PKIB, a Novel Target for Cancer Therapy
Abstract
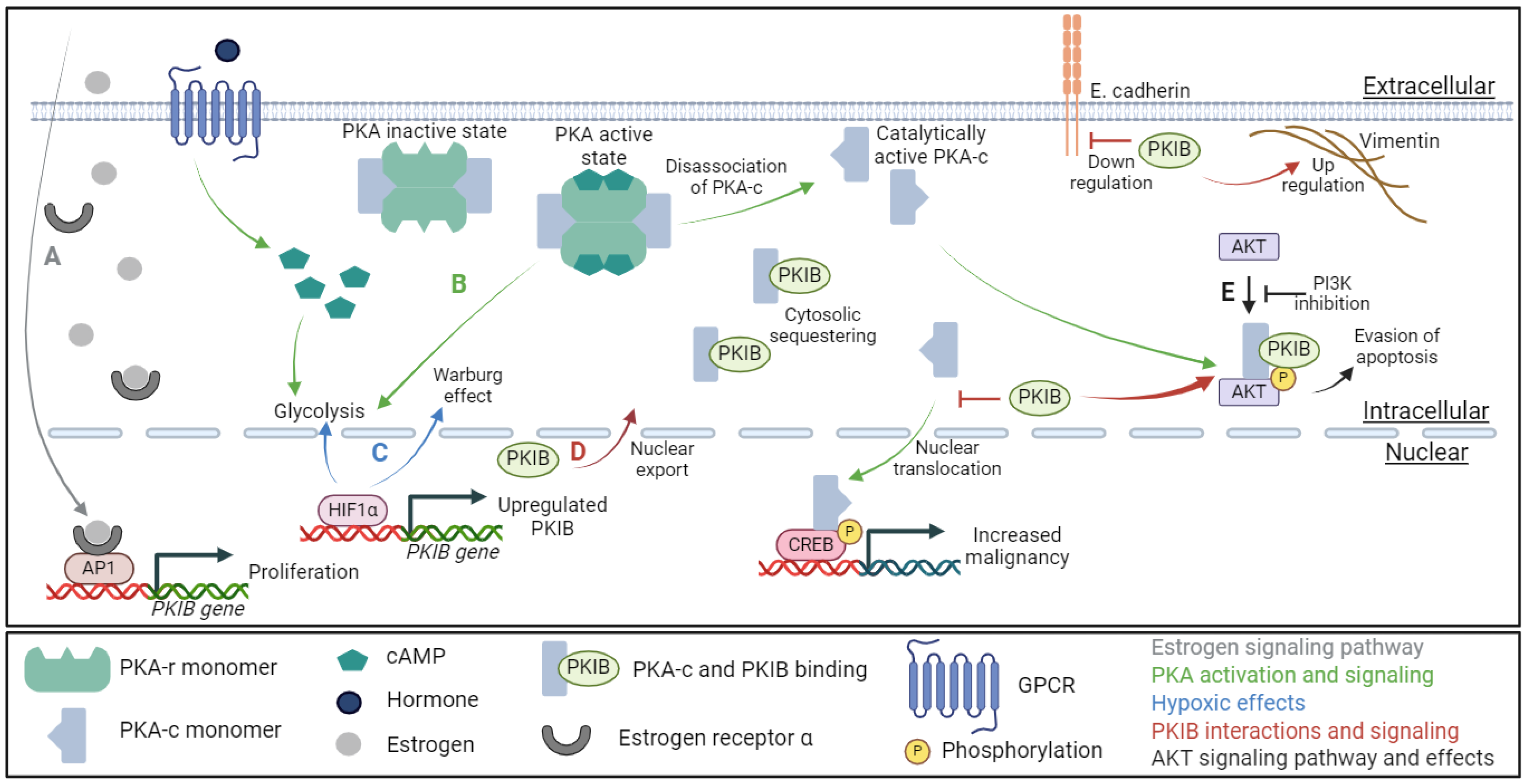
1. Introduction
2. The Regulation of PKA Activity by PKI
3. PKIB Impacts on Cancer
4. PKIB in Cytosolic Signaling
5. PKI in Cancer
6. PKIB Research with Indirect Cancer Impact
7. Discussion
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Taylor, S.S.; Kim, C.; Cheng, C.Y.; Brown, S.H.J.; Wu, J.; Kannan, N. Signaling through cAMP and cAMP-dependent Protein Kinase: Diverse Strategies for Drug Design. Biochim. Biophys. Acta 1784, 16, 2008. [Google Scholar] [CrossRef] [PubMed]
- Cadd, G.; McKnight, G.S. Distinct patterns of cAMP-dependent protein kinase gene expression in mouse brain. Neuron 1989, 3, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Mucignat-Caretta, C.; Caretta, A. Protein Kinase a Catalytic and Regulatory Subunits Interact Differently in Various Areas of Mouse Brain. Int. J. Mol. Sci. 2020, 21, 3051. [Google Scholar] [CrossRef] [PubMed]
- Caretta, A.; Mucignat-Caretta, C. Protein kinase a in cancer. Cancers 2011, 3, 913–926. [Google Scholar] [CrossRef] [PubMed]
- London, E.; Stratakis, C.A. The regulation of PKA signaling in obesity and in the maintenance of metabolic health. Pharmacol. Ther. 2022, 237, 108113. [Google Scholar] [CrossRef] [PubMed]
- Sossin, W.S.; Abrams, T.W. Evolutionary conservation of the signaling proteins upstream of cyclic AMP-dependent kinase and protein kinase C in gastropod mollusks. Brain Behav. Evol. 2009, 74, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ke, P.; Zhang, J.; Zhang, X.; Chen, X. Protein Kinase Inhibitor Peptide as a Tool to Specifically Inhibit Protein Kinase A. Front. Physiol. 2020, 11, 574030. [Google Scholar] [CrossRef] [PubMed]
- Smith, F.D.; Esseltine, J.L.; Nygren, P.J.; Veesler, D.; Byrne, D.P.; Vonderach, M.; Strashnov, I.; Eyers, C.E.; Eyers, P.A.; Langeberg, L.K.; et al. Local protein kinase A action proceeds through intact holoenzymes. Science 2017, 356, 1288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, Y.; Liu, J.; Chen, J.; Wang, J.; Hua, H.; Jiang, Y. cAMP-PKA/EPAC signaling and cancer: The interplay in tumor microenvironment. J. Hematol. Oncol. 2024, 17, 5. [Google Scholar] [CrossRef]
- Broggi, G.; Filetti, V.; Magro, G.; Morante, B.; Garro, R.; Ledda, C.; Rapisarda, V.; Lombardo, C.; Loreto, C.; Caltabiano, R. Immunohistochemical expression of cAMP in fluoroedenite-induced malignant pleural mesothelioma: Preliminary results. Mol. Med. Rep. 2023, 28, 132. [Google Scholar] [CrossRef]
