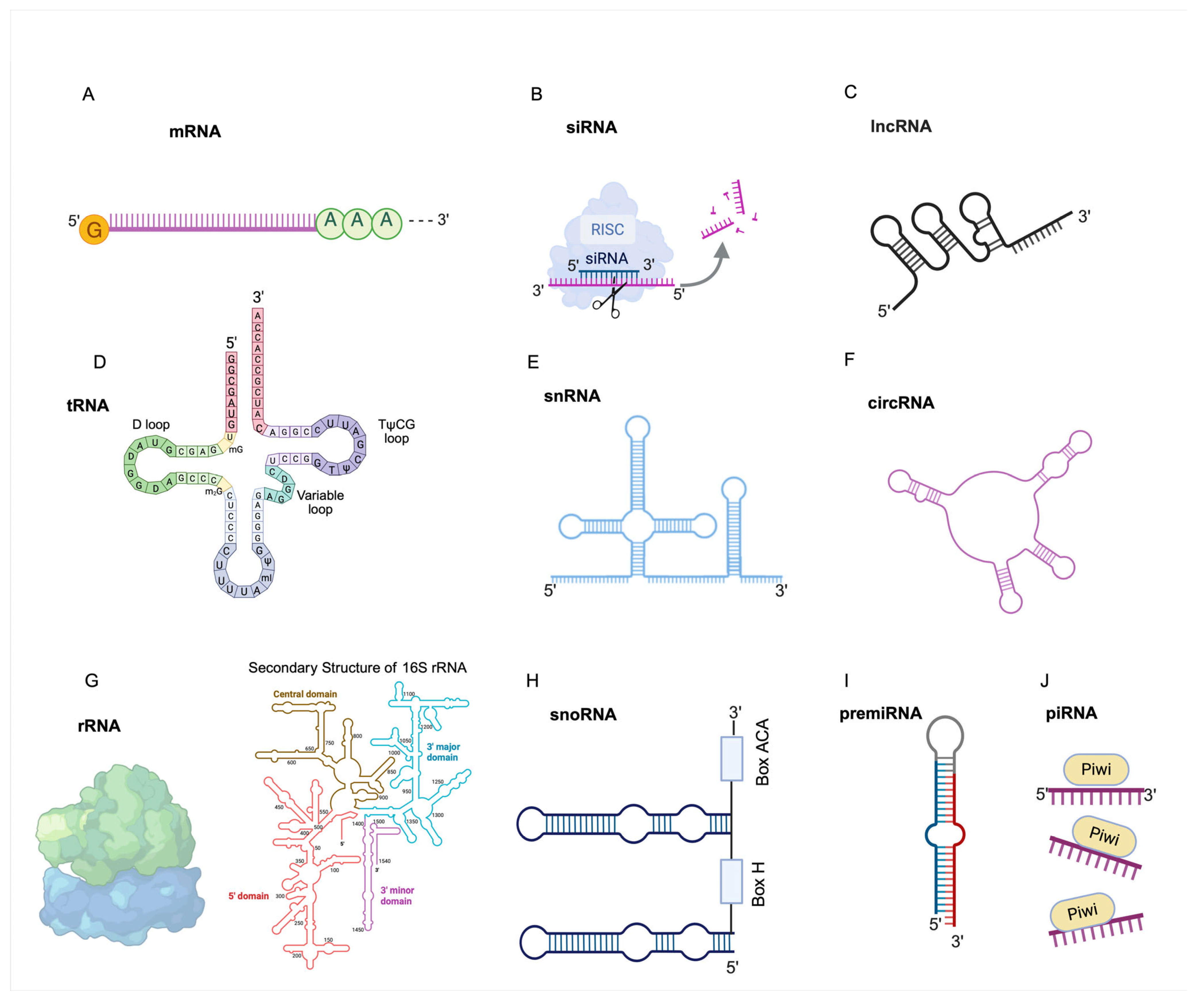
3. Transcriptome
- (A)
- Protein synthesis
- (B)
- Non-coding regulatory RNA
- (C)
- Small nucleolar RNA (snoRNA): These are small non-coding RNAs that are widely present in the nucleoli of eukaryotic cells [72]. SnoRNAs are mainly encoded by the intronic regions of both protein-coding and non-protein-coding genes [73,74,75,76]. SnoRNAs are generally classified into three main groups based on their structure and function. C/D box snoRNAs (SNORDs) are typically 60–90 nucleotides long and contain conserved C (RUGAUGA) and D (CUGA) motifs; H/ACA box snoRNAs (SNORAs) are usually 120–140 nucleotides in length and are defined by the presence of an H box (ANANNA) and a terminal ACA motif, and a third group, known as small Cajal body-specific RNAs (scaRNAs or SCARNAs), containing both C/D and H/ACA box motifs in diverse combinations [77,78,79].
- (D)
- Small nuclear RNAs (snRNA): These are a type of short RNA molecules found in the splicing speckles and Cajal bodies inside the nuclei of eukaryotic cells, which are synthesized by either RNA polymerase II (Pol II) or III [91,92,93,94,95]. Their primary role is to process heterogeneous nuclear RNA (hnRNA). Small nuclear RNAs (snRNAs) collaborate with specific proteins to form small nuclear ribonucleoproteins (snRNPs), which are essential components of the spliceosome. The core of the spliceosome is U-rich, U1 (164 nt), U2 (191nt), U4 (141nt), U5 (116 nt), and U6 (107 nt) snRNAs, each having a unique function in identifying and excising introns during the splicing process [96,97,98,99,100,101].
- (E)
- Long non-coding RNAs (long ncRNAs, lncRNA): These are a class of RNA transcripts longer than 200 nucleotides that exhibit little or no protein-coding potential [102]. These molecules can regulate gene expression through diverse mechanisms. Some lncRNAs act in cis, modulating the activity of nearby genes by recruiting chromatin-modifying complexes to specific genomic loci, thereby promoting either transcriptional activation or repression [103,104]. Others function in trans, influencing the expression of distant genes. A well-characterized example is HOTAIR (HOX transcript antisense RNA), which is transcribed from the HOXC locus but represses transcription at the HOXD locus through epigenetic silencing mechanisms [105]. Additionally, lncRNAs play structural roles in the nucleus, contributing to the organization of nuclear architecture and the formation of subnuclear domains such as paraspeckles and nuclear speckles, ultimately influencing gene expression [106,107]. Over 100,000 human lncRNAs have been identified [108,109,110]; however, the functions and biological significance of most of them remain largely uncharacterized.