- Zhang, H.; Kong, Q.; Wang, J.; Jiang, Y.; Hua, H. Complex roles of cAMP–PKA–CREB signaling in cancer. Exp. Hematol. Oncol. 2020, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Grieco, D.; Porcellini, A.; Avvedimento, E.V.; Gottesman, M.E. Requirement for cAMP-PKA pathway activation by M phase-promoting factor in the transition from mitosis to interphase. Science 1996, 271, 1718–1723. [Google Scholar] [CrossRef] [PubMed]
- Hochbaum, D.; Hong, K.; Barila, G.; Ribeiro-Neto, F.; Altschuler, D.L. Epac, in Synergy with cAMP-dependent Protein Kinase (PKA), Is Required for cAMP-mediated Mitogenesis. J. Biol. Chem. 2008, 283, 4464–4468. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.M.; Nakaseko, Y.; Samejima, I.; Kumada, K.; Yamada, H.; Michaelson, D.; Yanagida, M. 20S cyclosome complex formation and proteolytic activity inhibited by the cAMP/PKA pathway. Nature 1996, 384, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Sun, J.; Xie, S.; Chi, C.; Zhu, Y.; Pan, J.; Dong, B.; Huang, Y.; Xia, W.; Sha, J.; et al. PRKAR2B-HIF-1α loop promotes aerobic glycolysis and tumour growth in prostate cancer. Cell Prolif. 2020, 53, e12918. [Google Scholar] [CrossRef] [PubMed]
- Jin, N.; Ma, D.; Gu, J.; Shi, J.; Xu, X.; Iqbal, K.; Gong, C.-X.; Liu, F.; Chu, D. O-GlcNAcylation modulates PKA-CREB signaling in a manner specific to PKA catalytic subunit isoforms. Biochem. Biophys. Res. Commun. 2018, 497, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Neary, C.L.; Nesterova, M.; Yee, S.C.; Cheadle, C.; Becker, K.G.; Cho-Chung, Y.S. Protein kinase A isozyme switching: Eliciting differential cAMP signaling and tumor reversion. Oncogene 2004, 23, 8847–8856. [Google Scholar] [CrossRef] [PubMed]
- Farrow, B.; Rychahou, P.; Murillo, C.; O’Connor, K.L.; Iwamura, T.; Evers, B.M. Inhibition of pancreatic cancer cell growth and induction of apoptosis with novel therapies directed against protein kinase A. Surgery 2003, 134, 197–205. [Google Scholar] [CrossRef]
- Thomas, J.; Van Patten, S.M.; Howard, P.; Day, K.H.; Mitchell, R.D.; Sosnick, T.; Trewhella, J.; Walsh, D.A.; Maurer, R.A. Expression in Escherichia coli and Characterization of the Heat-stable Inhibitor of the CAMP-dependent Protein Kinase. J. Biol. Chem. 1991, 266, 10906–10911. [Google Scholar] [CrossRef]
- Collins, S.P.; Uhler, M.D. Characterization of PKIg, a Novel Isoform of the Protein Kinase Inhibitor of cAMP-Dependent Protein Kinase. 1997. Available online: http://www.jbc.org (accessed on 28 September 2023).
- Gammt, D.M.; Uhler, M.D. Isoform-specific Differences in the Potencies of Murine Protein Kinase Inhibitors Are Due to Nonconserved Amino-terminal Residues. J. Biol. Chem. 1995, 270, 7227–7232. [Google Scholar] [CrossRef]
- Scott, J.D.; Fischer, E.H.; Takio, K.; Demaille, J.G.; Krebs, E.G. Amino acid sequence of the heat-stable inhibitor of the cAMP-dependent protein kinase from rabbit skeletal muscle. Proc. Natl. Acad. Sci. USA 1985, 82, 5732–5736. [Google Scholar] [CrossRef] [PubMed]
- Dabanaka, K.; Chung, S.; Nakagawa, H.; Nakamura, Y.; Okabayashi, T.; Sugimoto, T.; Hanazaki, K.; Furihata, M. PKIB expression strongly correlated with phosphorylated Akt expression in breast cancers and also with triple-negative breast cancer subtype. Med. Mol. Morphol. 2012, 45, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Van Patten, S.M.; Donaldson, L.F.; McGuinness, M.P.; Kumar, P.; Alizadeh, A.; Griswold, M.D.; Walsh, D.A. Specific Testicular Cellular Localization and Hormonal Regulation of the PKIα and PKIβ Isoforms of the Inhibitor Protein of the cAMP-dependent Protein Kinase. J. Biol. Chem. 1997, 272, 20021–20029. [Google Scholar] [CrossRef] [PubMed]
- Hauer, J.A.; Barthe, P.; Taylor, S.S.; Parello, J.; Padilla, A. Two well-defined motifs in the cAMP-dependent protein kinase inhibitor (PKIα) correlate with inhibitory and nuclear export function. Protein Sci. 1999, 8, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Hazel, L.U.M.; Zengping, H.A.O.; Gayle, D.; Kumar, P.; Patterson, C.E.; Uhler, A.M.D. Vascular endothelial cells express isoforms of protein kinase A inhibitor. Am. J. Physiol.-Cell Physiol. 2002, 282, C59–C66. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pilger, J.; Mazur, A.; Monecke, P.; Schreuder, H.; Elshorst, B.; Bartoschek, S.; Langer, T.; Schiffer, A.; Krimm, I.; Wegstroth, M.; et al. A combination of spin diffusion methods for the determination of protein-ligand complex structural ensembles. Angew. Chem. Int. Ed. 2015, 54, 6511–6515. [Google Scholar] [CrossRef] [PubMed]
- Surveillance Research Program and National Cancer Institute SEER Incidence Data. Surveillance, Epidemiology, and End Results Cancer Statistics. Available online: https://seer.cancer.gov/ (accessed on 6 October 2023).