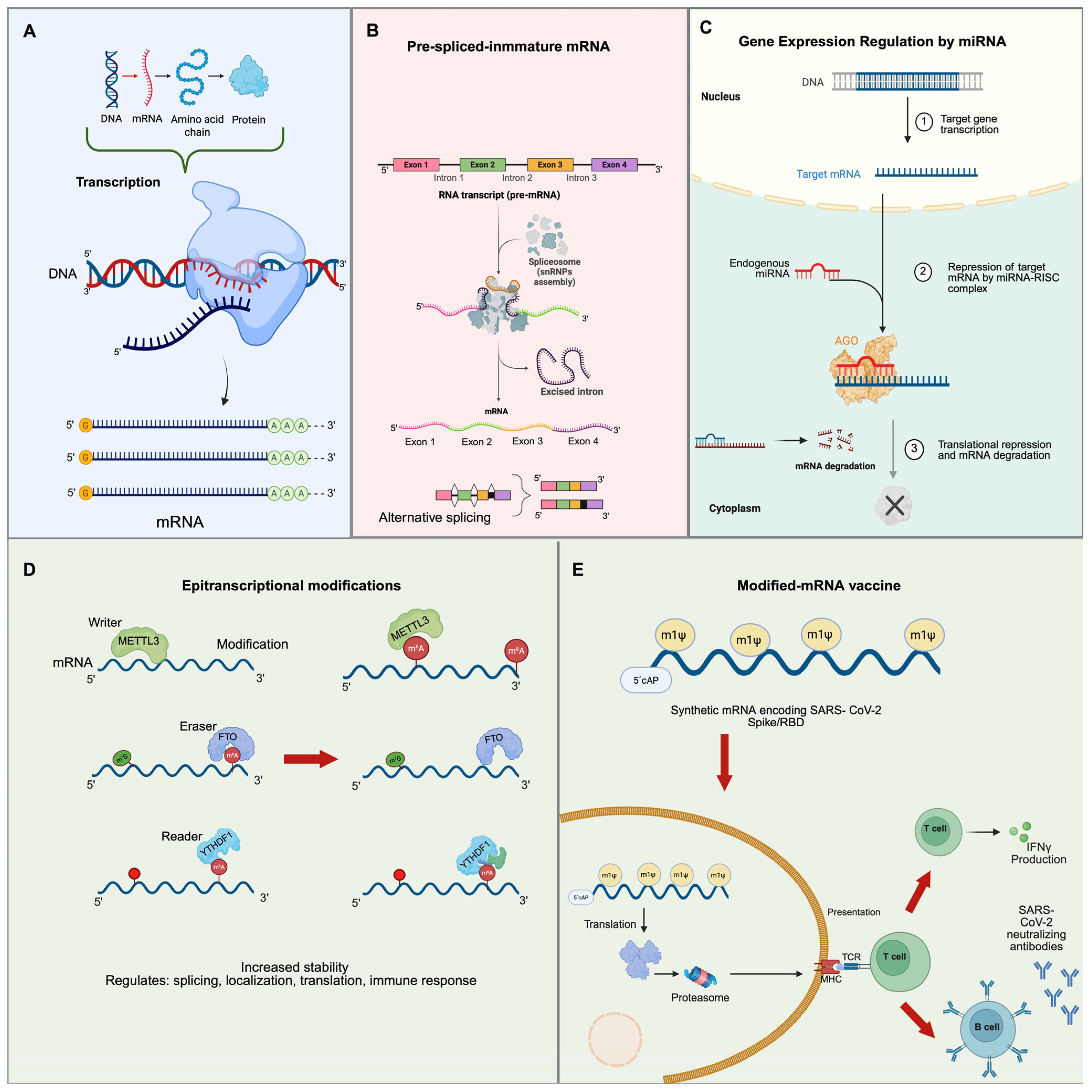
Splicing
4. MicroRNAs
4.1. Biogenesis of MicroRNAs
4.2. MicroRNAs as Therapeutic Agents
5. mRNA Vaccines
6. RNA Modifications (Epitranscriptional Modifications)
6.1. Writers
6.2. Erasers
6.3. Readers
6.4. Modified RNA and Immune Response
6.5. Modified mRNA Vaccines
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Joyce, G.F. RNA evolution and the origins of life. Nature 1989, 338, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Orgel, L.E. The origin of life—A review of facts and speculations. Trends Biochem. Sci. 1998, 23, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R. A model for the RNA-catalyzed replication of RNA. Proc. Natl. Acad. Sci. USA 1986, 83, 4360–4363. [Google Scholar] [CrossRef]
- Bartel, D.P.; Szostak, J.W. Isolation of new ribozymes from a large pool of random sequences. Science 1993, 261, 1411–1418. [Google Scholar] [CrossRef]
- Hager, A.J.; Szostak, J.W. Isolation of novel ribozymes that ligate AMP-activated RNA substrates. Chem. Biol. 1997, 4, 607–617. [Google Scholar] [CrossRef]
- Joyce, G.F. The antiquity of RNA-based evolution. Nature 2002, 418, 214–221. [Google Scholar] [CrossRef]
- Doudna, J.A.; Cech, T.R. The chemical repertoire of natural ribozymes. Nature 2002, 418, 222–228. [Google Scholar] [CrossRef]
- Atkins, J.F.; Gesteland, R.F.; Cech, T.R. The RNA World, 3rd ed.; I.K. International Publishing House Pvt. Limited: Delhi, India, 2006. [Google Scholar]
- Guerrier-Takada, C.; Altman, S. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science 1984, 223, 285–286. [Google Scholar] [CrossRef]
- Zaug, A.J.; Cech, T.R. The intervening sequence RNA of Tetrahymena is an enzyme. Science 1986, 231, 470–475. [Google Scholar] [CrossRef]
- Westheimer, F.H. Polyribonucleic acids as enzymes. Nature 1986, 319, 534–535. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Urey, H.C. On the Early Chemical History of the Earth and the Origin of Life. Proc. Natl. Acad. Sci. USA 1952, 38, 351–363. [Google Scholar] [CrossRef]
- Miller, S.L. A production of amino acids under possible primitive earth conditions. Science 1953, 117, 528–529. [Google Scholar] [CrossRef]
- Mills, D.R.; Peterson, R.L.; Spiegelman, S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc. Natl. Acad. Sci. USA 1967, 58, 217–224. [Google Scholar] [CrossRef]
- Chen, X.; Li, N.; Ellington, A.D. Ribozyme catalysis of metabolism in the RNA world. Chem. Biodivers. 2007, 4, 633–655. [Google Scholar] [CrossRef]
- Serganov, A.; Nudler, E. A decade of riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef]
- Paul, N.; Joyce, G.F. A self-replicating ligase ribozyme. Proc. Natl. Acad. Sci. USA 2002, 99, 12733–12740. [Google Scholar] [CrossRef]
- Forterre, P. The two ages of the RNA world, and the transition to the DNA world: A story of viruses and cells. Biochimie 2005, 87, 793–803. [Google Scholar] [CrossRef]
- Higgs, P.G.; Lehman, N. The RNA World: Molecular cooperation at the origins of life. Nat. Rev. Genet. 2015, 16, 7–17. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Ousset, M.J.; Pianciola, L.A.; Mazzeo, M.; Oteiza, J.M.; Jaureguiberry, M.S.; Venturino, A.; Barril, P.A. Improved SARS-CoV-2 RNA recovery in wastewater matrices using a CTAB-based extraction method. J. Virol. Methods 2024, 327, 114918. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Valdez, H.; Madrid-Marina, V.; Cohen, A. Phorbol esters and cAMP differentially regulate the expression of CD4 and CD8 in human thymocytes. BMC Immunol. 2002, 3, 1. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ostankovitch, M. Dynamic mechanisms in the life cycle of an RNA molecule. J. Mol. Biol. 2013, 425, 3747–3749. [Google Scholar] [CrossRef]
- Brenner, S.; Jacob, F.; Meselson, M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 1961, 190, 576–581. [Google Scholar] [CrossRef]
- Gros, F.; Hiatt, H.; Gilbert, W.; Kurland, C.G.; Risebrough, R.W.; Watson, J.D. Unstable ribonucleic acid revealed by pulse labelling of Escherichia coli. Nature 1961, 190, 581–585. [Google Scholar] [CrossRef]
- Nirenberg, M.W.; Matthaei, J.H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. USA 1961, 47, 1588–1602. [Google Scholar] [CrossRef]
- Perry, R.P.; Kelley, D.E. Existence of methylated messenger RNA in mouse L cells. Cell 1974, 1, 37–42. [Google Scholar] [CrossRef]
- Perry, R.P.; Kelley, D.E. Kinetics of formation of 5′ terminal caps in mRNA. Cell 1976, 8, 433–442. [Google Scholar] [CrossRef]
- Proudfoot, N.J.; Brownlee, G.G. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature 1976, 263, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Berget, S.M.; Moore, C.; Sharp, P.A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA 1977, 74, 3171–3175. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.T.; Gelinas, R.E.; Broker, T.R.; Roberts, R.J. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 1977, 12, 1–8. [Google Scholar] [CrossRef]
- Chen, S.; Jiang, Q.; Fan, J.; Cheng, H. Nuclear mRNA export. Acta Biochim. Biophys. Sin. 2024, 57, 84–100. [Google Scholar] [CrossRef]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef]
- Baptista, B.; Carapito, R.; Laroui, N.; Pichon, C.; Sousa, F. mRNA, a Revolution in Biomedicine. Pharmaceutics 2021, 13, 2090. [Google Scholar] [CrossRef]
- Berg, M.D.; Brandl, C.J. Transfer RNAs: Diversity in form and function. RNA Biol. 2021, 18, 316–339. [Google Scholar] [CrossRef]
- Lorenz, C.; Lünse, C.E.; Mörl, M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules 2017, 7, 35. [Google Scholar] [CrossRef]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef]
- Yu, X.; Xie, Y.; Zhang, S.; Song, X.; Xiao, B.; Yan, Z. tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics 2021, 11, 461–469. [Google Scholar] [CrossRef]
- Schimmel, P. The emerging complexity of the tRNA world: Mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018, 19, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Sekulovski, S.; Trowitzsch, S. Transfer RNA processing—From a structural and disease perspective. Biol. Chem. 2022, 403, 749–763. [Google Scholar] [CrossRef]
- Steitz, T.A.; Moore, P.B. RNA, the first macromolecular catalyst: The ribosome is a ribozyme. Trends Biochem. Sci. 2003, 28, 411–418. [Google Scholar] [CrossRef]
- Moore, P.B.; Steitz, T.A. The roles of RNA in the synthesis of protein. Cold Spring Harb. Perspect. Biol. 2011, 3, a003780. [Google Scholar] [CrossRef]
- Rodnina, M.V.; Wintermeyer, W. Recent mechanistic insights into eukaryotic ribosomes. Curr. Opin. Cell Biol. 2009, 21, 435–443. [Google Scholar] [CrossRef]
- Klinge, S.; Voigts-Hoffmann, F.; Leibundgut, M.; Ban, N. Atomic structures of the eukaryotic ribosome. Trends Biochem. Sci. 2012, 37, 189–198. [Google Scholar] [CrossRef]
- Aubert, M.; O’Donohue, M.F.; Lebaron, S.; Gleizes, P.E. Pre-Ribosomal RNA Processing in Human Cells: From Mechanisms to Congenital Diseases. Biomolecules 2018, 8, 123. [Google Scholar] [CrossRef]
- Ranganathan, K.; Sivasankar, V. MicroRNAs—Biology and clinical applications. J. Oral Maxillofac. Pathol. 2014, 18, 229–234. [Google Scholar] [CrossRef]
- Ferragut Cardoso, A.P.; Banerjee, M.; Nail, A.N.; Lykoudi, A.; States, J.C. miRNA dysregulation is an emerging modulator of genomic instability. Semin. Cancer Biol. 2021, 76, 120–131. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef]
- Dragomir, M.P.; Knutsen, E.; Calin, G.A. Classical and noncanonical functions of miRNAs in cancers. Trends Genet. 2022, 38, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Dumesic, P.A.; Madhani, H.D. Recognizing the enemy within: Licensing RNA-guided genome defense. Trends Biochem. Sci. 2014, 39, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Alshaer, W.; Zureigat, H.; Al Karaki, A.; Al-Kadash, A.; Gharaibeh, L.; Hatmal, M.M.; Aljabali, A.A.A.; Awidi, A. siRNA: Mechanism of action, challenges, and therapeutic approaches. Eur. J. Pharmacol. 2021, 905, 174178. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Li, L.C.; Okino, S.T.; Zhao, H.; Pookot, D.; Place, R.F.; Urakami, S.; Enokida, H.; Dahiya, R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA 2006, 103, 17337–17342. [Google Scholar] [CrossRef]
- Aravin, A.A.; Naumova, N.M.; Tulin, A.V.; Vagin, V.V.; Rozovsky, Y.M.; Gvozdev, V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001, 11, 1017–1027. [Google Scholar] [CrossRef]
- Jiang, M.; Hong, X.; Gao, Y.; Kho, A.T.; Tantisira, K.G.; Li, J. piRNA associates with immune diseases. Cell Commun. Signal. 2024, 22, 347. [Google Scholar] [CrossRef]
- Le Thomas, A.; Rogers, A.K.; Webster, A.; Marinov, G.K.; Liao, S.E.; Perkins, E.M.; Hur, J.K.; Aravin, A.A.; Tóth, K.F. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013, 27, 390–399. [Google Scholar] [CrossRef]
- Klenov, M.S.; Lavrov, S.A.; Korbut, A.P.; Stolyarenko, A.D.; Yakushev, E.Y.; Reuter, M.; Pillai, R.S.; Gvozdev, V.A. Impact of nuclear Piwi elimination on chromatin state in Drosophila melanogaster ovaries. Nucleic Acids Res. 2014, 42, 6208–6218. [Google Scholar] [CrossRef]
- Iwasaki, Y.W.; Murano, K.; Ishizu, H.; Shibuya, A.; Iyoda, Y.; Siomi, M.C.; Siomi, H.; Saito, K. Piwi Modulates Chromatin Accessibility by Regulating Multiple Factors Including Histone H1 to Repress Transposons. Mol. Cell 2016, 63, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Wang, P.J. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet. 2012, 8, e1003038. [Google Scholar] [CrossRef] [PubMed]
- Gou, L.T.; Dai, P.; Yang, J.H.; Xue, Y.; Hu, Y.P.; Zhou, Y.; Kang, J.Y.; Wang, X.; Li, H.; Hua, M.M.; et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014, 24, 680–700. [Google Scholar] [CrossRef]
- Duc, C.; Yoth, M.; Jensen, S.; Mouniée, N.; Bergman, C.M.; Vaury, C.; Brasset, E. Trapping a somatic endogenous retrovirus into a germline piRNA cluster immunizes the germline against further invasion. Genome Biol. 2019, 20, 127. [Google Scholar] [CrossRef]
- Goriaux, C.; Théron, E.; Brasset, E.; Vaury, C. History of the discovery of a master locus producing piRNAs: The flamenco/COM locus in Drosophila melanogaster. Front. Genet. 2014, 5, 257. [Google Scholar] [CrossRef]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Yi, Q.; Feng, J.; Lan, W.; Shi, H.; Sun, W. CircRNA and lncRNA-encoded peptide in diseases, an update review. Mol. Cancer 2024, 23, 214. [Google Scholar] [CrossRef]
- Dawoud, A.; Ihab Zakaria, Z.; Hisham Rashwan, H.; Braoudaki, M.; Youness, R.A. Circular RNAs: New layer of complexity evading breast cancer heterogeneity. Non-Coding RNA Res. 2023, 8, 60–74. [Google Scholar] [CrossRef]
- He, L.; Man, C.; Xiang, S.; Yao, L.; Wang, X.; Fan, Y. Circular RNAs’ cap-independent translation protein and its roles in carcinomas. Mol. Cancer 2021, 20, 119. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Nicoloso, M.; Caizergues-Ferrer, M.; Michot, B.; Azum, M.C.; Bachellerie, J.P. U20, a novel small nucleolar RNA, is encoded in an intron of the nucleolin gene in mammals. Mol. Cell. Biol. 1994, 14, 5766–5776. [Google Scholar] [CrossRef] [PubMed]
- Bachellerie, J.P.; Nicoloso, M.; Qu, L.H.; Michot, B.; Caizergues-Ferrer, M.; Cavaille, J.; Renalier, M.H. Novel intron-encoded small nucleolar RNAs with long sequence complementarities to mature rRNAs involved in ribosome biogenesis. Biochem. Cell Biol. 1995, 73, 835–843. [Google Scholar] [CrossRef]
- Nicoloso, M.; Qu, L.H.; Michot, B.; Bachellerie, J.P. Intron-encoded, antisense small nucleolar RNAs: The characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J. Mol. Biol. 1996, 260, 178–195. [Google Scholar] [CrossRef]
- Hirose, T.; Steitz, J.A. Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 2001, 98, 12914–12919. [Google Scholar] [CrossRef]
- Balakin, A.G.; Smith, L.; Fournier, M.J. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 1996, 86, 823–834. [Google Scholar] [CrossRef]
- Darzacq, X.; Jády, B.E.; Verheggen, C.; Kiss, A.M.; Bertrand, E.; Kiss, T. Cajal body-specific small nuclear RNAs: A novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002, 21, 2746–2756. [Google Scholar] [CrossRef]
- van der Werf, J.; Chin, C.V.; Fleming, N.I. SnoRNA in Cancer Progression, Metastasis and Immunotherapy Response. Biology 2021, 10, 809. [Google Scholar] [CrossRef]
- Schimmang, T.; Tollervey, D.; Kern, H.; Frank, R.; Hurt, E.C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989, 8, 4015–4024. [Google Scholar] [CrossRef]
- Baserga, S.J.; Yang, X.D.; Steitz, J.A. An intact Box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 1991, 10, 2645–2651. [Google Scholar] [CrossRef]
- Dunbar, D.A.; Wormsley, S.; Lowe, T.M.; Baserga, S.J. Fibrillarin-associated box C/D small nucleolar RNAs in Trypanosoma brucei. Sequence conservation and implications for 2′-O-ribose methylation of rRNA. J. Biol. Chem. 2000, 275, 14767–14776. [Google Scholar] [CrossRef] [PubMed]
- McKeegan, K.S.; Debieux, C.M.; Watkins, N.J. Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol. Cell. Biol. 2009, 29, 4971–4981. [Google Scholar] [CrossRef] [PubMed]
- Bizarro, J.; Charron, C.; Boulon, S.; Westman, B.; Pradet-Balade, B.; Vandermoere, F.; Chagot, M.E.; Hallais, M.; Ahmad, Y.; Leonhardt, H.; et al. Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J. Cell Biol. 2014, 207, 463–480. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Tien, A.L.; Fournier, M.J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 1997, 89, 565–573. [Google Scholar] [CrossRef]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; León-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef]