- Folkman, J. Angiogenesis and c-Jun. J. Natl. Cancer Inst. 2004, 96, 15–18. [Google Scholar] [CrossRef]
- Atsaves, V.; Leventaki, V.; Rassidakis, G.Z.; Claret, F.X. AP-1 Transcription Factors as Regulators of Immune Responses in Cancer. Cancers 2019, 11, 1037. [Google Scholar] [CrossRef]
- Gerhard, G.M.; Bill, R.; Messemaker, M.; Klein, A.M.; Pittet, M.J. Cancer Focus: Tumor-infiltrating dendritic cell states are conserved across solid human cancers. J. Exp. Med. 2020, 218-1, e20200264. [Google Scholar] [CrossRef]
- Lee, E.-S.; Son, D.-S.; Kim, S.-H.; Lee, J.; Jo, J.; Han, J.; Kim, H.; Lee, H.J.; Choi, H.Y.; Jung, Y.; et al. Prediction of Recurrence-Free Survival in Postoperative Non–Small Cell Lung Cancer Patients by Using an Integrated Model of Clinical Information and Gene Expression. Clin. Cancer Res. 2008, 14, 7397–7404. [Google Scholar] [CrossRef]
- Hoy, J.J.; Parra, N.S.; Park, J.; Kuhn, S.; Iglesias-Bartolome, R. Protein kinase A inhibitor proteins (PKIs) divert GPCR-Gαs-cAMP signaling towards EPAC and ERK activation and are involved in tumor growth. FASEB J. 2020, 34, 13900. [Google Scholar] [CrossRef] [PubMed]
- Lacalamita, A.; Piccinno, E.; Scalavino, V.; Bellotti, R.; Giannelli, G.; Serino, G. A gene-based machine learning classifier associated to the colorectal adenoma—Carcinoma sequence. Biomedicines 2021, 9, 1937. [Google Scholar] [CrossRef]
- Wan, R.; Yang, G.; Liu, Q.; Fu, X.; Liu, Z.; Miao, H.; Liu, H.; Huang, W. PKIB involved in the metastasis and survival of osteosarcoma. Front. Oncol. 2022, 12, 965838. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Furihata, M.; Tamura, K.; Uemura, M.; Daigo, Y.; Nasu, Y.; Miki, T.; Shuin, T.; Fujioka, T.; Nakamura, Y.; et al. Overexpressing PKIB in prostate cancer promotes its aggressiveness by linking between PKA and Akt pathways. Oncogene 2009, 28, 2849–2859. [Google Scholar] [CrossRef] [PubMed]
- Dou, P.; Zhang, D.; Cheng, Z.; Zhou, G.; Zhang, L. PKIB promotes cell proliferation and the invasion-metastasis cascade through the PI3K/Akt pathway in NSCLC cells. Exp. Biol. Med. 2016, 241, 1911. [Google Scholar] [CrossRef] [PubMed]
- Fei, L.; Wang, Y. microRNA-495 reduces visceral sensitivity in mice with diarrhea-predominant irritable bowel syndrome through suppression of the PI3K/AKT signaling pathway via PKIB. IUBMB Life 2020, 72, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-B.; Song, W.; Wang, Y.-Y.; Liu, M.-G.; Sun, M.-M.; Liu, H. Study on correlation between PKIB and pAkt expression in breast cancer tissues. Eur. Rev. Med. Pharmacol. Sci. Available online: https://www.europeanreview.org/article/12399 (accessed on 27 September 2023).
- Wu, V.H.; Yung, B.S.; Faraji, F.; Saddawi-Konefka, R.; Wang, Z.; Wenzel, A.T.; Song, M.J.; Pagadala, M.S.; Clubb, L.M.; Chiou, J.; et al. The GPCR–Gαs–PKA signaling axis promotes T cell dysfunction and cancer immunotherapy failure. Nat. Immunol. 2023, 24, 1318–1330. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, Z.; Schacht, T.; Herrmann, A.K.; Albrecht, P.; Lefkimmiatis, K.; Methner, A. Protein kinase inhibitor β enhances the constitutive activity of G-protein-coupled zinc receptor GPR39. Biochem. J. 2014, 462, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, V.; Davis, D.A.; Haque, M.; Huang, L.E.; Yarchoan, R. Differential Gene Up-Regulation by Hypoxia-Inducible Factor-1A A and Hypoxia-Inducible Factor-2A A in HEK293T Cells. Cancer Res. 2005, 65, 3299–3306. Available online: http://aacrjournals.org/cancerres/article-pdf/65/8/3299/2542551/3299-3306.pdf (accessed on 26 September 2023).