- Dragon, F.; Pogacić, V.; Filipowicz, W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 2000, 20, 3037–3048. [Google Scholar] [CrossRef]
- Pogacić, V.; Dragon, F.; Filipowicz, W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 2000, 20, 9028–9040. [Google Scholar] [CrossRef]
- Wang, C.; Meier, U.T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004, 23, 1857–1867. [Google Scholar] [CrossRef]
- Wajahat, M.; Bracken, C.P.; Orang, A. Emerging Functions for snoRNAs and snoRNA-Derived Fragments. Int. J. Mol. Sci. 2021, 22, 10193. [Google Scholar] [CrossRef]
- Jensen, E.G.; Hellung-Larsen, P.; Frederiksen, S. Synthesis of low molecular weight RNA components A, C and D by polymerase II in alpha-amanitin-resistant hamster cells. Nucleic Acids Res. 1979, 6, 321–330. [Google Scholar] [CrossRef]
- Matera, A.G.; Ward, D.C. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J. Cell Biol. 1993, 121, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Sadowski, C.L.; Henry, R.W.; Kobayashi, R.; Hernandez, N. The SNAP45 subunit of the small nuclear RNA (snRNA) activating protein complex is required for RNA polymerase II and III snRNA gene transcription and interacts with the TATA box binding protein. Proc. Natl. Acad. Sci. USA 1996, 93, 4289–4293. [Google Scholar] [CrossRef] [PubMed]
- Frey, M.R.; Bailey, A.D.; Weiner, A.M.; Matera, A.G. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol. 1999, 9, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Jády, B.E.; Darzacq, X.; Tucker, K.E.; Matera, A.G.; Bertrand, E.; Kiss, T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 2003, 22, 1878–1888. [Google Scholar] [CrossRef]
- Sontheimer, E.J.; Steitz, J.A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 1993, 262, 1989–1996. [Google Scholar] [CrossRef]
- Gozani, O.; Feld, R.; Reed, R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996, 10, 233–243. [Google Scholar] [CrossRef]
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. [Google Scholar] [CrossRef]
- Kondo, Y.; Oubridge, C.; van Roon, A.M.; Nagai, K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. eLife 2015, 4, e04986. [Google Scholar] [CrossRef]
- Wan, R.; Yan, C.; Bai, R.; Wang, L.; Huang, M.; Wong, C.C.; Shi, Y. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science 2016, 351, 466–475. [Google Scholar] [CrossRef]
- Mabin, J.W.; Lewis, P.W.; Brow, D.A.; Dvinge, H. Human spliceosomal snRNA sequence variants generate variant spliceosomes. RNA 2021, 27, 1186–1203. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef] [PubMed]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Naganuma, T.; Hirose, T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013, 10, 456–461. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, H.; Fang, S.; Kang, Y.; Wu, W.; Hao, Y.; Li, Z.; Bu, D.; Sun, N.; Zhang, M.Q.; et al. NONCODE 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016, 44, D203–D208. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Seifuddin, F.; Singh, K.; Suresh, A.; Judy, J.T.; Chen, Y.C.; Chaitankar, V.; Tunc, I.; Ruan, X.; Li, P.; Chen, Y.; et al. lncRNAKB, a knowledgebase of tissue-specific functional annotation and trait association of long noncoding RNA. Sci. Data 2020, 7, 326. [Google Scholar] [CrossRef]
- Scott, W.G.; Finch, J.T.; Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell 1995, 81, 991–1002. [Google Scholar] [CrossRef]
- Scott, W.G.; Murray, J.B.; Arnold, J.R.; Stoddard, B.L.; Klug, A. Capturing the structure of a catalytic RNA intermediate: The hammerhead ribozyme. Science 1996, 274, 2065–2069. [Google Scholar] [CrossRef] [PubMed]
- Doherty, E.A.; Herschlag, D.; Doudna, J.A. Assembly of an exceptionally stable RNA tertiary interface in a group I ribozyme. Biochemistry 1999, 38, 2982–2990. [Google Scholar] [CrossRef] [PubMed]
- Rangan, P.; Masquida, B.; Westhof, E.; Woodson, S.A. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc. Natl. Acad. Sci. USA 2003, 100, 1574–1579. [Google Scholar] [CrossRef] [PubMed]
- Gasser, C.; Gebetsberger, J.; Gebetsberger, M.; Micura, R. SHAPE probing pictures Mg2+-dependent folding of small self-cleaving ribozymes. Nucleic Acids Res. 2018, 46, 6983–6995. [Google Scholar] [CrossRef]
- Crow, M.; Gillis, J. Single cell RNA-sequencing: Replicability of cell types. Curr. Opin. Neurobiol. 2019, 56, 69–77. [Google Scholar] [CrossRef]
- Grosshans, H.; Filipowicz, W. Molecular biology: The expanding world of small RNAs. Nature 2008, 451, 414–416. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Wolfien, M.; Brauer, D.L.; Bagnacani, A.; Wolkenhauer, O. Workflow Development for the Functional Characterization of ncRNAs. Methods Mol. Biol. 2019, 1912, 111–132. [Google Scholar] [CrossRef]
- Liu, Q.; Fang, L.; Wu, C. Alternative Splicing and Isoforms: From Mechanisms to Diseases. Genes 2022, 13, 401. [Google Scholar] [CrossRef]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef]
- Bradley, R.K.; Anczuków, O. RNA splicing dysregulation and the hallmarks of cancer. Nat. Rev. Cancer 2023, 23, 135–155. [Google Scholar] [CrossRef] [PubMed]
- Supadmanaba, I.G.P.; Mantini, G.; Randazzo, O.; Capula, M.; Muller, I.B.; Cascioferro, S.; Diana, P.; Peters, G.J.; Giovannetti, E. Interrelationship between miRNA and splicing factors in pancreatic ductal adenocarcinoma. Epigenetics 2022, 17, 381–404. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Liu, M.; Stribinskis, V.; Klinge, C.M.; Ramos, K.S.; Colburn, N.H.; Li, Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008, 27, 4373–4379. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.S.; Su, J.L.; Cha, S.T.; Tarn, W.Y.; Wang, M.Y.; Hsu, H.C.; Lin, M.T.; Chu, C.Y.; Hua, K.T.; Chen, C.N.; et al. miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans. J. Clin. Investig. 2011, 121, 3442–3455. [Google Scholar] [CrossRef]
- Wang, D.; Sun, X.; Wei, Y.; Liang, H.; Yuan, M.; Jin, F.; Chen, X.; Liu, Y.; Zhang, C.Y.; Li, L.; et al. Nuclear miR-122 directly regulates the biogenesis of cell survival oncomiR miR-21 at the posttranscriptional level. Nucleic Acids Res. 2018, 46, 2012–2029. [Google Scholar] [CrossRef]
- Zampetaki, A.; Willeit, P.; Tilling, L.; Drozdov, I.; Prokopi, M.; Renard, J.M.; Mayr, A.; Weger, S.; Schett, G.; Shah, A.; et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J. Am. Coll. Cardiol. 2012, 60, 290–299. [Google Scholar] [CrossRef]
- Loyer, X.; Potteaux, S.; Vion, A.C.; Guérin, C.L.; Boulkroun, S.; Rautou, P.E.; Ramkhelawon, B.; Esposito, B.; Dalloz, M.; Paul, J.L.; et al. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ. Res. 2014, 114, 434–443. [Google Scholar] [CrossRef]
- Searles, C.D. MicroRNAs and Cardiovascular Disease Risk. Curr. Cardiol. Rep. 2024, 26, 51–60. [Google Scholar] [CrossRef]
- Liu, C.G.; Wang, J.L.; Li, L.; Wang, P.C. MicroRNA-384 regulates both amyloid precursor protein and β-secretase expression and is a potential biomarker for Alzheimer’s disease. Int. J. Mol. Med. 2014, 34, 160–166. [Google Scholar] [CrossRef]
- Liu, X.; He, F.; Pang, R.; Zhao, D.; Qiu, W.; Shan, K.; Zhang, J.; Lu, Y.; Li, Y.; Wang, Y. Interleukin-17 (IL-17)-induced microRNA 873 (miR-873) contributes to the pathogenesis of experimental autoimmune encephalomyelitis by targeting A20 ubiquitin-editing enzyme. J. Biol. Chem. 2014, 289, 28971–28986. [Google Scholar] [CrossRef]
- Cressatti, M.; Juwara, L.; Galindez, J.M.; Velly, A.M.; Nkurunziza, E.S.; Marier, S.; Canie, O.; Gornistky, M.; Schipper, H.M. Salivary microR-153 and microR-223 Levels as Potential Diagnostic Biomarkers of Idiopathic Parkinson’s Disease. Mov. Disord. 2020, 35, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Zhong, Z.; Hu, B.; Yan, S.; Yan, Y.; Zhang, J.; Shi, T.; Jiang, L.; Li, W.; Huang, W. MicroRNA-3473b regulates the expression of TREM2/ULK1 and inhibits autophagy in inflammatory pathogenesis of Parkinson disease. J. Neurochem. 2021, 157, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. [Google Scholar] [CrossRef]
- Tijsterman, M.; Ketting, R.F.; Plasterk, R.H. The genetics of RNA silencing. Annu. Rev. Genet. 2002, 36, 489–519. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Johnson, S.M.; Lin, S.Y.; Slack, F.J. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 2003, 259, 364–379. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Meyer, J.; Borkhardt, A.; Tuschl, T. New microRNAs from mouse and human. RNA 2003, 9, 175–179. [Google Scholar] [CrossRef]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- Landthaler, M.; Yalcin, A.; Tuschl, T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004, 14, 2162–2167. [Google Scholar] [CrossRef]
- Brownawell, A.M.; Macara, I.G. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol. 2002, 156, 53–64. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar] [CrossRef]
- Knight, S.W.; Bass, B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 2001, 293, 2269–2271. [Google Scholar] [CrossRef]
- Ketting, R.F.; Fischer, S.E.; Bernstein, E.; Sijen, T.; Hannon, G.J.; Plasterk, R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001, 15, 2654–2659. [Google Scholar] [CrossRef]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef]
- Nykänen, A.; Haley, B.; Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 2001, 107, 309–321. [Google Scholar] [CrossRef]
- Martinez, J.; Patkaniowska, A.; Urlaub, H.; Lührmann, R.; Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 2002, 110, 563–574. [Google Scholar] [CrossRef]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef]
- Mourelatos, Z.; Dostie, J.; Paushkin, S.; Sharma, A.; Charroux, B.; Abel, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002, 16, 720–728. [Google Scholar] [CrossRef]
- Hutvágner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060. [Google Scholar] [CrossRef]
- Pham, J.W.; Pellino, J.L.; Lee, Y.S.; Carthew, R.W.; Sontheimer, E.J. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 2004, 117, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W. RNAi: The nuts and bolts of the RISC machine. Cell 2005, 122, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Sontheimer, E.J. Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell Biol. 2005, 6, 127–138. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Provost, P.; Dishart, D.; Doucet, J.; Frendewey, D.; Samuelsson, B.; Rådmark, O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002, 21, 5864–5874. [Google Scholar] [CrossRef]
- Zhang, H.; Kolb, F.A.; Brondani, V.; Billy, E.; Filipowicz, W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002, 21, 5875–5885. [Google Scholar] [CrossRef]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Song, J.J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004, 305, 1434–1437. [Google Scholar] [CrossRef]
- Ma, J.B.; Yuan, Y.R.; Meister, G.; Pei, Y.; Tuschl, T.; Patel, D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 2005, 434, 666–670. [Google Scholar] [CrossRef]
- Rivas, F.V.; Tolia, N.H.; Song, J.J.; Aragon, J.P.; Liu, J.; Hannon, G.J.; Joshua-Tor, L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005, 12, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Haley, B.; Zamore, P.D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004, 11, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Tabara, H.; Yigit, E.; Siomi, H.; Mello, C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 2002, 109, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Ishizuka, A.; Siomi, H.; Siomi, M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004, 18, 1655–1666. [Google Scholar] [CrossRef]
- Tahbaz, N.; Kolb, F.A.; Zhang, H.; Jaronczyk, K.; Filipowicz, W.; Hobman, T.C. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004, 5, 189–194. [Google Scholar] [CrossRef]
- Jiang, F.; Ye, X.; Liu, X.; Fincher, L.; McKearin, D.; Liu, Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005, 19, 1674–1679. [Google Scholar] [CrossRef]
- Saito, K.; Ishizuka, A.; Siomi, H.; Siomi, M.C. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005, 3, e235. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Grishok, A.; Tabara, H.; Mello, C.C. Genetic requirements for inheritance of RNAi in C. elegans. Science 2000, 287, 2494–2497. [Google Scholar] [CrossRef]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef]
- Liu, Q.; Rand, T.A.; Kalidas, S.; Du, F.; Kim, H.E.; Smith, D.P.; Wang, X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 2003, 301, 1921–1925. [Google Scholar] [CrossRef] [PubMed]
- Tabara, H.; Sarkissian, M.; Kelly, W.G.; Fleenor, J.; Grishok, A.; Timmons, L.; Fire, A.; Mello, C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 1999, 99, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Ketting, R.F.; Haverkamp, T.H.; van Luenen, H.G.; Plasterk, R.H. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 1999, 99, 133–141. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.; Zapp, M.L.; Weng, Z.; et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Malone, C.D.; Zhou, R.; Stark, A.; Schlingeheyde, C.; Dus, M.; Perrimon, N.; Kellis, M.; Wohlschlegel, J.A.; Sachidanandam, R.; et al. An endogenous small interfering RNA pathway in Drosophila. Nature 2008, 453, 798–802. [Google Scholar] [CrossRef]
- Okamura, K.; Chung, W.J.; Ruby, J.G.; Guo, H.; Bartel, D.P.; Lai, E.C. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 2008, 453, 803–806. [Google Scholar] [CrossRef]
- Kawamura, Y.; Saito, K.; Kin, T.; Ono, Y.; Asai, K.; Sunohara, T.; Okada, T.N.; Siomi, M.C.; Siomi, H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 2008, 453, 793–797. [Google Scholar] [CrossRef]