- Blanchet, E.; Van de Velde, S.; Matsumura, S.; Hao, E.; LeLay, J.; Kaestner, K.; Montminy, M. Feedback inhibition of CREB signaling promotes beta cell dysfunction in insulin resistance. Cell Rep. 2015, 10, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Dahlman-Wright, K.; Qiao, Y.; Jonsson, P.; Gustafsson, J.Å.; Williams, C.; Zhao, C. Interplay between AP-1 and estrogen receptor α in regulating gene expression and proliferation networks in breast cancer cells. Carcinogenesis 2012, 33, 1684–1691. [Google Scholar] [CrossRef]
- Yang, P.; Yang, X.; Wang, D.; Yang, H.; Li, Z.; Zhang, C.; Zhang, S.; Zhu, J.; Li, X.; Su, P.; et al. PSMD14 stabilizes estrogen signaling and facilitates breast cancer progression via deubiquitinating ERα. Oncogene 2023, 43, 248–264. [Google Scholar] [CrossRef] [PubMed]
- Creevey, L.; Bleach, R.; Madden, S.F.; Toomey, S.; Bane, F.T.; Varešlija, D.; Hill, A.D.; Young, L.S.; McIlroy, M. Altered steroid milieu in AI-resistant breast cancer facilitates AR mediated gene-expression associated with poor response to therapy. Mol. Cancer Ther. 2019, 18, 1731–1743. [Google Scholar] [CrossRef] [PubMed]
- Siatis, K.E.; Giannopoulou, E.; Manou, D.; Sarantis, P.; Karamouzis, M.V.; Raftopoulou, S.; Fasseas, K.; Alzahrani, F.M.; Kalofonos, H.P.; Theocharis, A.D. Resistance to hormone therapy in breast cancer cells promotes autophagy and EGFR signaling pathway. Am. J. Physiol. Cell Physiol. 2023, 325, C708–C720. [Google Scholar] [CrossRef] [PubMed]
- Dahia, C.L.; Rao, A.J. Regulation of FSH receptor, PKIbeta, IL-6 and calcium mobilization: Possible mediators of differential action of FSH. Mol. Cell Endocrinol. 2006, 247, 73–81. [Google Scholar] [CrossRef]
- Doliba, N.M.; Rozo, A.V.; Roman, J.; Qin, W.; Traum, D.; Gao, L.; Liu, J.; Manduchi, E.; Liu, C.; Golson, M.L.; et al. α Cell dysfunction in islets from nondiabetic, glutamic acid decarboxylase autoantibody-positive individuals. J. Clin. Investig. 2022, 132, e156243. [Google Scholar] [CrossRef]
- Feyeux, M.; Bourgois-Rocha, F.; Redfern, A.; Giles, P.; Lefort, N.; Aubert, S.; Bonnefond, C.; Bugi, A.; Ruiz, M.; Deglon, N.; et al. Early transcriptional changes linked to naturally occurring Huntington’s disease mutations in neural derivatives of human embryonic stem cells. Hum. Mol. Genet. 2012, 21, 3883–3895. [Google Scholar] [CrossRef]
- Koldaeva, A.; Zhang, C.; Huang, Y.-P.; Reinert, J.K.; Mizuno, S.; Sugiyama, F.; Takahashi, S.; Soliman, T.; Matsunami, H.; Fukunaga, I. Generation and Characterization of a Cell Type-Specific, Inducible Cre-Driver Line to Study Olfactory Processing. J. Neurosci. 2021, 41, 6449–6467. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musket, A.; Moorman, J.P.; Zhang, J.; Jiang, Y. PKIB, a Novel Target for Cancer Therapy. Int. J. Mol. Sci. 2024, 25, 4664. https://doi.org/10.3390/ijms25094664
Musket A, Moorman JP, Zhang J, Jiang Y. PKIB, a Novel Target for Cancer Therapy. International Journal of Molecular Sciences. 2024; 25(9):4664. https://doi.org/10.3390/ijms25094664
Chicago/Turabian StyleMusket, Anna, Jonathan P. Moorman, Jinyu Zhang, and Yong Jiang. 2024. "PKIB, a Novel Target for Cancer Therapy" International Journal of Molecular Sciences 25, no. 9: 4664. https://doi.org/10.3390/ijms25094664
APA StyleMusket, A., Moorman, J. P., Zhang, J., & Jiang, Y. (2024). PKIB, a Novel Target for Cancer Therapy. International Journal of Molecular Sciences, 25(9), 4664. https://doi.org/10.3390/ijms25094664