- Zhou, R.; Hotta, I.; Denli, A.M.; Hong, P.; Perrimon, N.; Hannon, G.J. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol. Cell 2008, 32, 592–599. [Google Scholar] [CrossRef]
- Förstemann, K.; Tomari, Y.; Du, T.; Vagin, V.V.; Denli, A.M.; Bratu, D.P.; Klattenhoff, C.; Theurkauf, W.E.; Zamore, P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005, 3, e236. [Google Scholar] [CrossRef]
- Yang, N.; Kazazian, H.H., Jr. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 2006, 13, 763–771. [Google Scholar] [CrossRef]
- Tam, O.H.; Aravin, A.A.; Stein, P.; Girard, A.; Murchison, E.P.; Cheloufi, S.; Hodges, E.; Anger, M.; Sachidanandam, R.; Schultz, R.M.; et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 2008, 453, 534–538. [Google Scholar] [CrossRef]
- Watanabe, T.; Totoki, Y.; Toyoda, A.; Kaneda, M.; Kuramochi-Miyagawa, S.; Obata, Y.; Chiba, H.; Kohara, Y.; Kono, T.; Nakano, T.; et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 2008, 453, 539–543. [Google Scholar] [CrossRef]
- Sasidharan, R.; Gerstein, M. Genomics: Protein fossils live on as RNA. Nature 2008, 453, 729–731. [Google Scholar] [CrossRef] [PubMed]
- Jurj, A.; Fontana, B.; Varani, G.; Calin, G.A. Small molecules targeting microRNAs: New opportunities and challenges in precision cancer therapy. Trends Cancer 2024, 10, 809–824. [Google Scholar] [CrossRef] [PubMed]
- Salazar-León, J.R.-R.F.; Meneses-Acosta, A.; Merchant, H.; Lagunas-Martínez, A.; Meda, E.; Pita-López, M.L.; Bermúdez-Morales, V.; Madrid-Marina, V.; Gómez-Cerón, C.; Peralta-Zaragoza, O. Silencing of HPV16 E6 & E7 oncogenic activities by small interference RNA induces autophagy and apoptosis in human cervical cancer cells. J. Nucleic Acids Investig. 2011, 2, 59–69. [Google Scholar]
- Rosenson, R.S.; Gaudet, D.; Hegele, R.A.; Ballantyne, C.M.; Nicholls, S.J.; Lucas, K.J.; San Martin, J.; Zhou, R.; Muhsin, M.; Chang, T.; et al. Zodasiran, an RNAi Therapeutic Targeting ANGPTL3, for Mixed Hyperlipidemia. New Engl. J. Med. 2024, 391, 913–925. [Google Scholar] [CrossRef]
- Traber, G.M.; Yu, A.M. RNAi-Based Therapeutics and Novel RNA Bioengineering Technologies. J. Pharmacol. Exp. Ther. 2023, 384, 133–154. [Google Scholar] [CrossRef]
- Pascolo, S. Synthetic Messenger RNA-Based Vaccines: From Scorn to Hype. Viruses 2021, 13, 270. [Google Scholar] [CrossRef]
- Wang, F.; Kream, R.M.; Stefano, G.B. An Evidence Based Perspective on mRNA-SARS-CoV-2 Vaccine Development. Med. Sci. Monit. 2020, 26, e924700. [Google Scholar] [CrossRef]
- Shin, M.D.; Shukla, S.; Chung, Y.H.; Beiss, V.; Chan, S.K.; Ortega-Rivera, O.A.; Wirth, D.M.; Chen, A.; Sack, M.; Pokorski, J.K.; et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020, 15, 646–655. [Google Scholar] [CrossRef]
- Huang, X.; Tang, T.; Zhang, G.; Liang, T. Identification of tumor antigens and immune subtypes of cholangiocarcinoma for mRNA vaccine development. Mol. Cancer 2021, 20, 50. [Google Scholar] [CrossRef]
- Chekaoui, A.; Garofalo, M.; Gad, B.; Staniszewska, M.; Chiaro, J.; Pancer, K.; Gryciuk, A.; Cerullo, V.; Salmaso, S.; Caliceti, P.; et al. Cancer vaccines: An update on recent achievements and prospects for cancer therapy. Clin. Exp. Med. 2024, 25, 24. [Google Scholar] [CrossRef] [PubMed]
- Adamopoulos, P.G.; Athanasopoulou, K.; Daneva, G.N.; Scorilas, A. The Repertoire of RNA Modifications Orchestrates a Plethora of Cellular Responses. Int. J. Mol. Sci. 2023, 24, 2387. [Google Scholar] [CrossRef]
- Arzumanian, V.A.; Dolgalev, G.V.; Kurbatov, I.Y.; Kiseleva, O.I.; Poverennaya, E.V. Epitranscriptome: Review of Top 25 Most-Studied RNA Modifications. Int. J. Mol. Sci. 2022, 23, 13851. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Ke, S.; Pandya-Jones, A.; Saito, Y.; Fak, J.J.; Vågbø, C.B.; Geula, S.; Hanna, J.H.; Black, D.L.; Darnell, J.E., Jr.; Darnell, R.B. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017, 31, 990–1006. [Google Scholar] [CrossRef]
- Balacco, D.L.; Soller, M. The m6A Writer: Rise of a Machine for Growing Tasks. Biochemistry 2019, 58, 363–378. [Google Scholar] [CrossRef]
- Eyler, D.E.; Franco, M.K.; Batool, Z.; Wu, M.Z.; Dubuke, M.L.; Dobosz-Bartoszek, M.; Jones, J.D.; Polikanov, Y.S.; Roy, B.; Koutmou, K.S. Pseudouridinylation of mRNA coding sequences alters translation. Proc. Natl. Acad. Sci. USA 2019, 116, 23068–23074. [Google Scholar] [CrossRef]
- Martinez, N.M.; Su, A.; Burns, M.C.; Nussbacher, J.K.; Schaening, C.; Sathe, S.; Yeo, G.W.; Gilbert, W.V. Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing. Mol. Cell 2022, 82, 645–659.E9. [Google Scholar] [CrossRef]
- Davis, F.F.; Allen, F.W. Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem. 1957, 227, 907–915. [Google Scholar] [CrossRef]
- Cappannini, A.; Ray, A.; Purta, E.; Mukherjee, S.; Boccaletto, P.; Moafinejad, S.N.; Lechner, A.; Barchet, C.; Klaholz, B.P.; Stefaniak, F.; et al. MODOMICS: A database of RNA modifications and related information. 2023 update. Nucleic Acids Res. 2024, 52, D239–D244. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, X.; Yi, C. Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat. Methods 2016, 14, 23–31. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, Y.; Hang, D.; Meng, J.; Wei, Z. Detecting RNA modification using direct RNA sequencing: A systematic review. Comput. Struct. Biotechnol. J. 2022, 20, 5740–5749. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Śledź, P.; Jinek, M. Structural insights into the molecular mechanism of the m6A writer complex. eLife 2016, 5, e18434. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Nombela, P.; Miguel-López, B.; Blanco, S. The role of m6A, m5C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol. Cancer 2021, 20, 18. [Google Scholar] [CrossRef]
- Xue, C.; Zhao, Y.; Li, L. Advances in RNA cytosine-5 methylation: Detection, regulatory mechanisms, biological functions and links to cancer. Biomark. Res. 2020, 8, 43. [Google Scholar] [CrossRef]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012, 19, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, Y.; Sun, B.F.; Chen, Y.S.; Xu, J.W.; Lai, W.Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Cerneckis, J.; Cui, Q.; He, C.; Yi, C.; Shi, Y. Decoding pseudouridine: An emerging target for therapeutic development. Trends Pharmacol. Sci. 2022, 43, 522–535. [Google Scholar] [CrossRef]
- Safra, M.; Nir, R.; Farouq, D.; Vainberg Slutskin, I.; Schwartz, S. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 2017, 27, 393–406. [Google Scholar] [CrossRef]
- Carlile, T.M.; Martinez, N.M.; Schaening, C.; Su, A.; Bell, T.A.; Zinshteyn, B.; Gilbert, W.V. mRNA structure determines modification by pseudouridine synthase 1. Nat. Chem. Biol. 2019, 15, 966–974. [Google Scholar] [CrossRef]
- Borchardt, E.K.; Martinez, N.M.; Gilbert, W.V. Regulation and Function of RNA Pseudouridylation in Human Cells. Annu. Rev. Genet. 2020, 54, 309–336. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Zhou, K.; Wu, T.; Zhao, B.S.; Sun, M.; Chen, Z.; Deng, X.; Xiao, G.; Auer, F.; et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 2019, 567, 414–419. [Google Scholar] [CrossRef]
- Hao, J.D.; Liu, Q.L.; Liu, M.X.; Yang, X.; Wang, L.M.; Su, S.Y.; Xiao, W.; Zhang, M.Q.; Zhang, Y.C.; Zhang, L.; et al. DDX21 mediates co-transcriptional RNA m6A modification to promote transcription termination and genome stability. Mol. Cell 2024, 84, 1711–1726.E11. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhang, J.; Zhang, J.; Liu, B. SIRT7 is a deacetylase of N4-acetylcytidine on ribosomal RNA. Genome Instab. Dis. 2021, 2, 253–260. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Fu, L.; Guerrero, C.R.; Zhong, N.; Amato, N.J.; Liu, Y.; Liu, S.; Cai, Q.; Ji, D.; Jin, S.G.; Niedernhofer, L.J.; et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J. Am. Chem. Soc. 2014, 136, 11582–11585. [Google Scholar] [CrossRef]
- Huang, W.; Lan, M.D.; Qi, C.B.; Zheng, S.J.; Wei, S.Z.; Yuan, B.F.; Feng, Y.Q. Formation and determination of the oxidation products of 5-methylcytosine in RNA. Chem. Sci. 2016, 7, 5495–5502. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef]
- Wang, N.; Chen, R.X.; Deng, M.H.; Wei, W.S.; Zhou, Z.H.; Ning, K.; Li, Y.H.; Li, X.D.; Ye, Y.L.; Wen, J.H.; et al. m5C-dependent cross-regulation between nuclear reader ALYREF and writer NSUN2 promotes urothelial bladder cancer malignancy through facilitating RABL6/TK1 mRNAs splicing and stabilization. Cell Death Dis. 2023, 14, 139. [Google Scholar] [CrossRef]
- Liu, L.; Chen, Y.; Zhang, T.; Cui, G.; Wang, W.; Zhang, G.; Li, J.; Zhang, Y.; Wang, Y.; Zou, Y.; et al. YBX1 Promotes Esophageal Squamous Cell Carcinoma Progression via m5C-Dependent SMOX mRNA Stabilization. Adv. Sci. 2024, 11, e2302379. [Google Scholar] [CrossRef]
- Wei, S.; Dai, X.; Yuan, J.; He, S.; Shah, K.; Guo, S.; Duan, Z.; Murn, J.; Wang, Y. Quantitative Proteomics Identifies Profilin-1 as a Pseudouridine-Binding Protein. J. Am. Chem. Soc. 2025, 147, 1458–1462. [Google Scholar] [CrossRef]
- Ru, W.; Zhang, X.; Yue, B.; Qi, A.; Shen, X.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Insight into m6A methylation from occurrence to functions. Open Biol. 2020, 10, 200091. [Google Scholar] [CrossRef]
- Zhao, B.; Wang, W.; Zhao, Y.; Qiao, H.; Gao, Z.; Chuai, X. Regulation of Antiviral Immune Response by N 6-Methyladenosine of mRNA. Front. Microbiol. 2021, 12, 789605. [Google Scholar] [CrossRef]
- Gao, Y.; Fang, J. RNA 5-methylcytosine modification and its emerging role as an epitranscriptomic mark. RNA Biol. 2021, 18, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Zhang, J.; Liu, B.; Xu, J.; Cai, B.; Yang, H.; Straube, J.; Yu, X.; Ma, T. Biological roles of RNA m5C modification and its implications in Cancer immunotherapy. Biomark. Res. 2022, 10, 15. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Anderson, B.R.; Muramatsu, H.; Nallagatla, S.R.; Bevilacqua, P.C.; Sansing, L.H.; Weissman, D.; Karikó, K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010, 38, 5884–5892. [Google Scholar] [CrossRef]
- Durbin, A.F.; Wang, C.; Marcotrigiano, J.; Gehrke, L. RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio 2016, 7, e00833-16. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, Z.; Xue, M.; Zhao, B.S.; Harder, O.; Li, A.; Liang, X.; Gao, T.Z.; Xu, Y.; Zhou, J.; et al. N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 2020, 5, 584–598. [Google Scholar] [CrossRef]
- Kim, G.W.; Imam, H.; Khan, M.; Siddiqui, A. N6-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J. Biol. Chem. 2020, 295, 13123–13133. [Google Scholar] [CrossRef]
- Chen, S.; Kumar, S.; Espada, C.E.; Tirumuru, N.; Cahill, M.P.; Hu, L.; He, C.; Wu, L. N6-methyladenosine modification of HIV-1 RNA suppresses type-I interferon induction in differentiated monocytic cells and primary macrophages. PLoS Pathog. 2021, 17, e1009421. [Google Scholar] [CrossRef] [PubMed]
- Lichinchi, G.; Zhao, B.S.; Wu, Y.; Lu, Z.; Qin, Y.; He, C.; Rana, T.M. Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell Host Microbe 2016, 20, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.; Mason, C.E.; Rana, T.M. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016, 1, 16011. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Y.P.; Li, K.; Ye, Q.; Zhou, H.Y.; Sun, H.; Li, X.; Yu, L.; Deng, Y.Q.; Li, R.T.; et al. The m6A methylome of SARS-CoV-2 in host cells. Cell Res. 2021, 31, 404–414. [Google Scholar] [CrossRef]
- Gokhale, N.S.; McIntyre, A.B.R.; McFadden, M.J.; Roder, A.E.; Kennedy, E.M.; Gandara, J.A.; Hopcraft, S.E.; Quicke, K.M.; Vazquez, C.; Willer, J.; et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 2016, 20, 654–665. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Bogerd, H.P.; Kornepati, A.V.; Kang, D.; Ghoshal, D.; Marshall, J.B.; Poling, B.C.; Tsai, K.; Gokhale, N.S.; Horner, S.M.; et al. Posttranscriptional m6A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe 2016, 19, 675–685. [Google Scholar] [CrossRef]
- McIntyre, W.; Netzband, R.; Bonenfant, G.; Biegel, J.M.; Miller, C.; Fuchs, G.; Henderson, E.; Arra, M.; Canki, M.; Fabris, D.; et al. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 2018, 46, 5776–5791. [Google Scholar] [CrossRef]
- Courtney, D.G.; Tsai, K.; Bogerd, H.P.; Kennedy, E.M.; Law, B.A.; Emery, A.; Swanstrom, R.; Holley, C.L.; Cullen, B.R. Epitranscriptomic Addition of m5C to HIV-1 Transcripts Regulates Viral Gene Expression. Cell Host Microbe 2019, 26, 217–227.e6. [Google Scholar] [CrossRef]
- Tsai, K.; Jaguva Vasudevan, A.A.; Martinez Campos, C.; Emery, A.; Swanstrom, R.; Cullen, B.R. Acetylation of Cytidine Residues Boosts HIV-1 Gene Expression by Increasing Viral RNA Stability. Cell Host Microbe 2020, 28, 306–312.e6. [Google Scholar] [CrossRef]
- Burgess, H.M.; Depledge, D.P.; Thompson, L.; Srinivas, K.P.; Grande, R.C.; Vink, E.I.; Abebe, J.S.; Blackaby, W.P.; Hendrick, A.; Albertella, M.R.; et al. Targeting the m6A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes Dev. 2021, 35, 1005–1019. [Google Scholar] [CrossRef]
- Fleming, A.M.; Mathewson, N.J.; Howpay Manage, S.A.; Burrows, C.J. Nanopore Dwell Time Analysis Permits Sequencing and Conformational Assignment of Pseudouridine in SARS-CoV-2. ACS Cent. Sci. 2021, 7, 1707–1717. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Hui, H.; Bray, B.; Gonzalez, G.M.; Zeller, M.; Anderson, K.G.; Knight, R.; Smith, D.; Wang, Y.; Carlin, A.F.; et al. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep. 2021, 35, 109091. [Google Scholar] [CrossRef] [PubMed]
- Martinez Campos, C.; Tsai, K.; Courtney, D.G.; Bogerd, H.P.; Holley, C.L.; Cullen, B.R. Mapping of pseudouridine residues on cellular and viral transcripts using a novel antibody-based technique. RNA 2021, 27, 1400–1411. [Google Scholar] [CrossRef]
- Yang, S.L.; DeFalco, L.; Anderson, D.E.; Zhang, Y.; Aw, J.G.A.; Lim, S.Y.; Lim, X.N.; Tan, K.Y.; Zhang, T.; Chawla, T.; et al. Comprehensive mapping of SARS-CoV-2 interactions in vivo reveals functional virus-host interactions. Nat. Commun. 2021, 12, 5113. [Google Scholar] [CrossRef]
- Baek, A.; Lee, G.E.; Golconda, S.; Rayhan, A.; Manganaris, A.A.; Chen, S.; Tirumuru, N.; Yu, H.; Kim, S.; Kimmel, C.; et al. Single-molecule epitranscriptomic analysis of full-length HIV-1 RNAs reveals functional roles of site-specific m6As. Nat. Microbiol. 2024, 9, 1340–1355. [Google Scholar] [CrossRef]
- Morais, P.; Adachi, H.; Yu, Y.T. The Critical Contribution of Pseudouridine to mRNA COVID-19 Vaccines. Front. Cell Dev. Biol. 2021, 9, 789427. [Google Scholar] [CrossRef]
- Andries, O.; Mc Cafferty, S.; De Smedt, S.C.; Weiss, R.; Sanders, N.N.; Kitada, T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 2015, 217, 337–344. [Google Scholar] [CrossRef]
- Svitkin, Y.V.; Cheng, Y.M.; Chakraborty, T.; Presnyak, V.; John, M.; Sonenberg, N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017, 45, 6023–6036. [Google Scholar] [CrossRef]
- Chen, T.H.; Potapov, V.; Dai, N.; Ong, J.L.; Roy, B. N(1)-methyl-pseudouridine is incorporated with higher fidelity than pseudouridine in synthetic RNAs. Sci. Rep. 2022, 12, 13017. [Google Scholar] [CrossRef]
- Nance, K.D.; Meier, J.L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 748–756. [Google Scholar] [CrossRef]
- Roth, N.; Gergen, J.; Kovacikova, K.; Mueller, S.O.; Ulrich, L.; Schön, J.; Halwe, N.J.; Fricke, C.; Corleis, B.; Dorhoi, A.; et al. Assessment of Immunogenicity and Efficacy of CV0501 mRNA-Based Omicron COVID-19 Vaccination in Small Animal Models. Vaccines 2023, 11, 318. [Google Scholar] [CrossRef] [PubMed]
- Gote, V.; Bolla, P.K.; Kommineni, N.; Butreddy, A.; Nukala, P.K.; Palakurthi, S.S.; Khan, W. A Comprehensive Review of mRNA Vaccines. Int. J. Mol. Sci. 2023, 24, 2700. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, M.; Peng, X.; Yang, Y.; Chen, Q.; Liu, J.; She, Q.; Tan, J.; Lou, C.; Liao, Z.; et al. mRNA vaccine in cancer therapy: Current advance and future outlook. Clin. Transl. Med. 2023, 13, e1384. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Parkhouse, K.; Kirkpatrick, E.; McMahon, M.; Zost, S.J.; Mui, B.L.; Tam, Y.K.; Karikó, K.; Barbosa, C.J.; Madden, T.D.; et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018, 9, 3361. [Google Scholar] [CrossRef]
- Ye, Q.; Wu, M.; Zhou, C.; Lu, X.; Huang, B.; Zhang, N.; Zhao, H.; Chi, H.; Zhang, X.; Ling, D.; et al. Rational development of a combined mRNA vaccine against COVID-19 and influenza. NPJ Vaccines 2022, 7, 84. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Pelc, R.S.; Muramatsu, H.; Andersen, H.; DeMaso, C.R.; Dowd, K.A.; Sutherland, L.L.; Scearce, R.M.; Parks, R.; et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 2017, 543, 248–251. [Google Scholar] [CrossRef]
- Ramos da Silva, J.; Bitencourt Rodrigues, K.; Formoso Pelegrin, G.; Silva Sales, N.; Muramatsu, H.; de Oliveira Silva, M.; Porchia, B.; Moreno, A.C.R.; Aps, L.; Venceslau-Carvalho, A.A.; et al. Single immunizations of self-amplifying or non-replicating mRNA-LNP vaccines control HPV-associated tumors in mice. Sci. Transl. Med. 2023, 15, eabn3464. [Google Scholar] [CrossRef]
- Wollner, C.J.; Richner, M.; Hassert, M.A.; Pinto, A.K.; Brien, J.D.; Richner, J.M. A Dengue Virus Serotype 1 mRNA-LNP Vaccine Elicits Protective Immune Responses. J. Virol. 2021, 95, e02482-20. [Google Scholar] [CrossRef]
- Pardi, N.; LaBranche, C.C.; Ferrari, G.; Cain, D.W.; Tombácz, I.; Parks, R.J.; Muramatsu, H.; Mui, B.L.; Tam, Y.K.; Karikó, K.; et al. Characterization of HIV-1 Nucleoside-Modified mRNA Vaccines in Rabbits and Rhesus Macaques. Mol. therapy. Nucleic Acids 2019, 15, 36–47. [Google Scholar] [CrossRef]
- Friedrich, M.; Aigner, A. Therapeutic siRNA: State-of-the-Art and Future Perspectives. BioDrugs 2022, 36, 549–571. [Google Scholar] [CrossRef]
| Feature | Description |
|---|---|
| Genetic Information | RNA can store and transmit genetic information like DNA. Its sequence of nucleotides encodes information and can be replicated. |
| Catalytic Properties | Certain RNA molecules, known as ribozymes, can catalyze biochemical reactions. This suggests that RNA could facilitate critical metabolic processes even without proteins. |
| Self-Replication | Some RNA sequences can undergo self-replication under certain conditions, supporting the idea that RNA could have played a role in early life forms that required a means to reproduce. |
| Modification | Known Functions | Writers | Erasers | Readers |
|---|---|---|---|---|
| m6A | Regulates: splicing, stability localization, and translation of mRNA [241]. Immune response [242] | -METTL3-METTL14-WTAP-complex | -FTO -ALKBH5 | YTHDC1-2 YTHDF1-3 |
| m5C | Regulates: stability of tRNA; ribosome biogenesis; stability, localization, and translation of mRNA [243]. Immune response [244] | -NSUN1-7 -DNMT2 | -TET1-3, -ALKBH1 | ALYREF, YBX1 |
| Ψ | Regulates: stability of tRNA; ribosome biogenesis; translation of mRNA. Immune response [228] | -RNA-independent Pseudouridine synthases: PUS1, PUSL1, PUS3, TRUB1, TRUB2, PUS7, PUS7L PUS10, RPUSD1-4 -RNA-dependent Pseudouridine synthase: DKC1 | ? | PFN1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Campos, C.; Lanz-Mendoza, H.; Cime-Castillo, J.A.; Peralta-Zaragoza, Ó.; Madrid-Marina, V. RNA Through Time: From the Origin of Life to Therapeutic Frontiers in Transcriptomics and Epitranscriptional Medicine. Int. J. Mol. Sci. 2025, 26, 4964. https://doi.org/10.3390/ijms26114964
Martínez-Campos C, Lanz-Mendoza H, Cime-Castillo JA, Peralta-Zaragoza Ó, Madrid-Marina V. RNA Through Time: From the Origin of Life to Therapeutic Frontiers in Transcriptomics and Epitranscriptional Medicine. International Journal of Molecular Sciences. 2025; 26(11):4964. https://doi.org/10.3390/ijms26114964
Chicago/Turabian StyleMartínez-Campos, Cecilia, Humberto Lanz-Mendoza, Jorge A. Cime-Castillo, Óscar Peralta-Zaragoza, and Vicente Madrid-Marina. 2025. "RNA Through Time: From the Origin of Life to Therapeutic Frontiers in Transcriptomics and Epitranscriptional Medicine" International Journal of Molecular Sciences 26, no. 11: 4964. https://doi.org/10.3390/ijms26114964
APA StyleMartínez-Campos, C., Lanz-Mendoza, H., Cime-Castillo, J. A., Peralta-Zaragoza, Ó., & Madrid-Marina, V. (2025). RNA Through Time: From the Origin of Life to Therapeutic Frontiers in Transcriptomics and Epitranscriptional Medicine. International Journal of Molecular Sciences, 26(11), 4964. https://doi.org/10.3390/ijms26114964







